Abstract
SIRT1 is an NAD-dependent deacetylase and epigenetic regulator essential for normal mammalian development and homeostasis. Here we describe a human SIRT1 splice variant, designated SIRT1-Δ2/9, in which the deacetylase coding sequence is lost due to splicing between exons 2 and 9. This work aimed to determine if SIRT1-Δ2/9 is a novel functional product of the SIRT1 gene. Endogenous SIRT1-Δ2/9 protein was identified in human cell lysate by immunoblotting and splice variant-specific RNA interference (RNAi). SIRT1-Δ2/9 mRNA is bound by CUGBP2, which downregulates its translation. Using pulldown assays, we demonstrate that SIRT1-Δ2/9 binds p53 protein. SIRT1-Δ2/9 maintains basal p53 protein levels and supports p53 function in response to DNA damage, as evidenced by RNAi-mediated depletion of SIRT1-Δ2/9 prior to damage. In turn, basal p53 downregulates SIRT1-Δ2/9 RNA levels, while stress-activated p53 eliminates SIRT1-Δ2/9. Loss of wild-type (wt) p53 has been correlated with overexpression of SIRT1-Δ2/9 in a range of human cancers. Exogenous SIRT1-Δ2/9 protein associates with specific promoters in chromatin and can regulate cancer-related gene expression, as evidenced by chromatin immunoprecipitation analysis and RNAi/genomic array data. SIRT1 is of major therapeutic importance, and potential therapeutic drugs are screened against SIRT1 deacetylase activity. Our discovery of SIRT1-Δ2/9 identifies a new, deacetylase-independent therapeutic target for SIRT1-related diseases, including cancer.
INTRODUCTION
Mammalian SIRT1 belongs to the sirtuin family of proteins that was first identified and characterized in yeast and subsequently found to be highly conserved through evolution (2, 23, 43, 47, 51). The Saccharomyces cerevisiae homologue of SIRT1 is Sir2, which stabilizes yeast chromosomes and impacts yeast aging. In mammals, SIRT1 is an epigenetic regulator of normal development, gametogenesis, homeostasis, and aging-related processes (3, 22, 30, 34, 41, 54). Mammalian genes that fall within the scope of SIRT1 regulation include key genes linked, for example, with hormonal control of metabolism and insulin signaling (e.g., PGC-1α), the ability of cells to respond to stress (e.g., p53, Foxo, and p300), and the processing of amyloid precursor protein in neuronal cells of the brain (ADAM10) (6, 15, 17, 20, 31, 37, 42, 52, 53). These genes in turn link SIRT1 with disease processes, including diabetes, cancer, and neurodegeneration (4, 17, 54).
Given the multifunctional roles of SIRT1 in health and disease, it is not surprising that SIRT1 is now recognized as an important therapeutic target across a range of age-related diseases, and this is a strong driving force for understanding the pathways subject to SIRT1 activity. For example, with a mouse model of Alzheimer's disease, Guarente's group recently demonstrated that SIRT1 suppresses the production of β-amyloid protein and the formation of amyloid plaques in the brain. This is achieved via SIRT1-dependent transcriptional activation of α-secretase ADAM10, which is involved in the cellular cleavage of amyloid precursor protein (17). Activation of SIRT1 is thus identified as a viable strategy to combat Alzheimer's disease and possibly other neurodegenerative diseases.
Biochemically, SIRT1 functions as an NAD-dependent deacetylase. In this capacity it upregulates/downregulates the activities of target proteins, such as the transcription factor and tumor suppressor p53, transcriptional coactivator p300, and retinoic acid receptor β, a known regulator of ADAM10 (see above) (50). In the case of p53, there appears to be a regulatory feedback loop operating between stress-activated p53 and SIRT1 (9), and this may be important for balancing the p53 proapoptotic stress response versus cell survival and recovery from stress.
Use of SIRT1-deficient mice has proved an invaluable tool for exploring tissue-specific effects of SIRT1 during development (11, 14). However, different groups, using different SIRT1 gene modifications and/or deletions, have reported variable effects of SIRT1 deficiency upon the degree of embryonic viability and development (13, 34, 54). This may be explained in part at least by the recent discovery that SIRT1 pre-mRNA is subject to alternative splicing (33), as different experimental SIRT1 deletions may fail to completely eradicate all SIRT1 isoforms.
The first reported SIRT1 splice variant, SIRT1-Δ8, lacks exon 8 due to in-frame splicing between exons 7 and 9 of full-length SIRT1 (SIRT1-FL) RNA (33). The novel SIRT1-Δ8 isoform displays distinct differences in stress-sensitivity, RNA/protein stability, protein-protein interactions, and deacetylase activity compared with SIRT1-FL. Expression of SIRT1-Δ8 is stress responsive, p53 dependent, and conserved in mammals (33).
In the course of cloning SIRT1-Δ8, we discovered a second isoform of SIRT1, designated SIRT1-Δ2/9, generated by frameshift splicing between exon 2 and exon 9 of SIRT1 pre-mRNA. Here we present the evidence for SIRT1-Δ2/9 mRNA expression and show that it is regulated by p53. In turn, the SIRT1-Δ2/9 protein binds p53 protein and regulates basal levels of p53 under nonstress conditions; thus, SIRT1-Δ2/9 and p53 form an autoregulatory loop. This appears important in maintaining the p53 protein level at the basal threshold required for a p53-dependent apoptotic response following exposure of cells to genotoxic stress. Abnormal levels of SIRT1-Δ2/9 are evident in human cancer tissues, and we consider the possible implications of this newly discovered SIRT1 isoform in relation to normal function, disease, and SIRT1-based therapeutics.
MATERIALS AND METHODS
Cloning, sequencing, and plasmids.
Reverse transcription-PCR (RT-PCR) using 1F/10R primers (see Fig. 1A, below) on total RNA extracted from HCT116 cells identified ∼0.7-kbp fragments, which were cloned in the pCR2.1 vector (Invitrogen) and sequenced. SIRT1-Δ2/9 contained very high GC content, and this sequence was codon optimized, chemically synthesized with N-terminal Myc and His epitope tag sequences (Eurofins MWG Operon, Germany), and cloned in pcDNA3.1 (see Fig. S1 in the supplemental material). Mutant SIRT1-Δ2/9 plasmid was created using the codon-optimized Myc-His-SIRT1-Δ2/9-pcDNA3.1 construct (described above) that contained mutations in the CUGBP2 binding motif (see Fig. 2D, below) in the SIRT1-Δ2/9 splice junction (Eurofins MWG Operon, Germany). All clones were confirmed by sequencing (Eurofins MWG Operon, Germany). TP53 (exons 5 to 8) was amplified from tumors and normal tissues by TP53E5F/TP53E8R primers (see Table S2 in the supplemental material) and PCR products directly sequenced using the same primers.
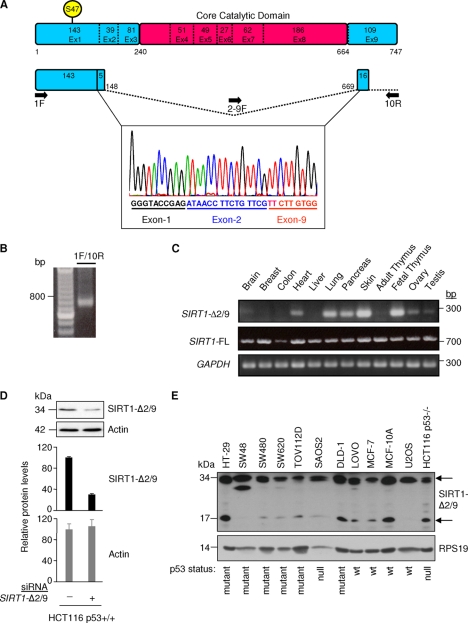
Fig 1.
Identification and tissue expression of a novel splice variant of SIRT1. (A) Schematic of SIRT1-FL, highlighting the exons, core catalytic domain, and serine 47. The sequence trace shows part of SIRT1 exon 1 and exon 2/9 splicing identified in HCT116 cells by 1F/10R primers. Splice donor and splice acceptor site sequences are as follows: exon 2 … ATAACCTTCTGTTCGgtgatgaaattatcactaatggttttcattcctgtgaaagtgatgaggaggatagagcctcacatgcaagctctagtgactggactccaaggccacggataggtaatcagtatctgtttttgccaccaaatcgttacattttccatggcgctgaggtatattcagactctgaagatgacgtcttatcctctagTTCTTGTGGCAGTAACAG … (italicized and nonitalicized sequences represent exon 2 and exon 9, respectively; uppercase sequence represents SIRT1-Δ2/9 RNA; splice donor and acceptor sites are underlined). (B) Agarose gel showing RT-PCR amplification products of SIRT1-Δ2/9 from HCT116 cells, using 1F/10R primers. (C) SIRT1-Δ2/9 mRNA expression in normal human tissue samples with 100 ng total RNA from each tissue using primer pair 2-9F/10R. Also shown is expression of SIRT1-FL, based on SEx4/10R primers and the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) control (primer sequences are shown in Table S1 in the supplemental material). (D) Detection of endogenous SIRT1-Δ2/9 protein. HCT116 cells were transfected with an siRNA targeting SIRT1-Δ2/9 specifically. The lysates were blotted with the indicated antibodies (upper panel); the graphs show quantification of Western blotting data by densitometry. (E) Expression of endogenous SIRT1-Δ2/9 in a panel of cell lines. Cell lysates were blotted for SIRT1 (N terminal) with the Epitomics antibody (upper panel) and RPS19 antibody (lower panel) as a loading control. Two molecular mass forms of SIRT1-Δ2/9 (34 kDa and 17 kDa) were detected (arrows) upon long exposure of the immunoblot. The N-terminal SIRT1 antibody also detected SIRT1-FL, and this is shown in Fig. S5 in the supplemental material (short exposure) for comparison with levels of SIRT1-Δ2/9 17-kDa and 34-kDa form detection.
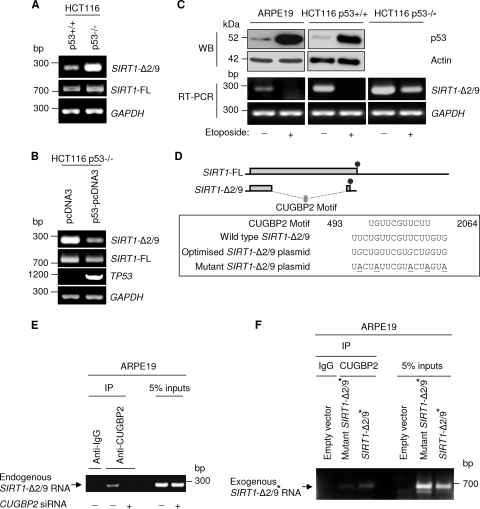
Fig 2.
Regulation of SIRT1-Δ2/9 expression. (A) p53 negatively regulates SIRT1-Δ2/9 mRNA expression. RT-PCR showed SIRT1-Δ2/9, SIRT1-FL, and GAPDH mRNA expression in HCT116 p53+/+ and HCT116 p53−/− cells. (B) Exogenous expression of p53 in HCT116 p53−/− cells downregulates SIRT1-Δ2/9 mRNA expression. RT-PCR showed mRNA expression of SIRT1-Δ2/9, SIRT1-FL, TP53, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) following exogenous expression of p53-pcDNA3. (C) Stress-activated p53 downregulates SIRT1-Δ2/9 mRNA expression. ARPE19, HCT116 p53+/+, and HCT116 p53−/− cells were treated with etoposide (see Materials and Methods); the levels of p53 protein were assessed by Western blotting with DO-1 antibody (upper panel), and the levels of SIRT1-2/9 RNA were determined by RT-PCR (lower panel). Actin and GAPDH were used as loading controls. (D) Schematic showing how exonic splicing of SIRT1-2/9 generates a novel putative CUGBP2-binding motif. Also shown are sequences of the 2/9 wild type, optimized, and mutant motifs. The codon-optimized nucleotides are shown in italics, and mutated nucleotides in the CUGBP2-binding site are underlined. Circles indicate stop codons, and the position of the motif sequence is based on NCBI sequence AF083106. (E) Endogenous SIRT1-Δ2/9 RNA coimmunoprecipitated with CUGBP2, but the interaction was abrogated following CUGBP2 knockdown in ARPE19 cells. IgG pulldown was used as a control. (F) In ARPE19 cells, exogenous SIRT1-Δ2/9 RNA coimmunoprecipitated with CUGBP2 (SIRT1-Δ2/9∗). This was attenuated following mutations of the CUGBP2-binding site in a mutant SIRT1-Δ2/9∗. Exogenous SIRT1-Δ2/9 RNA was detected using SEx-1/BGH-Rvs primers (see Fig. S1 in the supplemental material). SIRT1-Δ2/9∗ is a modified exogenous SIRT1-Δ2/9 protein (see the text).
RT-PCR and qRT-PCR.
Noncancer tissue RNA samples and paired noncancer/cancer tissue RNA samples were obtained from AMS Biotechnology Europe and Ambion, respectively. Total RNA from cell experiments was isolated using an RNeasy kit (Qiagen) and quantitated by UV spectroscopy (GeneSpecV). Standard RT-PCR was performed on a DNA Engine Dyad apparatus (MJ Research) using a one-step RT-PCR kit (Qiagen). For quantitative RT-PCR (qRT-PCR), reactions were run in quadruplicate on a DNA Engine Opticon (Bio-Rad) with a QuantiTect SYBR green RT-PCR kit (Qiagen). Sequences of the primers used for RT-PCR and qRT-PCR are provided in Table S1 of the supplemental material. The general cycling conditions were as follows: 50°C for 30 min, 94°C for 15 min, followed by the indicated thermal cycle (repeated for a number of cycles specific for each primer pair [see Table S2]) of 94°C for 10 s, annealing for 30 s, and 72°C extension for 30 s.
Cell culture and transfection.
Isogenic colorectal carcinoma cell lines HCT116 p53+/+ and HCT116 p53−/− and noncancer retinal epithelial cell line ARPE-19 were cultured as described previously (33). All other cell lines (HT29, SW48, SW480, SW620, TOV112D, SAOS-2, DLD-1, LoVo, MCF7, MCF10A, and U2OS) were cultured according to the corresponding ATCC guidelines. Transfection of small interfering RNA (siRNA) and plasmid expression constructs was as described previously (19, 20). SIRT1-FL siRNA (19) is within exon 8; CUGBP2 siRNA [sense, 5′-CGAAGAAAUGUAAUGAGAA(dTdT)-3′; antisense, 5′-UUCUCAUUACAUUUCUUCG(dTdT)-3′] and SIRT1-Δ2/9 siRNA [sense, 5′-CCUUCUGUUCGUUCUUGUG(dTdT)-3′; antisense, 5′-CACAAGAACGAACAGAAGG(dTdT)-3′; Dharmacon] are located across the alternate splice junction, at nucleotide (nt) position 498 in exon 2 and nt 2060 in exon 9 (based on the NM_012238 sequence). UV irradiation was performed as described previously (44), delivering 0 (control) or 10 J/m2 of UV irradiation. 5-Fluorouracil (5-FU) and etoposide were used at 75 μM and 30 μM, respectively. For combined siRNA transfection/UV or 5-FU treatment, UV or 5-FU was applied 33 h posttransfection and cells were harvested 24 h later. Apoptotic cells were identified by fluorescence-activated cell sorting (FACS) using annexin V-Fluos (Roche) following the protocol of the manufacturer.
Western blotting.
Protein preparation and Western blotting were performed as described previously (19). Antibodies were the following: anti-Sir2 residues 1 to 131 (07-131; Upstate Biostechnology; N terminus SIRT1 antibody) and anti-Sir2/SIRT1 (1054-1; Epitomics; N terminus antibody) for detection of endogenous and exogenous SIRT1-Δ2/9; anti-c-Myc (sc-40; Santa Cruz Biotechnolgoy) and anti-His (sc-803; Santa Cruz Biotechnolgoy) for detection of exogenous SIRT1-Δ2/9; anti-SIRT1 (sc-15404; Santa Cruz Biotechnology); anti-p53 (DO-1, sc-126; Santa Cruz Biotechnology); anti-p53 (FL393, sc-6243; Santa Cruz Biotechnology); anti-p53 phosphoserine 15 (9284S; Cell Signaling); anti-SIRT1 phosphoserine 47 (2314; Cell Signaling); anti-RPS19 (ab57643; Abcam); antiactin (monoclonal antibody 1501; Millipore). Actin and RPS19 were used as loading reference controls. Visualization of bound antibodies was obtained with enhanced chemiluminescence (Roche) with quantitation of signal intensity based on densitometry of signals within the linear range, using the Quantity One software (Bio-Rad).
Immunoprecipitation.
Exogenous SIRT1-Δ2/9 was expressed in ARPE19, HCT116 p53+/+, and DLD-1 cells for 24 h. A total of 12 × 106 cells were lysed in 300 μl of 2× immunoprecipitation (IP) buffer (150 mM NaCl, 50 mM Tris [pH 8.0], 1% Triton X-100, 20 mM imidazole, and 50 μl protease inhibitor cocktail [Roche]). Samples were sonicated with a Soniprep 150 sonicator (MSE UK Ltd.) at 2.5 μm in four 10-s bursts. Samples were then centrifuged at 4°C for 5 min, and supernatants were kept on ice and assayed for protein concentration. One hundred microliters of Ni-nitrotriacetic acid–agarose resin (Qiagen) was equilibrated with 1 ml of 3× IP buffer and centrifuged at 6,200 × g for 2 min at 4°C. Aliquots of 1.5 mg of protein lysate were added to the equilibrated resins and swirled for 1 h in a cold cabinet. Samples were washed 5 times in 1 ml IP buffer, and the bound proteins were eluted in 70 μl of elution buffer (150 mM NaCl, 50 mM Tris [pH 8.0], 1% Triton X-100, 250 mM imidazole, and protease inhibitor cocktail) by centrifuging at 10,000 × g for 1 to 2 min. SDS sample buffer was added to the eluates, and mixtures were heated at 90°C briefly and frozen to −70°C. For endogenous immunoprecipitation, whole-cell lysates from HCT116 p53+/+ cells were immunoprecipitated using anti-p53 (DO-1) antibody as described previously (1).
RNA-IP.
RNA immunoprecipitation (RNA-IP) was performed as described previously (39). For the exogenous SIRT1-Δ2/9 experiment, ARPE-19 cells were transfected with control vector, SIRT1-Δ2/9, or mutant SIRT1-Δ2/9 as described previously (20). Cell lysis was carried out using polysome lysis buffer and precleared with protein G-agarose for 3 h at 4°C. Protein-RNA complexes from 6 × 106 cells were captured overnight at 4°C with 5 μg of antibodies against CUGBP2 (HL1460; Sigma) or control mouse IgG (Upstate Biotechnology). Samples were washed and eluted, followed by proteinase K treatment at 55°C for 30 min. The IP-captured RNA was purified using an RNeasy kit (Qiagen) following the manufacturer's protocol. The purified RNA samples and input were subjected to RT-PCR using exogenous SIRT1-Δ2/9-specific primers (see Fig. S1 in the supplemental material). For the endogenous SIRT1-Δ2/9 experiment, ARPE-19 cells were transfected with control or CUGBP2-specific siRNA as described earlier. RNA-IP was performed using 8.7 × 106 cells as described above, and RT-PCR for SIRT1-Δ2/9 was carried out using endogenous SIRT1-Δ2/9-specific primers (see Fig. S1).
ChIP.
Chromatin immunoprecipitation (ChIP) was performed according to the manufacturer's protocol (EZ ChIP; Upstate Biotechnology) as described previously (1). Briefly, ARPE-19 cells were transfected with vector or SIRT1-Δ2/9 as described previously (19) for 24 h. A total of 1 × 107 cells from each transfection was cross-linked in 1% formaldehyde for 20 min and sonicated to shear the DNA to an average of ∼400 bp. Samples were precleared with protein G-agarose for 2 h at 4°C. Protein-DNA complexes in 2 × 106 cells were captured overnight at 4°C with 5 μg of antibody against His (H-15; Santa Cruz Biotechnology), p53 (DO-1; Santa Cruz Biotechnology), or corresponding control rabbit or mouse IgG (Upstate Biotechnology). Samples were washed, and cross-links removed by heating with proteinase K at 65°C overnight, followed by purification of DNA on the columns provided. The IP-captured DNA sample and sonicated genomic DNA extract (input) were subject to qRT-PCR. The target site in the IGFBP3 promoter was amplified using IGFBP3-pF and IGFBP3-pR, IGFBP3 intron target using IGFBP3-iF and IGFBP3-iR, and the heme oxygenase 1 (HO-1) promoter using HO-1pF and HO-1pR primers (see Table S1 in the supplemental material). ChIP enrichment results with specific antibodies were normalized to signals from the nonexpressed Goosecoid (GSC) control in the same sample and are expressed as the fold enrichment relative to IgG control (5).
Microarray.
ARPE19 cells were treated with control, SIRT1-Δ2/9, or SIRT1-FL siRNAs for 72 h, and RNA was extracted. Two micrograms of total RNA was processed for the microarray by using the Affymetrix GeneChip one-cycle target-labeling kit (Affymetrix, Santa Clara, CA) according to the manufacturer's recommended protocols. The resulting cRNA was hybridized to GeneChip human genome U133 Plus 2.0 arrays for 16 h at 45°C. The arrays were washed, stained, and scanned using the Affymetrix recommended protocols (Affymetrix, Santa Clara, CA). Raw data processing was performed by using the Affymetrix GCOS 1.2 software. MAS5 normalized data were collected and analyzed by using the GeenSpring GX10 Expression software (Agilent Technologies). Differentially expressed genes were identified by using a two-class t test, where the significance level was set at a P level of <0.05. Genes that were >2.0-fold up- or downregulated between groups were selected.
Statistical analysis.
All data were derived from at least three independent experiments, and statistical significance was determined using Student's two-tailed t test. P values of <0.05 were considered significant.
RESULTS
Identification of SIRT1-Δ2/9.
SIRT1-FL mRNA is represented schematically in Fig. 1A. Previously we reported a SIRT1 splice variant which lacked exon 8, termed SIRT1-Δ8 (33). In the course of cloning SIRT1-Δ8 (33), we reproducibly observed a 0.7-kb RNA species amplified by RT-PCR (Fig. 1B). Upon sequencing, this short species of SIRT1 RNA was found to represent nt 1 to 498/2060 to 2297 of SIRT1 with splicing between exon 2 (codon 148) and exon 9 (codon 669) (Fig. 1A). Upon detailed analysis of the SIRT1 genomic sequence, a potential splice donor within exon 2 and splice acceptor within exon 9 were identified at exactly the observed splice points required to generate SIRT1-Δ2/9 RNA (Fig. 1 legend). The exon 2/exon 9 splicing generated a codon frameshift in exon 9 (Fig. 1A) and we predicted a novel C-terminal amino acid sequence for the SIRT1-Δ2/9 protein (Fig. 1A; see also Fig. S1 in the supplemental material). The SIRT1-Δ2/9 coding sequence lacks the SIRT1 core catalytic deacetylatase domain, which stretches from codon 240 in exon 3 to codon 664 in exon 8 of SIRT1 (36): exon2-exon 9 splicing removed this domain (Fig. 1A).
SIRT1-Δ2/9 RNA was detectable at various levels in a range of human tissues (heart, lung, pancreas, skin, fetal thymus, ovary, prostate, testis) but was low/undetectable in other tissues (brain, breast, colon, liver, adult thymus) (Fig. 1C; data not shown for prostate). In contrast, SIRT1-FL RNA was clearly detectable in the same RNA samples from all tissues examined and showed little variation (Fig. 1C). This indicated that expression levels of SIRT1-Δ2/9 in normal human tissues are dependent upon tissue type and do not simply reflect SIRT1-FL expression levels. We concluded that SIRT1-Δ2/9 is widely expressed and is regulated independently from SIRT1-FL.
The above observations were made using RT-PCR amplification of SIRT1-Δ2/9 RNA, with the forward primer spanning the exon 2/exon 9 splice junction to confer specificity for the splice variant SIRT1-Δ2/9. This primer pair is selective for SIRT1-Δ2/9, gives good results for RT-PCR with a single product of the correct size (Fig. 1B), which was confirmed as SIRT1-Δ2/9 RNA by selective silencing using RNAi (see Fig. 4A, below). We also attempted quantitative RT-PCR, but unfortunately this proved unsuccessful, either with or without prior selection of poly(A)+ RNA by oligo(dT) column chromatography. Alternative reverse primers were also tested but again proved unsuccessful for quantitative RT-PCR of SIRT1-Δ2/9 RNA. The results observed using strictly defined conditions for RT-PCR (see Materials and Methods) were highly reproducible. SIRT1-FL is amplified by SIRT1-FL-specific primers to saturation after 34 rounds of PCR amplification, while amplification of SIRT1-Δ2/9 with SIRT1-Δ2/9-specifc primers was not observed until 44 cycles of PCR amplification. This suggested that SIRT1-Δ2/9 RNA is present in cells at low levels compared to SIRT1-FL.
Fig 4.
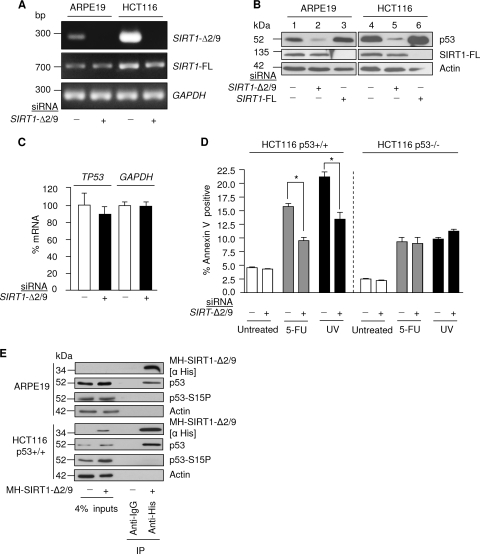
SIRT1-Δ2/9 maintains basal levels of p53 protein. (A) Expression of SIRT1-Δ2/9, SIRT1-FL, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNAs following SIRT1-Δ2/9 siRNA treatment in the indicated cell lines. (B) Ablation of SIRT1-Δ2/9 by RNAi causes depletion of p53 protein. Protein levels of p53, SIRT1-FL, and actin are shown. (C) qRT-PCR results, showing mRNA levels of TP53 and GAPDH in HCT116 p53+/+ cells following SIRT1-Δ2/9 depletion by siRNA. (D) SIRT1-Δ2/9 is required for induction of p53-dependent apoptosis following stress. Apoptosis was assessed by annexin V staining following treatment of cells with SIRT1-Δ2/9 siRNA with or without 5-FU or UV (see Materials and Methods); background levels of annexin V-positive cells were 4.5% and 2.5% in HCT116 p53+/+ and HCT116 p53−/− cells, respectively. ∗, P < 0.05. (E) Exogenous SIRT1-Δ2/9 interacts with wild-type p53. MH-SIRT1-Δ2/9 was exogenously expressed in ARPE19 and HCT116 p53+/+ cells, immunoprecipitated with anti-His antibody, and probed for SIRT1-Δ2/9, p53, and p53-S15P in both ARPE19 and HCT116 p53+/+ cell lines.
We next screened for expression of endogenous SIRT1-Δ2/9 protein with an antibody raised against residues 1 to 131 of SIRT1 and thus was predicted to detect SIRT1-Δ2/9 (Fig. 1A). A band of approximately 34 kDa was detected by immunoblotting of the HCT116 cell lysate (Fig. 1D). This band was reduced approximately 70% following selective silencing of SIRT1-Δ2/9 using splice-specific siRNA directed against the 2/9 RNA splice junction (Fig. 1D; selective silencing of SIRT1-Δ2/9 RNA is shown in Fig. 4A, below). These results indicated that endogenous SIRT1-Δ2/9 mRNA is translated into protein in human HCT116 cells.
Endogenous SIRT1-Δ2/9 protein was evident in a range of human cell lines of different origins after immunoblotting with two independent antibodies directed against the N terminus of SIRT1 (Upstate Biotechnology and Epitomics; see Materials and Methods) (Fig. 1E). Interestingly, two apparent molecular mass forms were observed, one at 17 kDa (Fig. 1E; the predicted mass for SIRT1-Δ2/9) and a second at 34 kDa (Fig. 1D and E). The latter 34-kDa band was reduced by SIRT1-Δ2/9 RNAi (Fig. 1D) and may represent a dimer of SIRT1-Δ2/9 or some other high-affinity complex containing SIRT1-Δ2/9 protein. SIRT1-FL was also detected in the same immunoblots (see Fig. S2 in the supplemental material) and was evident at much higher levels than SIRT1-Δ2/9. We estimate that SIRT1-Δ2/9 represents <1% to 10% of the full-length SIRT1 protein, depending upon the cell line.
Splicing of SIRT1-Δ2/9 RNA is downregulated by p53.
Comparison of isogenic clones of p53+/+ and p53−/− HCT116 human colorectal cancer cells revealed similar levels of SIRT1-FL RNA in p53+/+ and p53−/− cells but higher levels of SIRT1-Δ2/9 RNA in the p53−/− cells (Fig. 2A). This indicated that SIRT1-Δ2/9 exonic splicing may be suppressed in the presence of p53. This was confirmed by exogenous expression of p53 in the HCT116 p53−/− cells, which induced downregulation of SIRT1-Δ2/9 RNA levels (Fig. 2B). Exogenous p53 appeared to affect both SIRT1-FL and SIRT1-Δ2/9 RNA levels; however, the effect was more pronounced for SIRT1-Δ2/9 (Fig. 2B). Significantly, the above comparisons of p53+/+ and p53−/− HCT116 cells were performed in the absence of applied stress, leading us to conclude that nonactivated p53 can downregulate SIRT1-Δ2/9 expression under basal conditions of cell culture (i.e., in the absence of applied stress).
We were curious to determine if stress-induced activation of p53 caused any p53-dependent change in the downregulation of SIRT1-Δ2/9 RNA. This was examined by comparing SIRT1-Δ2/9 RNA levels in cells before and after exposure to the DNA-damaging agents 5-FU or etoposide (see Materials and Methods). In both cases, SIRT1-Δ2/9 RNA was undetectable after stress-induced activation of p53 (Fig. 2C, lower panel, etoposide results). The complete loss of SIRT1-Δ2/9 RNA was observed in both ARPE19 noncancer epithelial cells and also HCT116 colorectal cancer epithelial cells (Fig. 2C) and appeared to be p53 dependent (Fig. 2C, compare HCT116 p53+/+ with HCT116 p53−/−). Similar results were observed for 5-FU-treated cells (data not shown). Overall, these results indicated that basal p53 downregulates SIRT1-Δ2/9 RNA levels in the absence of applied stress, while stress-activated p53 eliminates SIRT1-Δ2/9 RNA.
SIRT1-Δ2/9 RNA is bound by the RNA-binding protein CUGBP2.
Analysis of the nucleotide sequence of SIRT1-Δ2/9 revealed a putative CUGBP2-binding site created by exonic splicing of exon 2/exon 9 (Fig. 2D). To determine whether CUGBP2 binds SIRT1-Δ2/9 RNA, we performed antibody pulldown experiments for CUGBP2 and looked for coimmunoprecipitated SIRT1-Δ2/9 RNA by RT-PCR in ARPE19 cells. Endogenous SIRT1-Δ2/9 RNA was clearly detectable in the CUGBP2 pull-down product (Fig. 2E). In contrast, no SIRT1-Δ2/9 RNA was evident in CUGBP2 pulldown assays when CUGBP2 was first selectively depleted by RNAi (Fig. 2E; selective knockdown of CUGBP2 is shown in Fig. 3E), indicating the specificity of the CUGBP2–SIRT1-Δ2/9 interaction.
Fig 3.
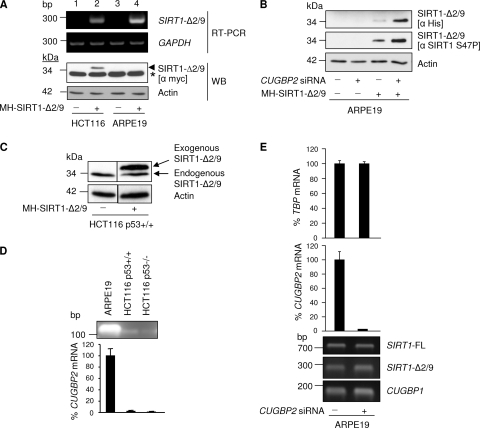
SIRT1-Δ2/9 protein is downregulated by CUGBP2. (A) Exogenous SIRT1-Δ2/9 protein is not detectable in ARPE19 cells. MH-SIRT1-Δ2/9 was exogenously expressed in HCT116 p53+/+ and ARPE19 cells. Cells were harvested after 24 h for RNA and protein. RT-PCR results for exogenous SIRT1-Δ2/9 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (upper panel) and Western blot analysis results for SIRT1-Δ2/9 (lower panel) are shown. ∗, a nonspecific band; arrowhead, SIRT1-Δ2/9 protein. (B) Downregulation of CUGBP2 by siRNA allows the expression of exogenous SIRT1-Δ2/9 protein in ARPE19 cells. CUGBP2 was silenced by siRNA prior to the exogenous expression of MH-SIRT1-Δ2/9 in ARPE19 cells. Protein samples were blotted for SIRT1-Δ2/9 and phosphorylated SIRT1 serine 47. (C) Detection of endogenous and exogenous SIRT1-Δ2/9. Total cell lysates from HCT116 p53+/+ cells treated with vector alone or MH-SIRT1-Δ2/9 plasmid were analyzed. The blots were probed with anti-SIRT1 residues 1 to 131 (α Sir2; catalog number 07-131; Upstate Biotechnology). (D) RT-PCR results showing mRNA expression of CUGBP2 (upper panel) and qRT-PCR results (lower panel) for the indicated cell lines. (E) CUGBP2 does not influence SIRT1-Δ2/9 or SIRT1-FL splicing. Expression levels of SIRT1-Δ2/9, SIRT1-FL, and CUGBP1 mRNAs (RT-PCR results [lower panel]) in CUGBP2 silenced ARPE19 cells (qRT-PCR results [upper panel]) are shown.
The above results indicate that cellular CUGBP2 binds cellular SIRT1-Δ2/9 RNA under basal conditions of culture. However, it remained formally possible that the CUGBP2 protein might bind at a site other than at the motif created at the 2/9 splice sequence of SIRT1-Δ2/9. In order to resolve this issue, we cloned SIRT1-Δ2/9 and analyzed CUGBP2 binding to an exogenous SIRT1-Δ2/9 transcript designed to exclude the 3′-extension in SIRT1-FL RNA (Fig. 2D). For technical reasons, it was necessary to introduce some conservative nucleotide substitutions in order to overcome cloning difficulties associated with the GC-rich nature of SIRT1-Δ2/9 (Fig. 2D; see also Fig. S1 in the supplemental material). This resulted in a modified 2/9 junctional motif (designated 2/9*), compared with the naturally occurring junctional CUGBP2 motif in endogenous SIRT1-Δ2/9 RNA (Fig. 2D, boxed sequences). Nonetheless, the modified junctional sequence retained characteristics of a CUGBP2-binding motif and was bound by endogenous CUGBP2, as indicated by the pulldown assay results (Fig. 2F). This indicates preservation of CUGBP2 binding by the modified SIRT1-Δ2/9 RNA clone.
We next employed conservative site-directed mutagenesis on the cloned SIRT1-Δ2/9* to disrupt the CUGBP2-binding nucleic acid sequence motif without affecting the SIRT1-Δ2/9* amino acid coding sequence. Importantly, the cloned SIRT1-Δ2/9* RNAs were amplified using primers specific to the exogenous RNA products (see Fig. S1 in the supplemental material). This avoided coamplification of endogenous SIRT1-Δ2/9 RNA bound to CUGBP2. Thus, the results shown in Fig. 2F are selective for the cloned SIRT1-Δ2/9* RNA, and endogenous SIRT1-Δ2/9 is not included. Binding by CUGBP2 was evident for the cloned SIRT1-Δ2/9* (Fig. 2F), but binding was largely abolished for the cloned SIRT1-Δ2/9* mutant containing the disrupted CUGBP2 motif (Fig. 2F, mutant SIRT1-Δ2/9*).
Overall the above results are strong evidence that the CUGBP2 protein binds an internal, novel CUGBP2-binding motif in SIRT1-Δ2/9 RNA and that this motif is created by 2/9 exonic splicing of the SIRT1 transcript.
Exogenous expression of the SIRT1-Δ2/9 protein.
When transfected into cells, the Myc-His-tagged SIRT1-Δ2/9 was transcribed and translated into a 34-kDa protein that was reactive with both anti-Myc antibody and anti-His antibody (Fig. 3A and B; note that the Myc antibody also detected a cross-reactive cellular protein running slightly ahead of Myc/His-tagged SIRT1-Δ2/9 on SDS-PAGE). In addition, the exogenous SIRT1-Δ2/9 was detected by the anti-SIRT1 S47P antibody (Fig. 3B, middle panel), indicating that exogenously expressed SIRT1-Δ2/9 protein was recognized and phosphorylated at serine 47 by endogenous cellular SIRT1 kinase. The endogenous 24-kDa SIRT1-Δ2/9 protein was also evident in the HCT116 cell lysate, running slightly ahead of the exogenous His-Myc-tagged SIRT1-Δ2/9 protein (Fig. 3C).
CUGBP2 downregulates SIRT1-Δ2/9 translation into protein.
RNA binding by CUGBP2 can regulate RNA turnover and/or suppress or enhance mRNA translation into protein. The discovery that CUGBP2 binds SIRT1-Δ2/9 RNA enabled us to resolve a puzzling observation related to exogenous SIRT1-Δ2/9 expression in different cell lines. Thus, exogenous SIRT1-Δ2/9 protein was detectable in the HCT116 cells but not in ARPE19 cells (Fig. 3A, WB panel, lane 2 versus lane 4), and yet good exogenous expression of SIRT1-Δ2/9 RNA was evident in both cell lines (Fig. 3A, upper panel, lanes 2 and 4). For some reason, ARPE19 cells appeared deficient for translation of SIRT1-Δ2/9 mRNA into protein.
Upon detailed analysis, it emerged that ARPE19 cells express high levels of CUGBP2 mRNA compared with HCT116 cells (Fig. 3D). This raised the possibility that the high levels of CUGPB2 in ARPE19 cells might suppress exogenous SIRT1-Δ2/9 mRNA translation, thus accounting for failure to detect exogenous SIRT1-Δ2/9 protein in these cells. If this were the case, we reasoned that RNAi depletion of CUGBP2 prior to exogenous expression of SIRT1-Δ2/9 might permit translation of the exogenous SIRT1-Δ2/9 RNA into protein. Indeed, this proved to be the case, and we found that selective silencing of CUGBP2 (Fig. 3E) allowed exogenous expression and accumulation of SIRT1-Δ2/9 protein in ARPE19 cells (Fig. 3B). Note that silencing did not affect CUGBP1 RNA levels (Fig. 3E), and our above observations are therefore specifically attributable to CUGBP2.
These results are important on three grounds. First, they demonstrate that different cell types are differentially programmed to downregulate the SIRT1-Δ2/9 protein via the RNA-binding protein CUGBP2 and that CUGBP2 suppresses SIRT1-Δ2/9 translation into protein. Second, regulation is highly selective for the SIRT1-Δ2/9 transcript of SIRT1, since it depends upon the unique junctional sequence generated by SIRT1-Δ2/9 exonic splicing; this sequence is absent from the other known forms of SIRT1, namely, SIRT1-FL and SIRT-Δ8 (see above and Fig. 2D). Third, the fact that expression of SIRT1-Δ2/9 is tightly regulated at two levels, namely, RNA splicing (downregulated by p53 [see above]) and mRNA translation (downregulated by CUGBP2 [this section]), is indicative of an important role(s) for SIRT1-Δ2/9 in vivo.
CUGBP2 does not affect SIRT1-Δ2/9 splicing.
It is well established that RNA-binding proteins can influence RNA splicing of pre-mRNAs. However, selective silencing of CUGBP2 by RNAi did not affect SIRT1-Δ2/9 RNA levels (Fig. 3E), and we conclude that the effects of CUGBP2 on the basal level of SIRT1-Δ2/9 expression are restricted to downregulation of SIRT1-Δ2/9 mRNA translation into protein (see above).
SIRT1-Δ2/9 maintains basal levels of p53 protein.
To explore the possible functions of SIRT1-Δ2/9, we designed an siRNA to target the exonic 2/9 mRNA splice junction. Good and selective SIRT1-Δ2/9 RNA knockdown was obtained (Fig. 4A). Interestingly and unexpectedly, depletion of SIRT1-Δ2/9 was accompanied by depletion of p53 protein levels (Fig. 4B, lanes 2 and 5 for ARPE19 and HCT116 cells, respectively). However, TP53 mRNA levels were unaffected (Fig. 4C). The effects were observed in ARPE19 noncancer epithelial cells and also HCT116 epithelial cancer cells, both cultured in the absence of applied stress (Fig. 4A and B), indicating that SIRT1-Δ2/9 sustains basal levels of p53 protein in both noncancer (ARPE19) and cancerous (HCT116) cells. As expected, RNAi-induced silencing of SIRT1-FL did not cause loss of p53 protein levels (Fig. 4B) (20).
SIRT1-Δ2/9 is linked with induction of p53-dependent apoptosis following stress.
Previously we demonstrated that the basal levels of p53 protein are critical for the ability of p53 to mount a stress response, with upregulation of target genes such as CDKN1A (p21) and MDM2, and that below a certain concentration threshold p53 protein may be activated in terms of posttranslational modifications but nonetheless be deficient for activating downstream stress response mediators (32). Our discovery that, in the absence of applied stress, SIRT1-Δ2/9 maintains basal p53 protein levels in cells (see above and Fig. 4) thus identifies SIRT1-Δ2/9 as a potential crucial player in the ability of cells to mount a p53-dependent response in the face of genotoxic and other stresses.
To test this possibility we next assessed the apoptotic response of HCT116 cells to UV irradiation and also to low doses of 5-FU. Parallel cultures of cells were employed. The control cells were intact. The experimental cells were depleted of SIRT1-Δ2/9 by RNAi before exposure to stress. A significant reduction in apoptosis was consistently observed in SIRT1-Δ2/9-depleted HCT116 p53+/+ cells (Fig. 4D). To determine whether this effect was p53 dependent, we compared isogenic clones of HCT116 p53+/+ and HCT116 p53−/− cells cultured in parallel and treated as above. To a lesser extent, stress treatments also induced p53-independent apoptosis in HCT116 p53−/− cells, as expected. Interestingly, SIRT1-Δ2/9 depletion failed to influence the apoptotic response in HCT116 p53−/− cells either exposed to UV or treated with 5-FU (Fig. 4D).
From the above results we conclude that SIRT1-Δ2/9 contributes toward the p53-dependent apoptotic response to stress. Since we also showed that SIRT1-Δ2/9 sustains basal p53 protein levels (see above), it seems reasonable to suggest that stabilization of p53 protein by SIRT1-Δ2/9 contributes toward p53-dependent apoptosis in response to stress.
Mutant p53 levels are not affected by SIRT1-Δ2/9.
Interestingly, mutant p53 protein levels appeared independent of SIRT1-Δ2/9, as evidenced by silencing of SIRT1-Δ2/9 in DLD-1 human colorectal cancer cells (see Fig. S3 in the supplemental material). The TP53 gene in DLD-1 cells is mutated at codon 241 to give p53 S241F. The resultant change from serine to phenylalanine is adjacent to the zinc-binding C242 residue and is predicted to disrupt p53 protein conformation and DNA-binding capacity. The observed lack of SIRT1-Δ2/9 effect on mutant p53 in DLD-1 cells therefore might have been due to loss of the wt p53 conformation with knock-on effects on molecular interactions. With this in mind, we next asked if SIRT1-Δ2/9 interacts physically with the p53 protein.
SIRT1-Δ2/9 protein binds p53 protein.
Protein-protein complexes containing SIRT1-Δ2/9 and wt p53 were detected by exogenous expression and immunoprecipitation of His-tagged SIRT1-Δ2/9, followed by immunoblotting for coprecipitated endogenous wt p53 protein (Fig. 4E, ARPE19 and HCT116 cells). A relatively high signal was observed for p53 that coimmunoprecipitated with overexpressed SIRT1-Δ2/9, suggesting stable complex formation between SIRT1-Δ2/9 and endogenous p53.
Under basal conditions of culture, HCT116 cells express a proportion of p53 protein which is phosphorylated at serine 15 (unpublished observations). Interestingly, p53 S15P appeared to be excluded from SIRT1-Δ2/9 complexes, indicating that SIRT1-Δ2/9 binding affinity may be selective for p53 lacking S15P (Fig. 4E).
A binding interaction between wild-type p53 and SIRT1-Δ2/9 was confirmed for endogenous proteins by using HCT116 cell lysates and pulldown of p53 protein followed by immunoblotting for SIRT1-Δ2/9 using an N-terminal monoclonal antibody (Epitomics) (see Fig. S5 in the supplemental material). Binding appeared preferential for the 34-kDa form of the SIRT1-Δ2/9 protein (see Fig. S5), similar to the results obtained with exogenously expressed SIRT1-Δ2/9 protein (Fig. 4E). Thus, complexing between endogenous p53 and SIRT1-Δ2/9 proteins is evident in HCT116 cells under normal conditions of culture, and future studies will seek to determine if such complexes contribute in whole or in part toward the ability of SIRT1-Δ2/9 to sustain basal p53 protein levels in human cells.
It is interesting that endogenous SIRT1-FL was not evident in complex with endogenous p53 (this work; see also Fig. S5 in the supplemental material), whereas Vaziri et al. (53) reported 0.0005% endogenous SIRT1-FL in complex with endogenous p53 in MCF7-L cells (MCF7 cells expressing an exogenous p21P-luc promoter). It is possible that this apparent discrepancy reflects cell type differences, and/or the presence of abnormal exogenous promoter expression in the MCF7-L cells, and/or differences in cell culture conditions. In addition, use of a polyclonal antibody against the C terminus of SIRT1 (53) may have allowed more sensitive detection below the threshold obtained with the SIRT1 N-terminal monoclonal antibody (this work). Whatever the explanation, it appears that wt p53 is preferentially complexed with SIRT1-Δ2/9 with little, if any, complexing with SIRT1-FL in HCT116 cells under basal conditions of cell culture (i.e., in the absence of applied stress).
In marked contrast to the results obtained with ARPE19 and HCT116 cells, which expressed wt p53, no SIRT1-Δ2/9–p53 protein complexes were evident following exogenous expression of SIRT1-Δ2/9 in DLD-1 cells (see Fig. S4 in the supplemental material). DLD-1 cells express mutant p53 S241F (see above), and we attributed lack of complex formation with SIRT1-Δ2/9 to the mutant nature of the p53 S241F protein. Thus, binding of p53 by SIRT1-Δ2/9 appears selective for the wild-type conformation of p53 protein.
SIRT1-Δ2/9 regulates expression of a subset of p53 target genes.
The above results demonstrate that SIRT1-Δ2/9 selectively complexes with wt p53 under basal conditions of cell culture and that binding is selective for a specific form of p53 protein (i.e., nonphosphorylated at serine 15). Given that p53 functions as a transcription factor and is able to recognize a range of target gene promoters, we next asked if SIRT1-Δ2/9 is linked with transcriptional regulation of p53 target genes. For this purpose, we first performed genomic RNA arrays before and after selective silencing of SIRT1-Δ2/9 by RNAi (using siRNA directed against the 2/9 junctional mRNA sequence [see above]). For comparison we also included, in parallel, experiments entailing selective silencing of SIRT1-FL using an siRNA directed against exon 8 of SIRT1 mRNA (20). Selective mRNA knockdown was obtained for SIRT1-Δ2/9 (without effect on SIRT1-FL RNA) (Fig. 4A) and enabled identification of SIRT1-Δ2/9-specific effects on gene expression.
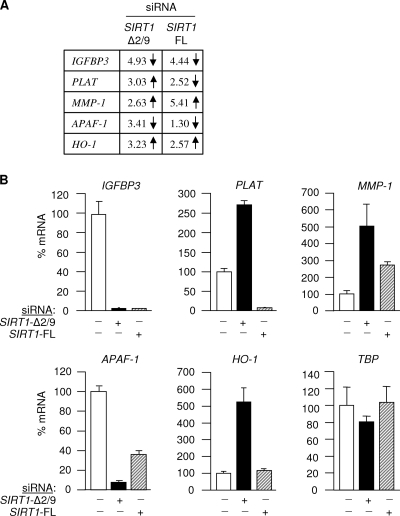
The genomic RNA array data indicated that SIRT1-Δ2/9 selectively governs the expression of a small group of genes in human cells (Fig. 5; see also the results described below). Since SIRT1-Δ2/9 lacks a deacetylase capacity, which is essential for epigenetic regulation of gene expression by full-length SIRT1, we concluded that SIRT1-Δ2/9 (i) represents a novel regulator of gene expression encoded by the SIRT1 gene and (ii) operates via a mechanism that is distinct from that of SIRT1-FL.
Fig 5.
SIRT1-Δ2/9 regulates a subset of p53 target genes. Fold change values and the direction of regulation of a subset of p53-responsive genes identified by SIRT1-Δ2/9 RNAi microarray in ARPE19 cells (A) and results when analyzed by qRT-PCR (B). TBP primers were used as a control.
Although the number of genes subject to SIRT1-Δ2/9 regulation is small, it was very interesting that a number of these genes are also known to be regulated by p53. We selected five p53-responsive genes from the SIRT1-Δ2/9 RNAi microarray for confirmation by quantitative RT-PCR, namely, IGFBP3, PLAT, MMP-1, APAF-1, and HO-1. IGFBP3 (insulin-like growth factor binding protein 3) is upregulated by p53 and regulates IGF-I availability for interaction with the IGF-1 receptor (7, 10). PLAT (tissue-type plasminogen activator) is a secreted serine protease that converts the proenzyme plasminogen to plasmin. p53 represses PLAT expression through a non-DNA-binding mechanism (29). Matrix metalloproteinase 1 (MMP-1; downregulated by p53 [49]) is involved in the breakdown of the extracellular matrix in normal physiological processes as well as in disease processes, such as metastasis. APAF-1 is a cytoplasmic protein that initiates apoptosis, and p53 directly regulates APAF-1 transcription via the two p53 consensus binding sites in the APAF-1 promoter (21). Finally, heme oxygenase 1 (HO-1) is a stress-inducible gene with antioxidant properties. The HO-1 promoter contains a p53 response element and is stimulated in a p53-dependent manner (35). The effects of SIRT1-Δ2/9 depletion on these p53-responsive genes are presented in Fig. 5A and B.
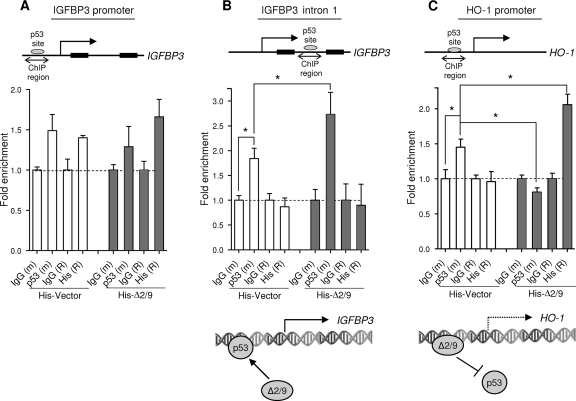
SIRT1-Δ2/9 binds specific p53 promoter targets and downregulates p53 binding.
Combined, the above observations demonstrate (i) that the SIRT1-Δ2/9 protein complexes with p53 protein and (ii) that SIRT1-Δ2/9 is linked with the regulated expression of a subset of established p53 target genes. Does SIRT1-Δ2/9 in some way direct p53 to a subset of target genes? Alternatively, when SIRT1-Δ2/9 downregulates p53 target genes, does SIRT1-Δ2/9 somehow block promoter transactivation by p53? To address these questions we asked if SIRT1-Δ2/9 can bind chromatin at p53 target promoters, using ChIP (see Materials and Methods). Two p53-related gene targets were selected. The first, IGFBP3, was strongly downregulated upon SIRT1-Δ2/9 silencing (Fig. 5A and B), indicating that SIRT1-Δ2/9 is required, directly or indirectly, to maintain IGFBP3 gene expression. The second, HO-1, is upregulated following SIRT1-Δ2/9 silencing, indicating that SIRT1-Δ2/9 normally suppresses HO-1 expression under basal conditions of cell culture.
In the case of the IGFBP3 gene there are two reported p53-binding sites (7, 10), the first in the promoter region and the second within intron 1 (Fig. 6A and B). The ChIP results showed that exogenous SIRT1-Δ2/9 upregulated p53 binding at the intron 1 binding site [Fig. 6B; p53(m), compare the His-vector control with His-Δ2/9] but had no significant effect on p53 binding at the conventional promoter p53-binding site [Fig. 6B; His(R), compare the His-vector control with His-Δ2/9]. From this, we concluded that SIRT1-Δ2/9 can influence p53 binding at specific target sequences in chromatin DNA. Since SIRT1-Δ2/9 itself was not also evident at the same site [Fig. 6B, His(R)], we suggest that p53 binding is somehow facilitated by SIRT1-Δ2/9 but does not involve stable SIRT1-Δ2/9–p53 complexes.
Fig 6.
Distinct regulation of p53 target genes by SIRT1-Δ2/9. Chromatin immunoprecipitation results showed enrichment of p53 and His-SIRT1-Δ2/9 on p53-binding sites located in the IGFBP3-promoter (A), IGFBP3 intron 1 (B), and the HO-1 promoter (C). The histograms show the fold enrichment relative to IgG controls (see Materials and Methods). The results for SIRT1-Δ2/9 specifically support the recruitment of p53 to the p53-binding site on IGFBP3 intron 1. In contrast, SIRT1-Δ2/9 binds to the HO-1 promoter and prevents the recruitment of p53 to this site. m and R indicate corresponding mouse and rabbit IgGs, respectively. ∗, P < 0.05.
In the case of HO-1 expression, which is downregulated by SIRT1-Δ2/9, a different picture emerged. Under basal, control conditions the endogenous p53 protein was bound at the promoter and presumably was responsible for basal HO-1 expression levels [Fig. 6C; p53(m), compare the His-vector control with His-Δ2/9]. However, p53 binding was lost following exogenous expression of SIRT1-Δ2/9 [Fig. 6C, His(R), compare the His-vector control with His-Δ2/9]. This is consistent with the apparent suppression of HO-1 by endogenous SIRT1-Δ2/9, as indicated by the 5-fold increase in HO-1 expression following selective SIRT1-Δ2/9 silencing (Fig. 5A and B) (see above). Importantly, loss of p53 binding at the HO-1 promoter correlated with de novo binding of exogenous His-SIRT1-Δ2/9 at this same site (Fig. 6C). This was particularly interesting and identified SIRT1-Δ2/9 as a chromatin-binding protein. It also raised the possibility that SIRT1-Δ2/9 may downregulate transcription factors such as p53 by occluding and therefore competing for specific DNA-binding sequences in chromatin. Future studies will explore this intriguing possibility in more detail.
It is important to remember that the above experiments were all performed in the absence of applied stress, and under conditions of RNAi we have demonstrated (1) an effective target for mRNA depletion without activating cellular stress responses. Overall, these observations indicate (i) that SIRT1-Δ2/9 can modulate the transcriptional activity of proteins such as p53, (ii) that such effects are specific for given regulatory target sequences in chromatin, and (iii) that SIRT1-Δ2/9 is itself a chromatin-binding protein and can result in either upregulation or downregulation of promoter activity.
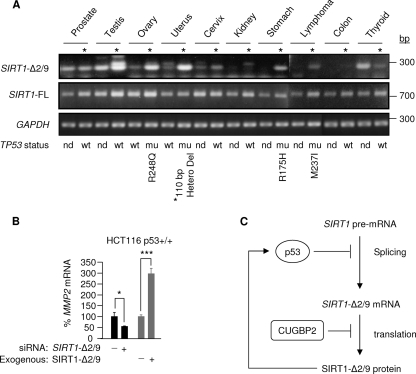
SIRT1-Δ2/9 is overexpressed in human cancer.
The expression of SIRT1-Δ2/9 is detectable in both human cancer and paired noncancer tissues. In cancer tissues from the testis, ovary, uterus, cervix, and stomach and lymphoma, SIRT1-Δ2/9 was overexpressed relative to paired noncancerous tissue (Fig. 7A). To investigate this further, we sequenced the p53 (exons 5 to 8) present in the human tissue samples. The results showed that loss of wt p53 (5 cases) correlated with elevated SIRT1-Δ2/9 levels in 4/5 cases (Fig. 7A). These new clinical data indicate that loss of functional p53 may lead to overexpression of SIRT1-Δ2/9 in human cancers. Only one in six of the wt p53 samples expressed high SIRT1-Δ2/9 levels; interestingly, (i) this was a testicular cancer, in which mutation of p53 is uncommon, and (ii) SIRT1 transcripts are known to be highly expressed in testicular tissue.
Fig 7.
SIRT1-Δ2/9 mRNA is overexpressed in cancer tissues. (A) SIRT1-Δ2/9, SIRT1-FL, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA expression in paired cancer versus adjacent normal control tissue samples, using 100 ng total RNA from each tissue with primer pairs 2-9F/10R for SIRT1-Δ2/9, SEx4/8Rvs for SIRT1-FL, and GAPDH (primer sequences are shown in Table S1 of the supplemental material). TP53 (exons 5 to 8) was sequenced (Materials and Methods), and details of individual mutations are indicated. wt, wild-type p53; nd, not determined; ∗, deletion from residues 188 to 224. (B) SIRT1-Δ2/9 regulates MMP2. The graph shows levels of MMP2 RNA following siRNA knockdown or overexpression of SIRT1-Δ2/9 in HCT116 cells. ∗, P < 0.05; ∗∗∗, P < 0.001. MMP2 RNA levels were determined by qRT-PCR. (C) Schematic model of SIRT1-Δ2/9 regulation. SIRT1-Δ2/9 mRNA splicing and translation of mRNA into protein are suppressed by p53 and CUGBP2, respectively, while SIRT1-Δ2/9 is required to maintain basal p53 protein levels, thus forming a feedback loop between SIRT1-Δ2/9 and p53.
Overexpression of SIRT1-Δ2/9 in human cancers is consistent with our observations with human cells that exhibited high expression levels of SIRT-Δ2/9 in human HCT116 cancer cells compared with ARPE19 noncancer cells (see above). Microarray analysis following silencing of SIRT1-Δ2/9 has identified a set of key genes involved in the initiation of cancer cell invasion and metastasis (Z. H. Shah and J. Milner, unpublished observations), including MCAM (3.5-fold downregulation) and TGFBR2 (3.2-fold upregulation) in cancer. In addition, we also identified MMP2 (matrix metalloproteinase 2) as a positively regulated target of SIRT1-Δ2/9 (Fig. 7B). This was demonstrated by (i) qPCR of MMP2 mRNA following SIRT1-Δ2/9 silencing, which resulted in a decrease in MMP2 mRNA levels, and (ii) SIRT1-Δ2/9 overexpression, which increased MMP2 mRNA expression levels (Fig. 7B). Since MMP2 has a strong role in cancer metastasis (40), these observations further support a functional link between SIRT1-Δ2/9 and cancer. Overall, these results indicate that the observed abnormal overexpression of SIRT1-Δ2/9 in human tumor samples (Fig. 7A) may contribute to cancer progression and malignancy.
DISCUSSION
The discovery that SIRT1 is subject to alternative RNA splicing (reference 33 and this study) has profound implications for our understanding of the functions of SIRT1 in health and disease and also for the development of therapeutic agents designed to target defined disease-related functions of SIRT1. The discovery of SIRT1 splice variants may also help resolve apparently conflicting observations reported for the effects of SIRT1 depletion on embryonic development (13, 14, 34, 54) and for the link between SIRT1 and cancer, in which SIRT1 has been identified as both a tumor suppressor and also as a cancer-specific survival factor (8, 18, 20, 25, 26, 27, 48, 54). Alternative splicing from a single gene is an effective mechanism for amplifying the functional diversity of that same gene and can lead to the production of proteins with either coordinated functions or opposing functions (for a recent review, see reference 16). Thus, it is possible that alternative splicing of SIRT1 may, for example, enable differing functions in cancer development and cancer cell survival. In the case of SIRT1-Δ2/9 the loss of the central catalytic core domain indicates that this particular variant must be functionally distinct from SIRT1-FL, an NAD-dependent deacetylase.
SIRT1-Δ2/9 expression is highly regulated.
The expression of SIRT1-Δ2/9 is highly regulated. First, SIRT1 pre-mRNA splicing between exons 2 and 9 is downregulated by p53, thus forming an autoregulatory feedback loop between p53 and SIRT1-Δ2/9 (see the schematic in Fig. 7C). Second, the translation of SIRT1-Δ2/9 mRNA into protein is downregulated by the RNA-binding protein CUGBP2 (Fig. 7C). The latter effect is particularly evident in ARPE19 pigmented retinal epithelial cells, which express high endogenous levels of CUGBP2 compared with HCT116 colorectal cancer cells. We further showed that binding by CUGBP2 is dependent, in whole or in part, upon the creation of a CUGBP2-binding motif at the 2/9 splice junction, thus providing exquisite selectivity for regulated translation of SIRT1-Δ2/9 over SIRT1-FL and SIRT1-Δ8 mRNA.
SIRT1-Δ2/9 enables p53 tumor suppressor functioning.
Under basal, nonstress conditions, the levels of p53 protein are maintained at low levels via a sophisticated mechanism involving subcellular localization of p53 and systems for targeting the p53 protein for proteosomal degradation (see reference 9 and other articles in the same issue of Nature Reviews Cancer, 2009). This results in a rapid turnover of p53 protein. Nonetheless, the basal levels of p53 protein are important. This is evident in p53+/− cells in which the basal expression level of p53 protein is 25% of that in p53+/+ cells (32). Interestingly, this differential is maintained following oncogenic stress, resulting in an attenuated p53 stress response and reduced p53-dependent apoptosis (32).
We have now identified SIRT1-Δ2/9 as a key player in maintaining basal p53 protein levels and have showed that depletion of SIRT1-Δ2/9 by RNAi results in reduced p53 protein levels in both noncancer cells (ARPE19) and cancerous cells (HCT116) (Fig. 4B). This effect is specific to SIRT1-Δ2/9, since p53 protein levels increase following selective depletion of SIRT1-FL (Fig. 4B) and are unaffected by selective depletion of SIRT1-Δ8 (33). Thus, alternative splicing generates SIRT1 variants, each with individual effects upon resting levels of p53 protein.
The ability of SIRT1-Δ2/9 to maintain basal p53 protein levels appears important for the ability of p53 to induce apoptosis in response to stress, since depletion of SIRT1-Δ2/9 reduces p53-dependent apoptosis in response to DNA-damaging agents (Fig. 4D). This is completely consistent with the correlation between low basal p53 levels and haplo-insufficiency (32) (see above) and identifies SIRT1-Δ2/9 as a critical enabler of p53 tumor suppressor functioning.
Regulation of p53: SIRT1-FL versus SIRT1-Δ2/9.
In this work we have presented evidence for a new SIRT1 splice variant, SIRT1-Δ2/9, that lacks the catalytic core domain of SIRT1-FL. Nonetheless, both SIRT1-FL and SIRT1-Δ2/9 recognize and interact, directly or indirectly, with the tumor suppressor p53 protein (introduction and this study). However, the nature and consequences of these interactions are fundamentally different. Thus, SIRT1-FL recognizes activated p53, which it targets as a substrate for deacetylation and hence inactivation. In this way, SIRT1-FL downregulates activated p53 and impacts upon the p53 stress response in a negative manner. Conversely, SIRT1-Δ2/9 impacts upon the p53 stress response in a positive way, and this probably reflects the ability of SIRT1-Δ2/9 to bind and to maintain basal levels of nonactivated p53 at a critical threshold for activation of the cellular stress response (see above).
Interestingly, SIRT1-Δ2/9 does not appear to bind p53-S15P and therefore presumably does not compete with SIRT1-FL for access to the activated p53-S15P protein. It is possible that SIRT1-FL and SIRT1-Δ2/9 cooperate in tuning the cellular stress response to the degree of stress-induced damage sustained by the cell.
Despite lacking SIRT1 deacetylase activity, SIRT1-Δ2/9 nonetheless regulates the expression of specific target genes, including a subset of p53 target genes. This raises the question of the mechanism of SIRT1-Δ2/9 function. Our observations indicate that, in part at least, the functioning of SIRT1-Δ2/9 involves highly selective interactions with specific protein-binding partners, manifest by its selective binding characteristics for p53 protein. Thus, SIRT1-Δ2/9 selectively binds wild-type but not mutant p53 and appears able to discriminate between p53 that is either nonphosphorylated or phosphorylated at serine 15 (being selective for the nonphosphorylated p53 form). This observation offers a possible mechanism of SIRT1-Δ2/9-mediated stabilization of p53; SIRT1-Δ2/9 may protect p53 from ubiquitinylation by the E3 ligase MDM2, which binds the N terminus of nonphosphorylated p53 (S15) and targets p53 for rapid degradation (12, 24, 28, 38). Stress-activated S15 phosphorylation of p53 (p53 S15P) reduces MDM2 binding and enables stabilization of activated p53 S15P as part of the cellular stress response (45, 46). We propose that, by protecting the nonphosphorylated form of p53 from MDM2 under basal conditions, SIRT1-Δ2/9 may interfere with p53 targeting by MDM2 and thus help maintain basal levels of p53 protein. In this context it is relevant that we were unable to detect MDM2 present in the SIRT1-Δ2/9–p53 protein complexes, as predicted by this hypothesis (data not shown). Future studies will characterize the molecular mechanism of p53 stabilization by SIRT1-Δ2/9 in detail.
SIRT1-Δ2/9 and malignancy.
SIRT1-Δ2/9 RNA was found to be overexpressed in a number of cancer tissues compared with the paired noncancer samples (Fig. 7A). In cancer, p53 tumor suppressor function is lost due either to mutation or aberrant regulation. SIRT1-Δ2/9 overexpression would be entirely consistent with loss of p53-mediated suppression of SIRT1 2/9 pre-mRNA splicing, resulting in SIRT1-Δ2/9 overexpression. This suggestion is strongly supported by the observed correlation between loss of wt p53 and overexpression of SIRT1-Δ2/9 in the human tissue samples (Fig. 7A). Significantly, RNAi in combination with genomic array data indicated that SIRT1-Δ2/9 is linked with the selective expression of genes known to be involved in malignant cell transformation and cancer cell invasion. Thus, it is possible that SIRT1-Δ2/9, once it escapes downregulation by p53, may promote cancerous changes and metastasis.
In summary, we report the discovery of SIRT1-Δ2/9, a new splice variant of the epigenetic regulator SIRT1. Future studies will determine if abnormal SIRT1-Δ2/9 overexpression in cancer identifies SIRT1-Δ2/9 as a potential marker for malignant transformation, a process in which it may play a causal role via its influence on the expression of transformation-associated genes. In this context, SIRT1-Δ2/9 also holds promise as a novel target for anticancer therapy. In normal cells, the highly regulated expression of SIRT1-Δ2/9 at the levels of pre-mRNA splicing (downregulated by p53) and mRNA translation (downregulated by CUGBP2) indicates that this newly discovered epigenetic regulator is likely to exert a major impact upon gene expression patterns during mammalian development and differentiation and, when deregulated, in disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bert Vogelstein (John Hopkins University) for the HCT116 p53+/+ and p53−/− isogenic clones and also Naveed Aziz and Peter Ashton of the Genomics and Bioinformatics Laboratories, Department of Biology, University of York, for helpful discussions.
Z.H.S., J.R.F., and J.M. conceived and designed the experiments. Z.H.S., J.R.F., S.U.A., S.J.A., and J.R.P.K. performed the experiments. J.M. wrote the manuscript.
This work was funded by a program grant to J.M. from Yorkshire Cancer Research.
Footnotes
Published ahead of print 28 November 2011
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Ahmed SU, Milner J. 2009. Basal cancer cell survival involves JNK2 suppression of a novel JNK1/c-Jun/Bcl-3 apoptotic network. PLoS One 4:e7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allison SL, Stadler K, Mandl CW, Kunz C, Heinz FX. 1995. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J. Virol. 69:5816–5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Asher G, et al. 2008. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134:317–328 [DOI] [PubMed] [Google Scholar]
- 4. Banks AS, et al. 2008. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 8:333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baxter EW, Cummings WJ, Fournier RE. 2005. Formation of a large, complex domain of histone hyperacetylation at human 14q32.1 requires the serpin locus control region. Nucleic Acids Res. 33:3313–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bouras T, et al. 2005. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J. Biol. Chem. 280:10264–10276 [DOI] [PubMed] [Google Scholar]
- 7. Bourdon JC, et al. 1997. Further characterisation of the p53 responsive element: identification of new candidate genes for trans-activation by p53. Oncogene 14:85–94 [DOI] [PubMed] [Google Scholar]
- 8. Bradbury CA, et al. 2005. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia 19:1751–1759 [DOI] [PubMed] [Google Scholar]
- 9. Brooks CL, Gu W. 2009. How does SIRT1 affect metabolism, senescence and cancer? Nat. Rev. Cancer 9:123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Buckbinder L, et al. 1995. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature 377:646–649 [DOI] [PubMed] [Google Scholar]
- 11. Chen D, et al. 2008. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 22:1753–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen J, Marechal V, Levine AJ. 1993. Mapping of the p53 and mdm-2 interaction domains. Mol. Cell. Biol. 13:4107–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheng HL, et al. 2003. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 100:10794–10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coussens M, Maresh JG, Yanagimachi R, Maeda G, Allsopp R. 2008. Sirt1 deficiency attenuates spermatogenesis and germ cell function. PLoS One 3:e1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Das C, Lucia MS, Hansen KC, Tyler JK. 2009. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459:113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. David CJ, Manley JL. 2010. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 24:2343–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Donmez G, Wang D, Cohen DE, Guarente L. 2010. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell 142:320–332 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18. Firestein R, et al. 2008. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One 3:e2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ford J, Ahmed S, Allison S, Jiang M, Milner J. 2008. JNK2-dependent regulation of SIRT1 protein stability. Cell Cycle 7:3091–3097 [DOI] [PubMed] [Google Scholar]
- 20. Ford J, Jiang M, Milner J. 2005. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 65:10457–10463 [DOI] [PubMed] [Google Scholar]
- 21. Fortin A, et al. 2001. APAF1 is a key transcriptional target for p53 in the regulation of neuronal cell death. J. Cell Biol. 155:207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ghosh HS. 2008. The anti-aging, metabolism potential of SIRT1. Curr. Opin. Invest. Drugs 9:1095–1102 [PubMed] [Google Scholar]
- 23. Haigis MC, Guarente LP. 2006. Mammalian sirtuins: emerging roles in physiology, aging, and calorie restriction. Genes Dev. 20:2913–2921 [DOI] [PubMed] [Google Scholar]
- 24. Haupt Y, Maya R, Kazaz A, Oren M. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296–299 [DOI] [PubMed] [Google Scholar]
- 25. Hida Y, Kubo Y, Murao K, Arase S. 2007. Strong expression of a longevity-related protein, SIRT1, in Bowen's disease. Arch. Dermatol. Res. 299:103–106 [DOI] [PubMed] [Google Scholar]
- 26. Huffman DM, et al. 2007. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 67:6612–6618 [DOI] [PubMed] [Google Scholar]
- 27. Knight JR, Milner J. 2012. SIRT1, metabolism and cancer. Curr. Opin. Oncol. 24:68–75 [DOI] [PubMed] [Google Scholar]
- 28. Kubbutat MH, Jones SN, Vousden KH. 1997. Regulation of p53 stability by Mdm2. Nature 387:299–303 [DOI] [PubMed] [Google Scholar]
- 29. Kunz C, Pebler S, Otte J, von der Ahe D. 1995. Differential regulation of plasminogen activator and inhibitor gene transcription by the tumor suppressor p53. Nucleic Acids Res. 23:3710–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lavu S, Boss O, Elliott PJ, Lambert PD. 2008. Sirtuins: novel therapeutic targets to treat age-associated diseases. Nat. Rev. Drug Discov. 7:841–853 [DOI] [PubMed] [Google Scholar]
- 31. Luo J, et al. 2001. Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107:137–148 [DOI] [PubMed] [Google Scholar]
- 32. Lynch CJ, Milner J. 2006. Loss of one p53 allele results in four-fold reduction of p53 mRNA and protein: a basis for p53 haplo-insufficiency. Oncogene 25:3463–3470 [DOI] [PubMed] [Google Scholar]
- 33. Lynch CJ, et al. 2010. SIRT1 undergoes alternative splicing in a novel auto-regulatory loop with p53. PLoS One 5:e13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McBurney MW, et al. 2003. The mammalian SIR2α protein has a role in embryogenesis and gametogenesis. Mol. Cell. Biol. 23:38–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meiller A, et al. 2007. p53-dependent stimulation of redox-related genes in the lymphoid organs of gamma-irradiated: mice identification of haeme-oxygenase 1 as a direct p53 target gene. Nucleic Acids Res. 35:6924–6934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Milne JC, et al. 2007. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450:712–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Motta MC, et al. 2004. Mammalian SIRT1 represses forkhead transcription factors. Cell 116:551–563 [DOI] [PubMed] [Google Scholar]
- 38. Oliner JD, et al. 1993. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 362:857–860 [DOI] [PubMed] [Google Scholar]
- 39. Peritz T, et al. 2006. Immunoprecipitation of mRNA-protein complexes. Nat. Protoc. 1:577–580 [DOI] [PubMed] [Google Scholar]
- 40. Radisky ES, Radisky DC. 2010. Matrix metalloproteinase-induced epithelial-mesenchymal transition in breast cancer. J. Mammary Gland Biol. Neoplasia 15:201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ramsey KM, et al. 2009. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324:651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rodgers JT, et al. 2005. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434:113–118 [DOI] [PubMed] [Google Scholar]
- 43. Rogina B, Helfand SL. 2004. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. U. S. A. 101:15998–16003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rubbi CP, Milner J. 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 22:6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shieh SY, Ikeda M, Taya Y, Prives C. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325–334 [DOI] [PubMed] [Google Scholar]
- 46. Siliciano JD, et al. 1997. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 11:3471–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sinclair DA, Guarente L. 1997. Extrachromosomal rDNA circles: a cause of aging in yeast. Cell 91:1033–1042 [DOI] [PubMed] [Google Scholar]
- 48. Stunkel W, et al. 2007. Function of the SIRT1 protein deacetylase in cancer. Biotechnol. J. 2:1360–1368 [DOI] [PubMed] [Google Scholar]
- 49. Sun Y, Wenger L, Rutter JL, Brinckerhoff CE, Cheung HS. 1999. p53 down-regulates human matrix metalloproteinase-1 (collagenase-1) gene expression. J. Biol. Chem. 274:11535–11540 [DOI] [PubMed] [Google Scholar]
- 50. Tippmann F, Hundt J, Schneider A, Endres K, Fahrenholz F. 2009. Up-regulation of the alpha-secretase ADAM10 by retinoic acid receptors and acitretin. FASEB J. 23:1643–1654 [DOI] [PubMed] [Google Scholar]
- 51. Tissenbaum HA, Guarente L. 2001. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410:227–230 [DOI] [PubMed] [Google Scholar]
- 52. van der Horst A, et al. 2004. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1). J. Biol. Chem. 279:28873–28879 [DOI] [PubMed] [Google Scholar]
- 53. Vaziri H, et al. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149–159 [DOI] [PubMed] [Google Scholar]
- 54. Wang RH, et al. 2008. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell 14:312–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.