Abstract
The mouse PERIOD1 (mPER1) protein, along with other clock proteins, plays a crucial role in the maintenance of circadian rhythms. mPER1 also provides an important link between the circadian system and the cell cycle system. Here we show that the circadian expression of mPER1 is regulated by rhythmic translational control of mPer1 mRNA together with transcriptional modulation. This time-dependent translation was controlled by an internal ribosomal entry site (IRES) element in the 5′ untranslated region (5′-UTR) of mPer1 mRNA along with the trans-acting factor mouse heterogeneous nuclear ribonucleoprotein Q (mhnRNP Q). Knockdown of mhnRNP Q caused a decrease in mPER1 levels and a slight delay in mPER1 expression without changing mRNA levels. The rate of IRES-mediated translation exhibits phase-dependent characteristics through rhythmic interactions between mPer1 mRNA and mhnRNP Q. Here, we demonstrate 5′-UTR-mediated rhythmic mPer1 translation and provide evidence for posttranscriptional regulation of the circadian rhythmicity of core clock genes.
INTRODUCTION
Circadian rhythms are produced by an endogenous clock system and are present in single-celled to complex organisms. The principal circadian pacemaker is located in the suprachiasmatic nucleus (SCN) of the hypothalamus in mammals (33, 44). The mammalian molecular circadian clock system is composed of feedback loops comprised of regulatory steps at the transcriptional, translational, and posttranslational levels (31). These regulatory steps must be coordinated properly for the fine-tuning of both amplitude and 24-h periodicity. In particular, posttranscriptional regulation plays an important role, although its mechanism is less well understood (17, 26, 29, 46, 47). One of the core clock genes in mammals, Period1 (Per1), was originally identified as a structural homologue of the Drosophila melanogaster circadian clock gene per (42). The transcription of Per1 is activated by the CLOCK-BMAL1 heterodimer (13, 20) and repressed by a complex containing PER and cryptochrome (CRY) proteins (28), thus comprising one of the core feedback loops. Although the molecular function of mPER1 has not yet been defined, it is an essential gene for the maintenance of circadian rhythm, because Per1 knockout mice show an altered period (2, 5, 52). mPER1 is thought to be involved in resetting the circadian oscillator (1) and to provide an important link between the circadian system and the cell cycle system, such as cell growth and DNA damage control (14). Interestingly, mouse Per1 (mPer1) expression is rhythmic, but the phase of protein expression is delayed 6 to 8 h relative to the mRNA in mouse SCN (10), indicating that mPER1 expression may be regulated at a posttranscriptional step. This time lag between the mRNA and protein expression profiles has also been observed in the Drosophila per gene (51), suggesting that these time lags may be important for the clock system. Until now, many researchers interested in circadian systems have focused on transcriptional and posttranslational regulatory steps, with minor efforts focused on posttranscriptional control, especially mRNA stability. Moreover, the role of translational control in circadian rhythmicity is not well understood. We hypothesized that circadian phase-specific translational regulation of mPer1 mRNA might be a novel mechanism for controlling mPER1 expression. One mechanism of translational regulation is an internal ribosomal entry site (IRES)-mediated system. IRESs recruit ribosomes directly in a cap-independent manner, in contrast to the canonical cap-dependent scanning model (12, 19, 43). Since the discovery of viral IRESs (22, 37), various cellular mRNAs have been shown to contain IRESs. IRES-mediated translation is used to regulate protein synthesis in certain physiological circumstances (41, 50), such as apoptosis, cell cycle, development, and differentiation. Furthermore, IRES-mediated translation is important to nocturnal arylalkylamine N-acetyltransferase (AANAT) protein synthesis in the rat pineal gland (25). In contrast to canonical cap-dependent translation, IRES-mediated translation can potentially be controlled in various ways, such as in the presence of IRES trans-acting factors (ITAFs), RNA secondary structures, RNA levels, and in some cases, iron (38). ITAF is thought to function as an RNA chaperone (38, 40). The binding of ITAF stabilizes a specific IRES RNA conformation that enables the binding of other factors or of the ribosome. Therefore, the binding of a specific combination of ITAFs on a target IRES in the 5′ untranslated region (UTR) could control the translation system.
MATERIALS AND METHODS
Plasmid constructions.
mPer1 5′-UTRs (e1A and e1B) were amplified from mPer1 cDNA using Pfu polymerase (Solgent) and confirmed by sequencing. The resulting products were cloned into the SalI/SmaI site of the intercistronic region of a pRF bicistronic vector containing Renilla luciferase (Rluc) in the first cistron and firefly luciferase (Fluc) in the second cistron (8, 23, 25). We used pRF, pHRF, and ΔCMV RF vector backbones (25). To create the deletion constructs pHRF144 and pHRF63, mPer1 5′-UTR fragments were amplified from pRFe1A and pRFe1B and then inserted into the SalI/SmaI site of the mock vector pHRF (25).
For the in vitro binding assay/UV cross-linking experiment, fragments of the mPer1 5′-UTR were amplified, and the PCR products were digested and subcloned into the EcoRI/XbaI site of the pSK′ vector (24) to generate pSK′-e1A, pSK′-e1B, pSK′-144, and pSK′-63.
To generate the bicistronic mRNA reporter for mRNA transfection, pCY2-RFe1A, pCY2-RFe1B, and pCY2-RF63 were constructed as follows: the 5′-UTRs of mPer1 were cut from pRFe1A and pRFe1B using SalI/BamHI and inserted into the SalI/BamHI site of pCY2-RF (6, 25).
Cell culture, isolation of embryonic fibroblasts, and drug treatment.
HEK 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM; WelGENE) with 10% fetal bovine serum (HyClone) and 1% antibiotics (WelGENE). NIH 3T3 cells were cultured in DMEM with 10% fetal bovine serum and 1% antibiotics and maintained in a humidified 95% air–5% CO2 incubator. Mouse embryonic fibroblasts (MEFs) were isolated from trypsin-EDTA-digested embryonic day 13.5 (E13.5) embryos (30). Primary MEFs were cultured in DMEM containing glutamine but not Na-pyruvate (HyClone), with 1% antibiotics, 1% glutamine (Gibco), 0.1% 2-mercaptoethanol (Gibco), and 10% fetal bovine serum. The circadian oscillation of NIH 3T3 cells was synchronized by treatment with 100 nM dexamethasone. After 2 h, the medium was replaced with complete medium (4, 29, 46, 47). To block the translation system, NIH 3T3 cells were treated with 20 nM rapamycin or 100 μg/ml cycloheximide (CHX) and then harvested at the indicated times.
Transient transfection and RNA interference.
For expression of the reporter constructs, HEK 293T and NIH 3T3 cells were seeded in 24-well plates at a density of 5 × 104 cells per well 1 day prior to transfection. Transfections were carried out using Metafectene (Biontex) according to the manufacturer's instruction. After incubation for 36 h, cells were harvested. The reporter mRNA transfection was performed as follows: NIH 3T3 cells were transiently transfected with 1 μg of the capped bicistronic reporter mRNA and incubated for 2 h. The medium was then exchanged for complete medium, and the cells were incubated for a further 4 h. In the study of time-dependent transfection, NIH 3T3 cells were treated with dexamethasone and transiently transfected with 2 μg of the capped bicistronic reporter mRNA at intervals by using Lipofectamine 2000 (Invitrogen) and incubated for 6 h prior to harvest. Cell lysates were then prepared and subjected to a luciferase assay or immunoblotting, Northern blotting, or immunoprecipitation.
Small interfering RNA (siRNA; hnQ_si) was designed for endogenous mouse heterogeneous nuclear ribonucleoprotein Q (mhnRNP Q) knockdown. Mutated siRNA (hnQ_si_m) had changes in 3 nucleotides of hnQ_si. The siRNA sequences are shown in Table S1 of the supplemental material. For siRNA transfection into NIH 3T3 cells, a microporator (Digital Bio/Invitrogen) was used as recommended by the manufacturer. After 12 h, dexamethasone treatment or reporter transfection was performed.
In vitro RNA synthesis, in vitro binding, UV cross-linking, and immunoprecipitation.
For in vitro binding assays, [32P]UTP-labeled RNA was transcribed from XbaI-linearized recombinant pSK′ vectors with T7 RNA polymerase (Promega). For mRNA transfection, the bicistronic constructs pCY2-RFe1A and pCY2-RFe1B were linearized with EcoRI, as previously reported (25). This plasmid contains a 20-nucleotide (nt)-long poly(A) stretch between the XhoI and EcoRI restriction sites. Reporter mRNA was generated in vitro from the linearized plasmid with SP6 RNA polymerase (Promega) in the presence of the ribo(m7G) cap analogue (Promega). To identify proteins specifically bound to the mPer1 5′-UTR, in vitro binding and UV cross-linking assays were performed as previously described (25). Briefly, equal amounts of labeled RNAs were incubated with 15 μg nuclear extracts or 30 μg cytoplasmic extracts of NIH 3T3 cell for 20 min. After incubation, the samples were UV irradiated on ice for 10 min with a CL-1000 UV cross-linker (UVP). Unbound RNA was digested with 5 μl RNase cocktail (RNase A and RNase T1). The reaction mixtures were analyzed by SDS-PAGE and autoradiography. For UV cross-linking and immunoprecipitation, RNase-digested lysates were incubated with specific antibodies or, for the negative control, preimmune serum. After overnight incubation, protein G-agarose beads (Amersham Bioscience) were added to the sample, which was further incubated for 3 h. Washed beads were analyzed by SDS-PAGE and autoradiography.
Reporter assay, RNA quantification, and immunoprecipitation–reverse transcription (RT)-PCR (IP-PCR).
The luciferase assay was performed as previously described (25). The ratios between Renilla and firefly luciferase activities (FLUC/RLUC) were calculated. The ratio for the empty vector pRF was set to 1.
mRNA levels of endogenous or reporter plasmids were detected by quantitative real-time PCR using a MyiQ single-color real-time detector system (Bio-Rad) or a StepOnePlus real-time PCR system (Applied Biosystems) with the SYBR green mixture (Takara), as described previously (29, 46, 47). Specific primer pairs for mPer1, mTbp, and firefly luciferase were used for quantitative real-time PCR (the primer sequences are shown in Table S2 of the supplemental material).
For IP-RT, we used a slightly modified method from that previously reported (32, 48). The cytoplasmic extract was obtained as described previously (24). Immunoprecipitation was performed under RNase-free conditions and carried out in immunoprecipitation buffer containing 125 mM KCl, 20 mM HEPES (pH 7.4), 0.5 mM EDTA, 0.05% NP-40, 0.5 mM dithiothreitol, RNasin (Promega), and protease inhibitor cocktail (Calbiochem). RNA was extracted from the washed protein G-agarose bead pellet with an RNA isolation solution (Molecular Research Center). Reverse transcription and quantitative real-time PCR were performed as described above.
Immunoblot analyses.
Immunoblot analyses were performed with polyclonal anti-PER1, polyclonal anti-hnRNP Q (anti-SYNCRIP-N), monoclonal anti-hnRNP Q (Sigma [for immunoprecipitation]), polyclonal anti-phospho-4EBP (Cell Signaling), polyclonal anti-actin (Santa Cruz Biotechnology), monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH; Millipore), and monoclonal anti-14-3-3ζ (Santa Cruz Biotechnology) as primary antibodies. Horseradish peroxidase-conjugated species-specific secondary antibodies (KPL) were visualized using a SUPEX ECL solution kit (Neuronex) and a LAS-4000 chemiluminescence detection system (Fuji Film), and the acquired images were analyzed using Image Gauge (Fuji Film) according to the manufacturer's instructions.
Ribosomal profiling.
Control or hnRNP Q-specific siRNA-trans-fected NIH 3T3 cells were treated with cycloheximide (100 μg/ml) for 5 min at 37°C and then harvested. Cell extracts were subjected to sucrose gradient analysis, as previously described (9, 35). Total RNA of each fraction was purified using TRI reagent (Molecular Research Center) and subjected to real-time PCR analysis for quantification.
Statistical analyses.
All quantitative data are presented as means ± standard errors of the means (SEM). To compare results between more than two groups, we used a one-way analysis of variance with a post hoc Tukey's honestly significant difference test (IGOR Software). The criterion for statistical significance was set at a P level of <0.05.
RESULTS
Existence of an IRES element in mPer1 mRNA.
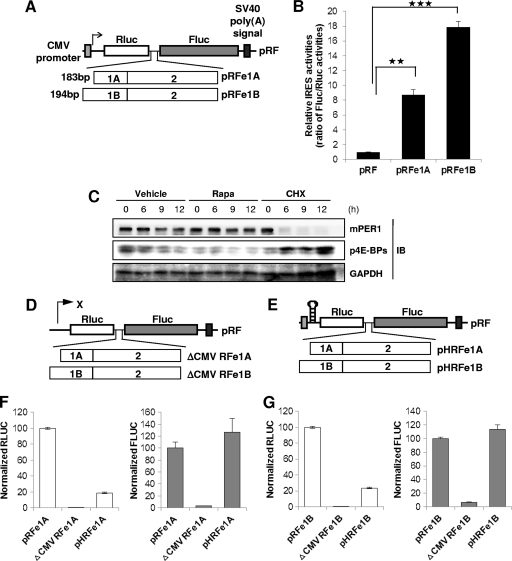
IRES-dependent translation is mainly modulated by 5′-UTRs (12, 19, 43). Interestingly, mPer1 has two forms of 5′-UTRs, 183 bp (e1A) and 194 bp (e1B) (Fig. 1A), both consisting of two exons. The first exons of the 5′-UTRs are different from each other, but the second exons, which include the start codon, are the same. Both types of 5′-UTRs are the result of alternative promoter usage, but the functional differences between the two are unknown (49). To investigate the existence of an IRES in mPer1 mRNA, we inserted the 5′-UTRs of mPer1 into a bicistronic reporter vector (Fig. 1A) (22, 25, 37, 43). The vector contained a cytomegalovirus (CMV) promoter for direct transcription of a bicistronic RNA encoding Renilla luciferase (Rluc) in the first cistron and firefly luciferase (Fluc) in the second cistron. The translation of Rluc from the first cistron is served by cap-dependent translation, while the translation of Fluc reflects the IRES activity of the inserted intergenic sequences. This system is considered the gold standard in finding IRES elements. Both 5′-UTRs (e1A and e1B) enhanced the translation of Fluc more than 8-fold compared to the control vector (Fig. 1B). These results suggest that mPer1 mRNA contains a potential IRES element within its 5′-UTR. We inhibited the mammalian target of rapamycin (mTOR) pathway in NIH 3T3 cells (11, 24). Rapamycin induces hypophosphorylation of eukaryotic initiation factor 4E-binding proteins (4E-BPs), causing inhibition of canonical cap-dependent translation (15, 18, 36, 39). When we treated NIH 3T3 cells with rapamycin, the phospho-4E-BP (p4E-BP) level was decreased, with no significant changes in the level of mPER1 protein (Fig. 1C). However, the general protein biosynthesis inhibitor CHX induced a dramatic decrease in mPER1 protein level. These results suggest that an alternative translational system, other than cap-dependent translation, may be involved in maintaining the mPER1 protein level. The results also indicate that IRES-mediated translation plays a role in mPER1 production.
Fig 1.
mPer1 has an IRES element. (A) Schematic diagram of bicistronic reporter plasmids containing the full-length 5′-UTRs of mPer1. The pRF bicistronic reporter plasmid (pRF), Renilla luciferase, and firefly luciferase are shown. SV40, simian virus 40. (B) HEK 293T cells were transiently transfected with bicistronic reporter plasmids. The ratio of the empty vector pRF was set to 1 (n = 5). ∗∗, P = 0.005639; ∗∗∗, P = 0.0005754. (C) Rapamycin (Rapa) or CHX-treated NIH 3T3 cells were harvested at the indicated time points (in h); then, the protein levels were checked by immunoblotting (IB). (D) Bicistronic vector system with no CMV promoter. (E) Bicistronic vector that harbors a hairpin and loop. (F and G) The CMV promoter-deleted and hairpin-inserted reporter constructs were transfected into HEK 293T cells, and a luciferase assay was performed. The activities of the pRF vector containing the full-length 5′-UTRs, as calculated based on the FLUC/RLUC ratio, were set to 100. The results are expressed as the mean ± SEM of five independent experiments.
Confirmation of an IRES element in mPer1 mRNA.
Recently, the notion of IRES-mediated translation of eukaryotic mRNA has been challenged on the basis of the methods typically used for the identification of IRES elements (27, 45). The result shown in Fig. 1B suggests that the 5′-UTRs of mPer1 mRNA may enhance the read-through of ribosomes through the intergenic region, contain IRES elements that enhance the translation of Fluc from the bicistronic mRNA by internal initiation, or contain a cryptic promoter or splicing acceptor site that creates a monocistronic transcript of Fluc. To exclude any cryptic promoter activity of the mPer1 5′-UTR, we removed the CMV promoter from bicistronic reporter constructs (Fig. 1D). Because RLUC and FLUC activities were measured from cells transfected with promoterless bicistronic constructs, the expression levels of Rluc and Fluc were almost zero (Fig. 1F and G, ΔCMV RFe1A and ΔCMV RFe1B). Based on these results, we confirmed that the 5′-UTRs of mPer1 do not contain cryptic promoters. To determine whether the effect of the mPer1 5′-UTRs on the translation of the second cistron is due to ribosome reinitiation, a synthetic hairpin loop was inserted upstream of Rluc in both pHRFe1A and pHRFe1B (Fig. 1E). The insertion of a hairpin loop reduced the expression of the first cistron, Rluc, by 80 to 90% compared to pRFe1A and pRFe1B, but the activity of the second cistron, Fluc, produced by pHRFe1A and pHRFe1B was not affected (Fig. 1F and G, pHRFe1A and pHRFe1B). This argues against ribosome reinitiation as a possible mechanism. We also confirmed the IRES of mPer1 in the mouse cell line NIH 3T3. 5′-UTRs of mPer1 enhanced the second cistron Fluc (see Fig. S1A in the supplemental material). The insertion of a hairpin loop also reduced Rluc but not Fluc, which are produced by pHRFe1A and pHRFe1B (see Fig. S1B, C, D, and E in the supplemental material). Noncryptic promoter activities of mPer1 5′-UTRs were also confirmed in NIH 3T3 cells (see Fig. S1F and G). The RNA levels of bicistronic reporters shown by quantitative RT-PCR or Northern blotting confirmed that the induction of Fluc translation was not caused by altered mRNA stability, transcription, or the presence of cryptic promoter activity or splice acceptors that produced monocistronic products (see Fig. S2A and B). From these results, we conclude that the 5′-UTRs of mPer1 mRNA have IRESs that can directly initiate translation.
Existence of rhythmic IRES activity.
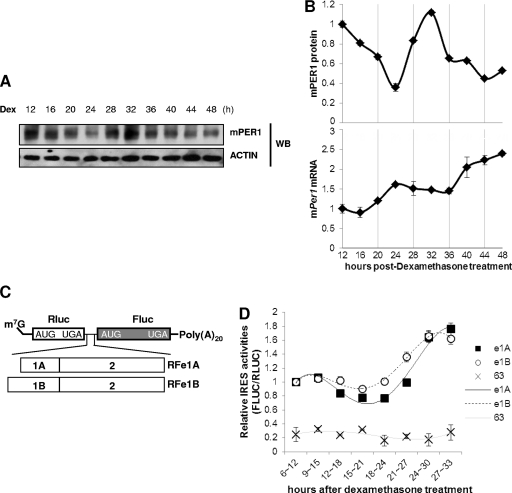
To verify the function of the mPer1 IRES under physiological conditions with circadian rhythm, dexamethasone was applied to achieve synchronization of circadian time in NIH 3T3 mouse fibroblasts (3, 29, 46, 47). In dexamethasone-treated NIH 3T3 cells, we confirmed the oscillation of mRNA and protein and showed the time lag to be approximately 8 h (Fig. 2A and B). To rule out any transcriptional or posttranscriptional effects of the 5′-UTR, we generated a bicistronic reporter mRNA (25) containing a cap structure for each construct, RFe1A and RFe1B (Fig. 2C). The relative IRES activities of constructs containing the 5′-UTRs were higher than that of the reporter lacking a 5′-UTR (data not shown). When we transiently transfected reporter mRNAs to dexamethasone-treated NIH 3T3 cells at certain time intervals, the IRES-mediated translation of mPer1 seemed to be regulated rhythmically (Fig. 2D). This time-dependent transfection did not change the endogenous circadian rhythm (see Fig. S3 in the supplemental material). From these results, we suggest that there is a rhythmic IRES-mediated translation of mPer1 mRNA that may be closely related to the circadian expression of mPER1 protein.
Fig 2.
Existence of rhythmic IRES activity. (A) The oscillation pattern of the mPER1 protein as shown by Western blotting (WB). NIH 3T3 cells were treated with dexamethasone; then, cells were subjected to immunoblotting at the indicated time points. (B) In the case of the mPER1 protein profile, the data from panel A were quantified. By using the same cell extracts as those for which results are shown in panel A, mPer1 mRNA levels were checked by quantitative real-time PCR. (C) Schematic diagram of the bicistronic mRNA reporter of mPer1 5′-UTRs; 7-methyl-guanosine (m7G) and the 20-nt-long poly(A) tail [poly(A)20] are shown. (D) NIH 3T3 cells were treated with 100 nM dexamethasone and transiently transfected with bicistronic mRNA reporters (full-length Per1 5′-UTRs e1A or e1B or a truncated Per1 5′-UTR which has no IRES activity and no interaction with hnRNP Q [construct 63]) for 6 h at the indicated times and subjected to luciferase assays. Relative IRES activities were calculated and plotted for e1A and e1B IRESs and for construct 63, which has no IRES activity.
mhnRNP Q specifically interacts with the mPer1 5′-UTR.
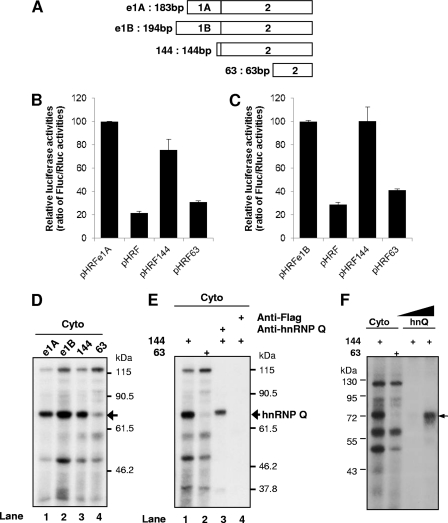
To determine the cis-acting element of the mPer1 5′-UTR that is responsible for IRES activity, we generated reporter constructs containing a truncated 5′-UTR of mPer1 mRNA (Fig. 3A). Because mPer1 5′-UTRs contain two exons that have the same second exon but different first exons, we made the first exon-deleted construct containing the second exon and only 10 nt of the first exon (Fig. 3A, construct 144). We also deleted 80 nt from the 144 constructs on the basis of the RNA secondary structure (Fig. 3A, construct 63) and then inserted the truncated 5′-UTRs of mPer1 into the pHRF vector. The pHRF63 deletion construct exhibited 60 to 70% decrease in Fluc activity compared to controls (pHRFe1A and pHRF e1B), yet the Rluc activity was similar among all constructs (Fig. 3B and C). These results suggest that the region between the 144 and 63 constructs in the mPer1 5′-UTR is important for IRES function.
Fig 3.
hnRNP Q specifically interacts with the mPer1 5′-UTR. (A) Schematic diagram of the serially deleted mutation strategy. (B) Each bicistronic deletion construct derived from the e1A full-length 5′-UTR was transfected into HEK 293T cells. The graph shows the relative luciferase activities derived from the FLUC/RLUC ratio. The activities of pHRFe1A were set at 100. (C) Results for an experiment similar to that shown in panel B, but with the 5′-UTR derived from e1B. (D) 5′-UTRs transcribed in vitro were subjected to in vitro binding and UV cross-linking with a CHO-K1 cytoplasmic extract. The arrow indicates the 68-kDa protein, showing differential binding. (E) Cytoplasmic extracts labeled by UV cross-linking with radiolabeled 5′-UTRs of mPer1 were subjected to immunoprecipitation with antibodies against hnRNP Q (lane 3) or Flag (lane 4) as a control and then separated by SDS-PAGE for autoradiography. (F) Cytoplasmic extracts or purified hnRNP Q were used for UV cross-linking. All results are expressed as the mean ± SEM of five independent experiments.
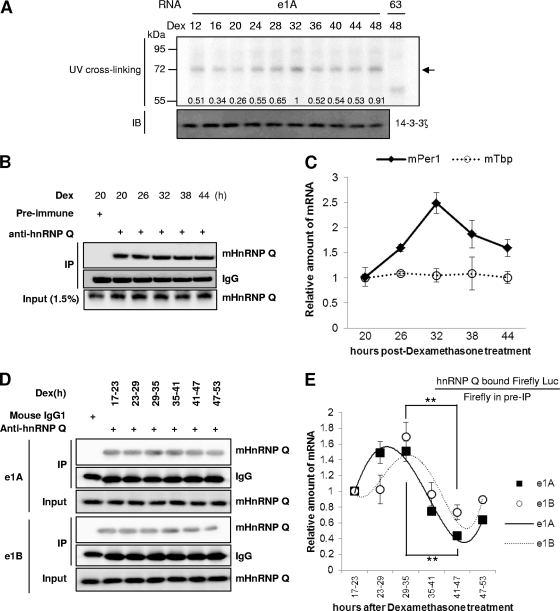
Generally, IRES-mediated translation requires ITAFs that regulate ribosome recruitment, which ultimately determines cap-independent translation efficiency (41, 43). We assumed that ITAFs might bind to the cis-acting element for IRES function of the mPer1 5′-UTR. To analyze the relationship between mPer1 IRES function and the binding patterns of cellular proteins, we performed UV cross-linking assays. All 5′-UTRs, wild-type (e1A and e1B) and deleted constructs (144 and 63) bound to some cytoplasmic cellular proteins in a similar manner (Fig. 3D). Among them, a 68-kDa protein (p68) showed strong binding to full-length e1A, e1B, and deletion construct 144, but not to deletion construct 63 (Fig. 3D, lane 4), which showed weak IRES activity. The binding patterns of p68 were also similar when nuclear protein extracts were used for the UV cross-linking assay (see Fig. S4A in the supplemental material). We previously reported the function of mhnRNP Q as an ITAF and 3′-UTR-binding protein involved in IRES-mediated translation and mRNA degradation in the expression of AANAT (24, 25). To test whether p68 is mhnRNP Q, immunoprecipitation of UV cross-linked proteins with radiolabeled, in vitro-transcribed 144 and 63 RNA (Fig. 3E, lanes 1 and 2) was performed using anti-mhnRNP Q antibody. mhnRNP Q was immunoprecipitated from the UV cross-linked proteins by its specific antibody (Fig. 3E, lane 3). No bands were detected when anti-Flag antibody was used as a negative control (Fig. 3E, lane 4). To confirm the direct interaction between hnRNP Q and the 5′-UTR of mPer1, radiolabeled 5′-UTR and purified hnRNP Q were subjected to in vitro binding and UV cross-linking. Purified hnRNP Q protein was shown to directly interact with the mPer1 5′-UTR (Fig. 3F). We also confirmed that the binding of mhnRNP Q to the 5′-UTR of mPer1 mRNA is specific, because the binding was markedly reduced by the addition of mPer1 5′-UTR 144 RNA competitors (see Fig. S4B in the supplemental material). Knockdown of hnRNP Q decreased direct binding between hnRNP Q and the mPer1 5′-UTR (see Fig. S4C). On the basis of these results, we concluded that the 68-kDa protein is mhnRNP Q and directly binds to the mPer1 5′-UTR.
mhnRNP Q as an IRES trans-acting factor of mPer1 mRNA.
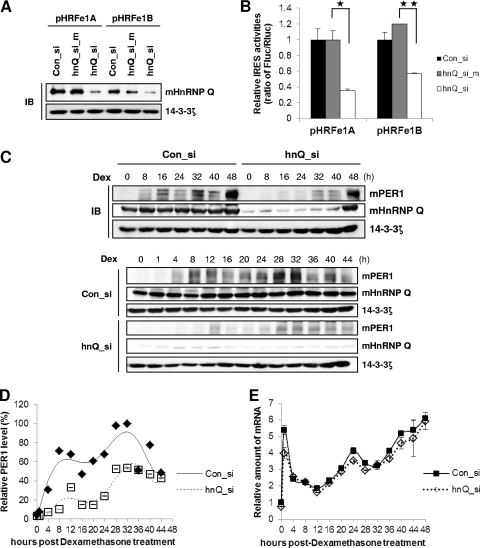
mhnRNP Q is involved in various aspects of mRNA metabolism, such as pre-mRNA splicing (34), mRNA degradation (16, 24), and cellular IRES-mediated translation (25). Because mhnRNP Q strongly bound to the wild-type 5′-UTR and the 144 region of mPer1 mRNA, but not the truncated 63 region, which exhibited insignificant IRES activity, we thought that mhnRNP Q might be one of the ITAFs of mPer1 mRNA. To validate the role of mhnRNP Q in mPer1 mRNA translation, we used a knockdown system employing siRNA against mhnRNP Q (hnQ_si) and a mutated siRNA (hnQ_si_m) that did not decrease mhnRNP Q levels. NIH 3T3 cells were transiently transfected with control siRNA (Con_si), hnQ_si_m, and hnQ_si; 12 h later, reporter constructs containing the 5′-UTR of mPer1 were transfected. hnQ_si markedly decreased mhnRNP Q protein levels, but Con_si and hnQ_si_m did not (Fig. 4A). Knockdown of the mhnRNP Q level decreased both forms of IRES activity of mPer1 mRNA (Fig. 4B). We confirmed the effects and concentration dependencies of hnQ_si and hnQ_si_m siRNAs (see Fig. S5 in the supplemental material). Thus, mhnRNP Q may play an important role as an ITAF in activating IRES-mediated translation of mPer1.
Fig 4.
hnRNP Q as an IRES trans-acting factor of mPer1 mRNA. (A) The cell extracts (10 μg) used for the experiment summarized in panel B were subjected to Western blotting. (B) siRNAs that target mhnRNP Q (hnQ_si), mutated mhnRNP Q siRNA (hnQ_si_m), and control siRNA (Con_si) were transfected with bicistronic reporters to NIH 3T3 cells. The activities of transfected cells with Con_si were set to 1 (n = 4). For pHRFe1A, P = 0.04551; for pHRFe1B, P = 0.007797. (C) NIH 3T3 cells were transfected with siRNAs targeting mhnRNP Q or control siRNA by microporation, incubated for 12 h, treated with dexamethasone, and harvested at the indicated time points. Then, immunoblotting (IB) was performed with the indicated antibodies. (D) The relative levels of mPER1 in panel C, normalized to 14-3-3ζ, were calculated and plotted. The mPER1 level at 36 h, normalized to 14-3-3ζ, was set to 100. (E) Total RNA was prepared from the harvested cells for which data are shown in panel D, and then RT and real-time PCR were performed with an mPer1-specific primer. Error bars represent the mean ± SEM of triplicate measurements.
To analyze the physiological role of IRES-mediated translation of mPer1 by mhnRNP Q in circadian expression, we used the knockdown approach in NIH 3T3 cells. Reduction of mhnRNP Q resulted in lower mPER1 protein levels (Fig. 4C and D), suggesting that mhnRNP Q is critical for mPer1 mRNA translation. Previously, we showed that hnRNP Q functions as a 3′-UTR-binding factor important for mRNA stability (24). Thus, we determined whether reduced mPER1 levels due to knockdown of mhnRNP Q were caused by mPer1 mRNA degradation. When mhnRNP Q was decreased, the oscillation pattern, amplitude, and phase of mPer1 mRNA were not dramatically changed (Fig. 4E). These data imply that decreased mPER1 protein due to decreased mhnRNP Q is mediated by reduced translational activity rather than transcriptional or posttranscriptional modulation. Furthermore, the decreased IRES activity due to reduced mhnRNP Q mediated the diminished and delayed mPER1 expression. Taken together, these results imply that the IRES activity of mPer1 by mhnRNP Q allows robust expression of mPER1 in relation to the circadian period.
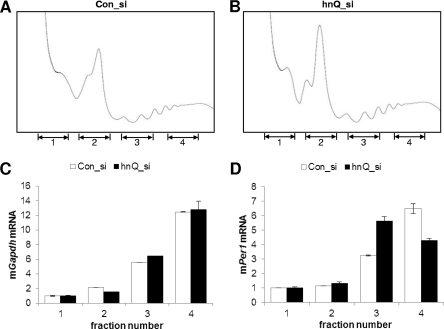
The importance of hnRNP Q in mPer1 mRNA translation was further investigated by analyzing the distribution patterns of control mGapdh mRNA and mPer1 mRNA in ribosome profiles with or without hnRNP Q knockdown (Fig. 5A and B). The overall profiles of ribosomes in sucrose gradient analyses were not altered by decreased hnRNP Q. This was reflected in the levels of a control mGapdh mRNA. The distribution pattern of mGapdh mRNA was not changed by knockdown of hnRNP Q (Fig. 5C). On the other hand, a shift of mPer1 mRNA from heavy polysome (fraction 4) to light polysome (fraction 3), reflecting a reduction in mPer1 mRNA translation, was observed in cells transfected with siRNA against hnRNP Q (Fig. 5D). These results suggest that hnRNP Q plays an important role in the translation of mPer1 mRNA.
Fig 5.
Knockdown of hnRNP Q results in redistribution of mPer1 RNAs in a ribosomal profile. (A and B) NIH 3T3 cells were transfected with control (Con_si) or hnRNP Q-specific siRNA (hnQ_si). After 24 h of incubation, the cells were treated with cycloheximide. Then, the ribosomal distributions in sucrose density gradients were analyzed in cell extracts. RNA samples were purified from fractions in the sucrose gradient. (C and D) Distribution of mRNA in sucrose gradients. The amounts of mGapdh mRNA (C) and mPer1 mRNA (D) across the gradient were analyzed by real-time PCR, and the relative amounts of RNA in each fraction are depicted by corresponding bars in the graphs.
Rhythmic binding between mhnRNP Q and mPer1 mRNA.
As shown in Fig. 2D, the IRES-mediated translation rate followed the circadian period. mhnRNP Q levels could be rhythmic to allow circadian IRES activity, but the level of mhnRNP Q was relatively constant (Fig. 4C). In fact, the mhnRNP Q levels in the cytosol and the nucleus were unchanged during the circadian time (see Fig. S6 in the supplemental material). We wondered if the interaction between mhnRNP Q and mPer1 mRNA is rhythmic. To test our hypothesis, we performed a UV cross-linking assay with dexamethasone-treated cell extracts. The binding affinity was correlated with the mPER1 protein phase (Fig. 6A). To confirm the rhythmic interaction between mhnRNP Q and mPer1 mRNA, we performed IP-RT (32, 48) by using mhnRNP Q antibody after dexamethasone treatment of NIH 3T3 cells. The immunoprecipitated mhnRNP Q levels and input levels were the same during their respective circadian time frames (Fig. 6B). Interestingly, mPer1 mRNA, which coimmunoprecipitated with mhnRNP Q, changed with dexamethasone treatment time, but mTbp mRNA did not (Fig. 6C). In addition, mPer1 mRNAs, which were coimmunoprecipitated, had peak levels at 32 h, which was also the peak time of mPER1 protein expression. mPer1 mRNA was not immunoprecipitated using preimmune serum (data not shown), indicating that mhnRNP Q rhythmically binds to mPer1 mRNA in a specific manner and mediates time-dependent translational activity. However, treatment with dexamethasone causes mPer1 mRNA to oscillate, and abundant mPer1 mRNA at peak time (24 h) may bind to more mhnRNP Q compared to less mPer1 mRNA at trough time. To exclude the possibility that rhythmic binding between mPer1 mRNA and mhnRNP Q was affected by changing mPer1 mRNA levels, we used reporter mRNAs (Fig. 2C) to maintain constant mRNA levels. We transiently transfected dexamethasone-treated NIH 3T3 cells at the indicated times with the 5′-UTR of mPer1 mRNA reporters and then performed IP-RT. The levels of immunoprecipitated mhnRNP Q were the same at all time points (Fig. 6D). Nevertheless, coimmunoprecipitated mRNA reporter levels were different at each time point (Fig. 6E). The time point showing higher levels of reporter mRNAs that coimmunoprecipitated with mhnRNP Q correlated with the mPER1 peak time. Therefore, the results suggest that rhythmic binding between mhnRNP Q and mPer1 mRNA leads to circadian time-dependent IRES-mediated translation regardless of mPer1 mRNA or mhnRNP Q protein levels.
Fig 6.
Rhythmic binding of mhnRNP Q to mPer1 mRNA. (A) Dexamethasone-treated cytoplasmic cell extracts of NIH 3T3 cells were subjected to in vitro binding and UV cross-linking with the radiolabeled mPer1 144 5′-UTR. The cytoplasmic cell extracts used for UV cross-linking were also subjected to immunoblotting (IB), using 14-3-3ζ as a control. The band intensities were quantified. (B) NIH 3T3 cells were treated with dexamethasone, and cytosolic extracts were prepared. Immunoprecipitation (IP) was performed using anti-hnRNP Q antibody and preimmune serum as a control. (C) The coimmunoprecipitated mRNAs with mhnRNP Q shown in panel B were analyzed by real-time PCR. (D) NIH 3T3 cells were treated with dexamethasone, and bicistronic mRNA reporters that harbor 5′-UTRs of mPer1 were transfected at the indicated time points and subjected to immunoprecipitation. (E) Two-thirds (based on volume) of the washed beads from the immunoprecipitation in panel D were used for total RNA preparation. Then, the RNA level was quantified by real-time PCR with Fluc-specific primers and normalized to the Fluc value obtained in the preimmunoprecipitation experiment. The relative numerical values at 17 to 23 h were set to 1 (n = 4). Means and SEM (error bars) are shown. ∗∗, P < 0.005.
Functional role of mhnRNP Q.
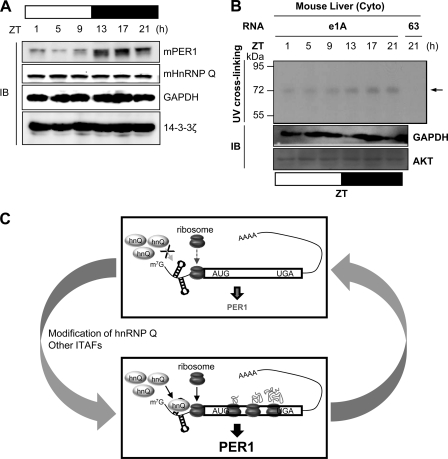
To analyze the relationship between mhnRNP Q and mPer1 mRNA in primary tissues, we determined the protein levels of mhnRNP Q and mPER1 in mouse liver. mPER1 protein levels were rhythmic to circadian time, but mhnRNP Q levels were relatively constant across time points (Fig. 7A). Cytosolic fractions of mouse liver were used for in vitro binding and UV cross-linking with the radiolabeled 5′-UTR of mPer1. Surprisingly, binding between hnRNP Q and e1A RNA correlated with the mPER1 protein phase as the circadian rhythm (Fig. 7B). These results suggest that rhythmic binding between mhnRNP Q and mPer1 mRNA leads to circadian time-dependent IRES-mediated translation regardless of mPer1 mRNA or mhnRNP Q protein levels and is as relevant in cultured cell lines as in animal tissues.
Fig 7.
The functional role of mhnRNP Q and a summary model. Mice were sacrificed at the indicated times (n = 6 for each time interval). (A) Liver extracts were subjected to immunoblotting (IB) with the indicated antibodies. (B) Cytoplasmic extracts of liver were subjected to in vitro binding and UV cross-linking with a radiolabeled 5′-UTR e1A or construct 63; immunoblotting was also performed. (C) Proposed model for rhythmic translation of mPer1 as a key regulatory mechanism of circadian mPER1 expression. mhnRNP Q, alone or with other ITAFs, strongly binds to mPer1 mRNA and accelerates IRES-mediated translation of mPer1. At other times, modifications in mhnRNP Q or its association with other ITAFs may inhibit interaction between mhnRNP Q and mPer1 mRNA. The weak binding of mhnRNP Q to mPer1 mRNA could not enhance cap-independent translation of mPer1, and only canonical cap-dependent translation could produce mPER1. For this reason, the mPER1 protein level is low.
DISCUSSION
Periodicity of transcription is essential for maintaining the cycle of a molecular clock system. Posttranscriptional and posttranslational regulations also appear to be important for fine-tuning such a system. In the expression of mPer1, there is a time lag between the mRNA oscillation pattern and the protein expression profile. To explain this phenomenon, posttranscriptional regulation was considered part of the mechanism. Recently, the importance of posttranscriptional regulation in the bio-clock system has emerged (7, 24–26, 29, 46, 47). Indeed, it has been reported that circadian-regulated rat AANAT expression is controlled by IRES-mediated translation (25). Translation initiation, in concert with transcription, could be a critical step for gene expression. We focused on the translation initiation step for posttranscriptional regulation of the bio-clock system, using mPer1 as an example. mPER1 is an important clock protein that is part of the core feedback loop in the circadian rhythm system. mPER1 is thought to be essential for maintaining circadian rhythm and phase resetting. PER1 is also linked to cell cycle regulation and cancer.
In the present study, we found that the 5′-UTR of mPer1 contained an IRES element and that its translation rate was circadian time dependent. As other clock genes also have rhythmic mRNA and protein profiles with time lag, we also checked their IRES activities: mouse cryptochrome 1 (mCry1), mouse Period2 (mPer2), and mouse Period3 (mPer3) (see Fig. S7 in the supplemental material). mPer1 5′-UTRs have high IRES activities, but not mCry1, mPer2, or mPer3. In the cases of mPer2 and mPer3, IRES activities were slightly increased above controls but were much lower than for mPer1. However, these results must be confirmed by other experiments. We think that some other mechanisms also can modulate their expression and can create time lag oscillation of clock genes. It has been reported that LARK regulates mPer1 translation through binding to the 3′-UTR of mPer1 (26). mPER1 expression is regulated via 3′- and 5′-UTR-mediated translation. To clarify the mechanism of rhythmic mPER1 expression, more information, such as activity of LARK protein under the condition of mhnRNP Q knockdown, the collective effects of LARK and mhnRNP Q on mPER1, expression time, and the binding pattern of hnRNP Q and LARK are required.
mhnRNP Q directly bound to the 5′-UTR of mPer1 mRNA, while a construct that could not bind to mhnRNP Q had little IRES activity. Indeed, knockdown of mhnRNP Q inhibited the IRES activity of mPer1 and resulted in decreased mPER1 protein expression without changing mPer1 mRNA levels. We examined the cycling of an mPer2-dsLuc reporter in dexamethasone-treated cells, with control siRNA or hnRNP Q siRNA transfection (see Fig. S8A in the supplemental material). Knockdown of hnRNP Q, which reduces mPER1 protein, did not change the period of mPer2-dsLuc activity. To check the effects of hnRNP Q in detail, we also observed endogenous mRNA profiles of clock genes. A decrease in the hnRNP Q level did not change the mouse D site albumin promoter-binding protein (mDbp) or mCry1 mRNA rhythm (see Fig. S8B and C). However, knockdown of hnRNP Q slightly reduced the mRNA level of mouse nuclear receptor subfamily 1 (mNr1d1) and mPer2 (see Fig. S8D and E). The possibility that hnRNP Q modulates other clock genes should be considered. We found that hnRNP Q could directly bind to the 3′-UTR of mCry1 (data not shown). As hnRNP Q binds to mRNA of mPer1 and other clock genes, the knockdown of hnRNP Q or mPer1 may lead to a different outcome. To understand the function of hnRNP Q in the overall clock system, further studies of the core clock protein levels and the relationship between hnRNP Q and other clock genes would be necessary.
At first, we thought that the mhnRNP Q protein level might follow a circadian rhythm because it was shown to be rhythmic in rat pineal glands (25). However, rather than the mhnRNP Q protein itself exhibiting circadian rhythm, it was the interaction between mhnRNP Q and mPer1 mRNA that was rhythmic, and their binding was strongest at peak mPER1 levels. It is not known why the expression profile of mhnRNP Q is different from previous results with rat pineal glands (25). mhnRNP Q may oscillate in the pineal gland but not in other tissues or cells. A further study to elucidate the mechanism of rhythmic mhnRNP Q binding to mPer1 mRNA is needed. The secondary structure of mPer1 mRNA at different circadian times or the interaction of mhnRNP Q with other unknown ITAFs in a time-dependent manner may affect the rhythmic binding of mhnRNP Q to mPer1 mRNA. Posttranslational modification of mhnRNP Q, such as phosphorylation, may also have an effect on the rhythmic interaction. mhnRNP Q may be phosphorylated on a tyrosine residue. It has been shown that the binding of RNA to mhnRNP Q specifically inhibited mhnRNP Q phosphorylation (21). We assume that rhythmic phosphorylation of mhnRNP Q may be one of the mechanisms allowing a time-dependent interaction between mhnRNP Q and mPer1 mRNA.
Because mPer1 has two 5′-UTR forms as a result of alternative promoter usage, any functional differences between the 5′-UTRs should be clarified. Based on transfection with DNA reporters (Fig. 1B), the two 5′-UTRs have different IRES activities. However, the IRES activities were similar, and the difference was small when mRNA reporters were transfected (data not shown). We think the discrepancy between the DNA and RNA transfection activities is due to a low cryptic promoter activity or a posttranscriptional effect.
Based on our data, the oscillation of mPER1 levels can be explained as illustrated in our model (Fig. 7C). mhnRNP Q binds to preexisting mPer1 mRNAs. Then, mhnRNP Q facilitates the recruitment of ribosomes, which accelerates IRES-mediated translation of mPer1; mPER1 protein then reaches peak levels. However, modifications in mhnRNP Q or a change in the ITAF complex may prevent mhnRNP Q binding to mPer1 mRNAs. This may bring about inefficient cap-independent translation, causing the mPER1 protein level to decrease. The present study revealed a new mechanism of rhythmic IRES-mediated mPer1 translation that may be an important step in the regulation of the circadian clock. This system may induce fine-tuning of the expression time and amplitude of mPER1 regardless of mRNA oscillation. A similar circadian time-dependent translation may be functional in other core clock genes. From these results, we suggest that the physiological circadian rhythm is generated by a very complex rhythmic molecular system that comprises time-dependent transcriptional, rhythmic posttranscriptional, and modulated translational and posttranslational regulation. This work may help to explain the time gap between mRNA and protein regulation of amplitude and phase. Our study may also ultimately give insight into the tightly and finely regulated molecular system of the circadian clock.
Supplementary Material
ACKNOWLEDGMENTS
We thank Choogon Lee for kindly providing anti-PER1 antibody, K. Mikoshiba for the anti-SYNCRIP-N antibody, and Kyungjin Kim and Jiwon Lee for the Per2-dsLuc plasmid.
This work was supported by grants from the National Research Foundation of Korea (20100030089, 20100002146, and 20100019706), the Brain Korea 21 program, the World Class University program (R31-10105), and the Regional Core Research Program/Anti-aging and Well-being Research Center, funded by the Korean Ministry of Education, Science, and Technology.
Footnotes
Published ahead of print 28 November 2011
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Albrecht U, Zheng B, Larkin D, Sun ZS, Lee CC. 2001. mPer1 and mper2 are essential for normal resetting of the circadian clock. J. Biol. Rhythms 16:100–104 [DOI] [PubMed] [Google Scholar]
- 2. Bae K, et al. 2001. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30:525–536 [DOI] [PubMed] [Google Scholar]
- 3. Balsalobre A, et al. 2000. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289:2344–2347 [DOI] [PubMed] [Google Scholar]
- 4. Balsalobre A, Damiola F, Schibler U. 1998. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93:929–937 [DOI] [PubMed] [Google Scholar]
- 5. Cermakian N, Monaco L, Pando MP, Dierich A, Sassone-Corsi P. 2001. Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the Period1 gene. EMBO J. 20:3967–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen CY, et al. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107:451–464 [DOI] [PubMed] [Google Scholar]
- 7. Cheng HY, et al. 2007. MicroRNA modulation of circadian-clock period and entrainment. Neuron 54:813–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cho S, Kim JH, Back SH, Jang SK. 2005. Polypyrimidine tract-binding protein enhances the internal ribosomal entry site-dependent translation of p27Kip1 mRNA and modulates transition from G1 to S phase. Mol. Cell. Biol. 25:1283–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cho S, et al. 2007. BiP internal ribosomal entry site activity is controlled by heat-induced interaction of NSAP1. Mol. Cell. Biol. 27:368–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Field MD, et al. 2000. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron 25:437–447 [DOI] [PubMed] [Google Scholar]
- 11. Gastel JA, Roseboom PH, Rinaldi PA, Weller JL, Klein DC. 1998. Melatonin production: proteasomal proteolysis in serotonin N-acetyltransferase regulation. Science 279:1358–1360 [DOI] [PubMed] [Google Scholar]
- 12. Gebauer F, Hentze MW. 2004. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 5:827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gekakis N, et al. 1998. Role of the CLOCK protein in the mammalian circadian mechanism. Science 280:1564–1569 [DOI] [PubMed] [Google Scholar]
- 14. Gery S, et al. 2006. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol. Cell 22:375–382 [DOI] [PubMed] [Google Scholar]
- 15. Gingras AC, Raught B, Sonenberg N. 2004. mTOR signaling to translation. Curr. Top. Microbiol. Immunol. 279:169–197 [DOI] [PubMed] [Google Scholar]
- 16. Grosset C, et al. 2000. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 103:29–40 [DOI] [PubMed] [Google Scholar]
- 17. Harms E, Kivimae S, Young MW, Saez L. 2004. Posttranscriptional and posttranslational regulation of clock genes. J. Biol. Rhythms 19:361–373 [DOI] [PubMed] [Google Scholar]
- 18. Hay N, Sonenberg N. 2004. Upstream and downstream of mTOR. Genes Dev. 18:1926–1945 [DOI] [PubMed] [Google Scholar]
- 19. Hellen CU, Sarnow P. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593–1612 [DOI] [PubMed] [Google Scholar]
- 20. Hida A, et al. 2000. The human and mouse Period1 genes: five well-conserved E-boxes additively contribute to the enhancement of mPer1 transcription. Genomics 65:224–233 [DOI] [PubMed] [Google Scholar]
- 21. Hresko RC, Mueckler M. 2002. Identification of pp68 as the tyrosine-phosphorylated form of SYNCRIP/NSAP1. A cytoplasmic RNA-binding protein. J. Biol. Chem. 277:25233–25238 [DOI] [PubMed] [Google Scholar]
- 22. Jang SK, et al. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim JH, et al. 2003. Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc mRNA in a cell cycle phase-dependent manner. Mol. Cell. Biol. 23:708–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim TD, et al. 2005. Rhythmic serotonin N-acetyltransferase mRNA degradation is essential for the maintenance of its circadian oscillation. Mol. Cell. Biol. 25:3232–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim TD, et al. 2007. Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev. 21:797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kojima S, et al. 2007. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc. Natl. Acad. Sci. U. S. A. 104:1859–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kozak M. 2001. New ways of initiating translation in eukaryotes? Mol. Cell. Biol. 21:1899–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kume K, et al. 1999. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98:193–205 [DOI] [PubMed] [Google Scholar]
- 29. Kwak E, Kim TD, Kim KT. 2006. Essential role of 3′-untranslated region-mediated mRNA decay in circadian oscillations of mouse Period3 mRNA. J. Biol. Chem. 281:19100–19106 [DOI] [PubMed] [Google Scholar]
- 30. Kwon MC, et al. 2008. Crif1 is a novel transcriptional coactivator of STAT3. EMBO J. 27:642–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lowrey PL, Takahashi JS. 2004. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu. Rev. Genomics Hum. Genet. 5:407–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ma S, Liu G, Sun Y, Xie J. 2007. Relocalization of the polypyrimidine tract-binding protein during PKA-induced neurite growth. Biochim. Biophys. Acta 1773:912–923 [DOI] [PubMed] [Google Scholar]
- 33. Moore RY, Eichler VB. 1972. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 42:201–206 [DOI] [PubMed] [Google Scholar]
- 34. Mourelatos Z, Abel L, Yong J, Kataoka N, Dreyfuss G. 2001. SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J. 20:5443–5452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Paek KY, Kim CS, Park SM, Kim JH, Jang SK. 2008. RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J. Virol. 82:12082–12093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pause A, et al. 1994. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371:762–767 [DOI] [PubMed] [Google Scholar]
- 37. Pelletier J, Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320–325 [DOI] [PubMed] [Google Scholar]
- 38. Pilipenko EV, et al. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 14:2028–2045 [PMC free article] [PubMed] [Google Scholar]
- 39. Pyronnet S, Pradayrol L, Sonenberg N. 2000. A cell cycle-dependent internal ribosome entry site. Mol. Cell 5:607–616 [DOI] [PubMed] [Google Scholar]
- 40. Song Y, et al. 2005. Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA 11:1809–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stoneley M, Willis AE. 2004. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene 23:3200–3207 [DOI] [PubMed] [Google Scholar]
- 42. Tei H, et al. 1997. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature 389:512–516 [DOI] [PubMed] [Google Scholar]
- 43. Vagner S, Galy B, Pyronnet S. 2001. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2:893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van den Pol AN, Dudek FE. 1993. Cellular communication in the circadian clock, the suprachiasmatic nucleus. Neuroscience 56:793–811 [DOI] [PubMed] [Google Scholar]
- 45. Wang Z, Weaver M, Magnuson NS. 2005. Cryptic promoter activity in the DNA sequence corresponding to the pim-1 5′-UTR. Nucleic Acids Res. 33:2248–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Woo KC, et al. 2010. Circadian amplitude of cryptochrome 1 is modulated by mRNA stability regulation via cytoplasmic hnRNP D oscillation. Mol. Cell. Biol. 30:197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Woo KC, et al. 2009. Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res. 37:26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xie J, Lee JA, Kress TL, Mowry KL, Black DL. 2003. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc. Natl. Acad. Sci. U. S. A. 100:8776–8781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yamaguchi S, et al. 2000. The 5′ upstream region of mPer1 gene contains two promoters and is responsible for circadian oscillation. Curr. Biol. 10:873–876 [DOI] [PubMed] [Google Scholar]
- 50. Yang DQ, Halaby MJ, Zhang Y. 2006. The identification of an internal ribosomal entry site in the 5′-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene 25:4613–4619 [DOI] [PubMed] [Google Scholar]
- 51. Zeng H, Hardin PE, Rosbash M. 1994. Constitutive overexpression of the Drosophila period protein inhibits period mRNA cycling. EMBO J. 13:3590–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zheng B, et al. 2001. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell 105:683–694 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.