Abstract
Lafora disease (LD), an inherited and fatal neurodegenerative disorder, is characterized by increased cellular glycogen content and the formation of abnormally branched glycogen inclusions, called Lafora bodies, in the affected tissues, including neurons. Therefore, laforin phosphatase and malin ubiquitin E3 ligase, the two proteins that are defective in LD, are thought to regulate glycogen synthesis through an unknown mechanism, the defects in which are likely to underlie some of the symptoms of LD. We show here that laforin's subcellular localization is dependent on the cellular glycogen content and that the stability of laforin is determined by the cellular ATP level, the activity of 5′-AMP-activated protein kinase, and the affinity of malin toward laforin. By using cell and animal models, we further show that the laforin-malin complex regulates cellular glucose uptake by modulating the subcellular localization of glucose transporters; loss of malin or laforin resulted in an increased abundance of glucose transporters in the plasma membrane and therefore excessive glucose uptake. Loss of laforin or malin, however, did not affect glycogen catabolism. Thus, the excessive cellular glucose level appears to be the primary trigger for the abnormally higher levels of cellular glycogen seen in LD.
INTRODUCTION
Glucose is an essential metabolite in living systems. However, the regulatory roles of glucose in cellular physiological pathways and the mechanisms by which cells respond to changes in the intracellular levels of glucose are not fully understood (26). Dysregulation in these processes is thought to underlie the pathology of a few disorders that are associated with cytoplasmic glycogen inclusions (50). One such disorder is Lafora disease (LD), a heritable and fatal neurodegenerative disorder characterized by progressive myoclonus epilepsy and other neurological deficits, including ataxia and dementia (17, 41). A hallmark of LD is the presence of Lafora bodies—insoluble and abnormally branched intracellular glycogen inclusions called polyglucosan—in neurons, muscle, liver, and other tissues (16, 17, 51, 52). LD is caused by defects in the gene EPM2A, which encodes a dual-specificity protein phosphatase named laforin, or the NHLRC1 gene, which encodes an E3 ubiquitin ligase named malin (6, 15, 20, 32). Laforin harbors a carbohydrate-binding domain (CBD) that binds to glycogen and Lafora bodies, both in vitro and in vivo (5, 18, 49). Thus, a role for laforin in carbohydrate metabolism and in the disposition of Lafora bodies was proposed (5, 18, 49). Besides Lafora bodies, glycogen content has also been found at higher levels in animals that were deficient for laforin or malin (11, 43). Intriguingly, the glycogen reserve in LD animal models shows a higher phosphate content (11, 43), and laforin has been shown to dephosphorylate glycogen (43, 44). A recent report suggested that glycogen phosphorylation possibly represents an error in a catalytic step in glycogen synthesis and that its removal by laforin could be a damage control mechanism (45). Since laforin and malin are known to function as a complex (14, 19, 39, 46), it has been proposed that laforin and malin, as nonredundant partners, regulate multiple steps in glycogen metabolism (14, 43, 46, 48). However, whether the laforin-malin complex regulates glycogenesis or glycogenolysis, or both of these processes, is yet to be resolved. For example, a role for laforin and malin in regulating the cellular level of glycogen synthase (GS) (48) and R5/PTG (subunit of protein phosphate 1) has been proposed (14, 46), but the cellular levels (and activities) of GS and R5/PTG were found to be unaltered in laforin- and malin-deficient mouse models (11, 44). Thus, the specific pathway through which the laforin-malin complex is able to regulate glycogen metabolic process is yet to be unequivocally established. We show here that laforin could be a glucose sensor, and its subcellular localization and stability are determined by the intracellular level of glucose metabolites. We further show that laforin and malin negatively regulate glucose uptake by modulating the subcellular localization of glucose transporters and that the loss of laforin or malin results in an excessive buildup of glycogen, as seen in LD.
MATERIALS AND METHODS
Cell culture transfections and animal models.
All experiments were carried out in COS-7 cells unless otherwise stated. COS-7, Neuro2a, HepG2, HEK293T, and HeLa cells were obtained from the National Centre for Cell Sciences (India) and were grown in Dulbecco's modified Eagle's medium (DMEM) with 25 mM glucose and 10% (vol/vol) fetal calf serum. For glucose starvation, cells were grown in DMEM without glucose but in the presence of serum. Cell were transfected using Lipofectamine 2000 (Invitrogen Inc.) or the Polyfect reagent (Qiagen India) as recommended by the manufacturer. Muscle tissues of laforin-deficient mice and their wild-type littermates (16) were obtained from Kazuhiro Yamakawa (RIKEN Brain Science Institute, Japan). For the starvation studies, adult mice (Swiss albino) were used.
Expression constructs and chemicals.
The mammalian expression constructs that code for the wild-type or the mutant forms of laforin and malin have been described (19, 33, 36). The shRNA knockdown constructs (RNA interference [RNAi]) for laforin, malin, and the control RNAi vector were purchased from Open Biosystems USA and have been validated in our previous studies (19, 36). The constructs that code for the wild-type and the dominant negative form of 5′-AMP-activated protein kinase (AMPK) were obtained from Addgene (plasmid ID numbers 15991 and 15992, respectively). Green fluorescent protein (GFP)-tagged expression constructs for Glut1 and Glut3 were generously provided by Juan P. Bolanos (Universitario de Salamanca, Spain). All chemicals were purchased from Sigma-Aldrich Pvt. Ltd. (India) unless stated otherwise.
Glucose deprivation.
For glucose deprivation, the cells, after transfection with the desired mammalian expression constructs wherever applicable, were first allowed to grow in glucose (25 mM) containing DMEM for at least 15 h and then shifted to DMEM without glucose for the next 12 or 24 h, as indicated below.
Pharmacological treatments.
Cells were treated with the following pharmacological agents: l-glucose (25 mM), mannitol (25 mM), 2-deoxy-glucose (2-DOG; 25 mM), 3-O-methyl glucose (3-OMG; 25 mM), cycloheximide (1 mg/ml), cytochalasin B (20 μg/ml), sodium azide (10 mM), mannoheptulose (20 mM), forskolin (100 μM), glucosamine (10 mM), diazo-oxo-norleucine (DON; 40 μM), benzyl-2-acetamido-2-deoxy-α-d-galactopyranoside (BADGP; 10 mM), MG132 (20 μM), metformin (10 mM), compound C (40 μM), and 2-[N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl) amino]-2-deoxy-d-glucose (2-NBDG; 50 μM). The duration of treatment ranged from 12 to 24 h.
Immunostaining and antibodies.
Cells, grown on a gelatin-coated sterile glass coverslip, were processed for immunofluorescence microscopy as described previously (19, 33). Briefly, cells were fixed with 4% paraformaldehyde, permeabilized with Triton X-100 (0.05%) and subsequently incubated with primary and secondary antibodies as per the suppliers' instructions. For staining the nuclei, cells were treated with 10 μM 4′,6-diamidino-2-phenylindole (DAPI) for 5 min. Immunofluorescence images were captured using a high-end fluorescence microscope (Nikon Eclipse 80i; 40× oil objective; Nikon, Japan) and were processed using the Media Cybernetics ImagePro Express image analysis and deconvolution software. The following antibodies were used in the present study: anti-Myc and anti-GFP (Roche India); anti-AMPKα, anticaveolin, and anti-flotillin 2 (Cell Signaling Technology); anti-Glut1, anti-Glut3, and anti-Glut4 (Millipore India); anti-β-O-linked N-acetylglucosamine and antitubulin (Sigma-Aldrich India Pvt. Ltd.); anti-transferrin receptor antibody (BD Transduction Laboratories). Anti-laforin antibody was described in our previous study (36). Antiglycogen antibody was a generous gift from Otto Baba (Tokyo Medical and Dental University, Japan). Secondary antibodies were obtained from Jackson ImmunoResearch Inc.
Cell counting.
Laforin's nuclear localization was scored by viewing cells with a fluorescence microscope in a blinded manner. For each condition/treatment, at least 300 cells were counted, and the data are presented as the percentage of cells having nuclear localization of protein.
Pulldown assay.
In vivo protein-protein interactions were established by using Ni-affinity resins as described previously (13, 19). Briefly, the lysate of cells that coexpressed His-tagged malin and GFP-tagged laforin was incubated with Ni-affinity resin (Sigma-Aldrich) for 2 h at 4°C and processed according to the manufacturer's instructions. The whole-cell lysate and pulldown fraction were detected by immunoblotting using specific antibodies.
Immunoblotting.
For immunoblotting, protein samples were separated through SDS-PAGE gels and transferred onto a nitrocellulose paper (MDI, India). After blocking with 5% nonfat dry milk powder, the membranes were processed for sequential incubations with primary and secondary antibodies. Immunoreactive proteins on the filters were visualized using a chemiluminscence detection kit (SuperSignal West PICO; Pierce). Signal intensities of the blots were measured using the NIH ImageJ software.
AMPK activity assay.
AMPK activity was measured by using a peptide substrate (SAMS) purchased from Upstate, Millipore, as previously described (10, 38). Briefly, the cell were lysed by sonication in an ice-cold lysis buffer at 4°C (50 mM Tris-HCl [pH 7.4], 250 mM mannitol, 1 mM sodium pyrophosphate, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol [DTT], 0.1 mM benzamidine, 0.1 mM phenylmethylsulfonyl fluoride [PMSF], 5 μg/ml soybean trypsin inhibitor, and 1% Triton X-100) supplemented with phosphatase (Roche Diagnostics) as well as protease inhibitor cocktail (Sigma-Aldrich India Pvt. Ltd.). The lysate was centrifuged (14,000 × g for 5 min at 4°C), and the supernatant was used for the kinase assay. For the kinase assay, an equal amount of protein (50 μg) was added to a reaction mixture containing 5 μl assay buffer (62.5 mM Na-HEPES [pH 7.0], 62.5 mM NaF, 62.5 mM NaCl, 1.25 mM EDTA, 1.25 mM EGTA, 1.25 mM sodium pyrophosphate, 0.1 mM PMSF, 1 mM DTT, 0.1 mM benzamidine, 5 μg/ml soybean trypsin inhibitor), 5 μl of 1 mM SAMS peptide, and 5 μl of 1 mM AMP. Finally, [γ-32]ATP (300 to 500 cpm/pmol) was added along with 5 mM MgCl2, and the reaction was carried out at 37°C for 10 min. Twenty-microliter aliquots were spotted on a phophocellulose paper (P81; Whatman) and washed 4 times with 1% H3PO4 (500 ml) and once with acetone, then air dried and counted in a scintillation counter (PerkinElmer Life Sciences). The AMPK activity is reported as pmol/min/mg of protein.
Glucose uptake assay.
A glucose uptake assay was performed essentially as described earlier (34). Transfected cells at 36 h posttransfection were incubated with Krebs-Ringer-HEPES (KRH) buffer for 1 to 2 h, and then 50 μM 2-NBDG (Invitrogen) was added for 10 min at 37°C. The cells were then washed and lysed by sonication, and the lysate was centrifuged at 12,000 × g for 5 min at 4°C. A small aliquot of the lysate was saved for protein estimation (Bradford method). The fluorescence intensity of the remaining cell lysate was measured using a fluorescence spectrometer (LS 55; Perkin-Elmer). As a control, the fluorescence levels of the lysate from cells treated with an inhibitor of glucose uptake, cytochalasin B (10 μM), were measured. Fluorescence values were normalized based on protein estimation data and are presented as the fold change.
Plasma membrane fractionation.
Plasma membrane fractionation from cultured cells was performed essentially as described elsewhere (2). At 36 h posttransfection, cells were first washed twice with phosphate-buffered saline and then scraped into detergent-free cell lysis buffer (10 mM Tris [pH 7.5], 137 mM NaCl, 2 mM phenylmethylsulfonyl fluoride) and a phosphatase and protease inhibitor cocktail. Cells were left on ice for 10 min and then sonicated to disrupt plasma membranes. Cell lysate was cleared first by low-speed centrifugation as 12,000 × g for 5 min, and then the cleared lysate was separated by ultracentrifugation at 100,000 × g (Sorvall MTX 150 microultracentrifuge; rotor S55-S) for 1 h. Supernatants were saved, and pellets were washed twice with lysis buffer and then resuspended in an equal amount of lysis buffer containing 1% Triton X-100.
For fractionation of the plasma membrane from mouse soleus skeletal muscle tissue, the protocol of Dombrowski et al. (12) was followed. Briefly, 500 mg of muscle tissue was homogenized in buffer A (10 mM sodium bicarbonate [pH 7.0], 0.25 mM sucrose, 5 mM sodium azide, and 100 μM phenylmethylsulfonyl fluoride). The homogenate was centrifuged at 1,300 × g for 10 min, supernatant was saved, and the resulting pellet was homogenized in buffer A. The homogenate was centrifuged at 1,300 × g for 10 min. The supernatant fractions from both steps were combined and centrifuged at 9,000 × g for 10 min, and the resulting supernatant was centrifuged at 190,000 × g for 1 h. The pellet fraction from this step was dissolved in sucrose-free buffer A and loaded on top of discontinuous sucrose density gradients (25, 32, and 35% [wt/wt]). The gradient was centrifuged at 150,000 × g for 16 h, and the plasma membrane fractions were collected from the top 25% of the gradient, diluted in buffer A, and centrifuged at 190,000 × g for 1 h. The final pellet was resuspended in buffer A. Samples were resolved by SDS-PAGE and immunoblotted as described above.
Proteasome activity assay.
Proteasome activity in cell extract was quantified using a fluorogenic proteasome substrate, Suc-Leu-Leu-Val-Tyr-AMC (Calbiochem), as per the manufacturer's instructions. Briefly, 10 μg of protein was incubated with the proteasome substrate in assay buffer for 2 h at 50°C, and fluorescence intensity was measured using a spectrofluorometer (excitation, 380 nm; emission, 460 nm). Assays were performed in triplicates.
Glycogen estimation.
Cellular glycogen content was estimated as described previously (42). The cells were suspended in 100 μl of ice-cold 30% KOH and heated at 100°C for 20 min. A small aliquot of the sample was saved for protein estimation, and the rest was spotted onto a filter paper (Whatman 31-ET CHR). The paper was washed in ice-cold 66% ethanol for 10 min, followed by two washes in the same solution at room temperature, dried overnight at 37°C, and then incubated in amyloglucosidase (0.5 mg/ml in 0.02 M sodium acetate [pH 4.8]) for 2 h at 37°C. The released glucose was measured by using a glucose estimation kit (ERBA Diagnostic Mannheim Gmbh Ltd.), and the quantity of glycogen is presented as the amount of released glucose per mg of total protein.
Statistical analysis.
Data were analyzed by a two-tailed, unpaired Student's t test using GraphPad software. Differences were considered significant at a P level of <0.05. Experiments were performed at least in triplicates, and scoring of cells (∼200 cells per set) was done in a blinded manner.
RESULTS
Subcellular localization of laforin, but not that of malin, is regulated by the extracellular glucose level.
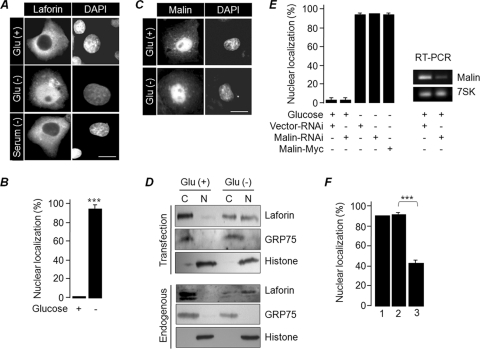
To explore whether glucose availability modulates the subcellular localization of laforin and malin, GFP-tagged laforin and malin were transiently expressed in COS-7 cells, and their subcellular localizations were evaluated by growing the cells for 24 h in a medium with glucose (25 mM) or no glucose. Laforin was exclusively localized in the cytoplasm of the cells grown in glucose-rich medium, while a significant population of cells grown in the glucose-free medium showed laforin in the nuclear compartment (Fig. 1A and B). The glucose starvation-induced nuclear translocation of laforin was found to be independent of the fusion tags used and the cell lines employed (data not shown), and it was not induced by serum starvation (Fig. 1A). The glucose-dependent localization of laforin was also noted in an earlier study (7). Malin did not show any difference in the subcellular localization when the cells were maintained in the glucose-free medium (Fig. 1C). The nuclear translocation of laforin was further confirmed by fractionating the cellular components into nuclear and cytoplasmic fractions; we detected laforin in the nuclear fraction of the glucose-starved cells (Fig. 1D). Similar observations were made when endogenous laforin was tested in the fractionated samples using an anti-laforin antibody (Fig. 1D). The antilaforin antibody, whose specificity was established previously (36, 39), did not work in the immunofluorescence staining.
Fig 1.
Subcellular localization of laforin is regulated by the extracellular glucose level. (A) Representative images showing the subcellular localization of GFP-tagged laforin in cells grown in the presence/absence of glucose or serum, as indicated. (B) The percentage of cells showing nuclear localization of laforin (as shown in panel A) when grown in the presence or absence of glucose. (C) Representative images showing subcellular localization GFP-tagged malin in cells grown in the presence/absence of glucose. (D) The cytoplasmic (C) and nuclear (N) fractions of cells grown in the presence (+) or absence (-) of glucose were immunoblotted to show enrichment of GFP-tagged laforin (upper panel) or endogenous laforin (lower panel) in the nuclear compartment. (E) The percentage of cells with nuclear localization of laforin when coexpressed with the indicated RNAi vector. The gel on the right shows RNAi-mediated reduction in the level of malin RNA, as detected by a semiquantitative PCR. Amplification of 7SK RNA served as a control. (F) Cells expressing GFP-tagged laforin were grown in glucose-free medium for 12 h and were then transferred to medium without glucose (bar 1), glucose-free medium with only cycloheximide added (bar 2), or medium with cycloheximide and glucose added (bar 3). The percentage of cells showing nuclear localization was scored at the end of 12 h of incubation. Bars in panels A and C, 10 μm. ***, P < 0.0005.
We next checked whether the presence of malin modulated the glucose-dependent nuclear translocation of laforin. Neither the loss of malin nor its coexpression prevented laforin's nuclear translocation when the cells were grown in the glucose-free medium (Fig. 1E). We also checked whether the nuclear translocation of laforin was a reversible process. For this, cells expressing GFP-laforin were first grown in glucose-free medium for 12 h and then transferred to a medium containing either cycloheximide (at 1 mg/ml; a translational blocker) and glucose (25 mM) or a glucose-free medium containing only cycloheximide. There was a significant reduction in the number of cells in which laforin localized in the nucleus in the set that was transferred to the medium containing glucose compared to the set that was maintained in glucose-free medium (Fig. 1F). Thus, the glucose-dependent nuclear translocation of laforin appears to be a reversible process. The nucleus-to-cytoplasm movement of laforin is independent of CRM1/exportin 1 protein, as leptomycin B treatment did not inhibit the translocation (data not shown).
The intracellular glycogen level regulates laforin's subcellular localization.
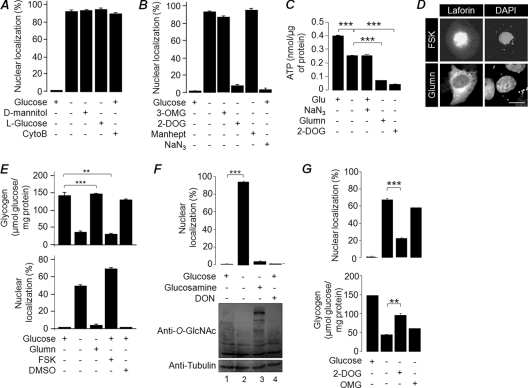
To establish the mechanism by which glucose is able to modulate the subcellular localization of laforin, we first checked whether glucose uptake is required for the cytoplasmic retention of laforin. For this, cells transiently expressing laforin were incubated in a medium containing l-glucose (an analog of d-glucose that does not enter the cell) or in glucose-containing medium supplemented with cytochalasin B (an inhibitor of glucose transport) for 24 h. Both these treatments resulted in the nuclear translocation of laforin in a significant number of cells (Fig. 2A), suggesting that glucose entry into the cell is a prerequisite for the retention of laforin in the cytoplasm. Supplementing the glucose-free medium with d-mannitol (24 h) did not prevent laforin from translocating into the nucleus (Fig. 2A), suggesting that laforin's nuclear translocation is not due to the osmolarity difference.
Fig 2.
The intracellular glycogen level regulates laforin's subcellular localization. Nuclear localization of laforin when cells were incubated in medium containing glucose, d-mannitol, l-glucose, or cytochalasin B (CytoB) (A) or with glucose, 3-OMG, 2-DOG, mannoheptulose (manhept), sodium azide (NaN3), or no addition (B) in the presence/absence of glucose is shown. (C) Bar diagram showing ATP levels in cells treated with glucosamine (Glumn), 2-DOG, or sodium azide in the presence/absence of glucose (Glu), as indicated. (D) Representative images showing the subcellular localization of GFP-tagged laforin in cells treated with forskolin (FSK) in the presence of glucose (top panel) or with glucosamine (Glumn) in the absence of glucose (lower panel). Bar, 10 μm. (E) Bar diagram showing the glycogen content in cells transiently expressing GFP-tagged laforin after various treatments as indicated (top panel) or the nuclear localization of laforin (lower panel). (F) Bar diagram showing nuclear localization of laforin (upper panel) or the level of glycosylated protein (immunoblot with anti-O-GlcNAc antibody; lower panel) in cells treated with glucose, glucosamine, or DON. (G) Nuclear localization of laforin (upper panel) or glycogen content (lower panel) in cells treated with glucose or glucose analogues (DOG or OMG). ***, P < 0.0005; **, P < 0.005.
Glucose, upon entry into the cell, gets converted into glucose-6-phophate via phosphorylation, and this in turn is routed into several metabolic pathways. We were therefore interested in checking whether phosphorylatable glucose could be the signal for laforin's retention in the cytoplasm. For this, cells transiently expressing laforin were maintained in medium containing either 3-OMG (25 mM; a nonphosphorylated glucose analogue) or 2-DOG (25 mM; a phosphorylated glucose analogue) for 24 h, and the localization of laforin was scored. As shown in Fig. 2B, 2-DOG but not 3-OMG was able to retain laforin in the cytoplasm. To confirm further whether phosphorylation of glucose is essential for laforin to stay within the cytoplasm, the laforin-expressing cells were treated with mannoheptulose (20 mM; 24 h), a potent and specific inhibitor of hexokinase activity (9). Addition of mannoheptulose did not alter laforin's translocation into the nucleus, suggesting that the observed effect of 2-DOG is due to its physiological role, which is independent from its phosphorylation status (Fig. 2B). Taken together, these results suggest that laforin's subcellular localization is independent of glucose phosphorylation (see below).
Phosphorylated glucose has several metabolic fates within the cell, and we wanted to check which of these pathways could regulate laforin's nuclear localization. Addition of sodium azide (for 12 h), an established inhibitor of oxidative phosphorylation and ATP production (1), did not alter laforin's subcellular localization (Fig. 2B), although it significantly reduced the cellular ATP level, which was comparable to the condition in which cells were deprived of glucose (Fig. 2C). Consistent with a previous report (53), a reduced ATP level was also observed when the cells were treated with 2-DOG (Fig. 2C), suggesting that laforin's translocation into the nucleus is not dependent on cellular ATP levels. Next, we checked whether the cellular glycogen level modulated laforin's nuclear localization. For this, laforin-expressing cells were treated with forskolin (12 h), a drug that promotes glycogen degradation (3, 8). In a parallel experiment, transfected cells were maintained in glucose-free medium supplemented with glucosamine (12 h). Glucosamine is a key metabolite of the hexosamine biosynthetic pathway (HBP), and supplementing the glucose-free medium with glucosamine is known to result in normal levels of glycogen, even under glucose deprivation conditions in vitro (30). Glucosamine treatment led to the retention of laforin in the cytoplasmic compartment in a majority of cells, even when they were maintained in glucose-free medium (Fig. 2D and E). On the other hand, addition of forskolin resulted in laforin translocating to the nucleus even when the cells were grown in glucose-containing medium (Fig. 2D and E). Thus, a higher glycogen level appears to restrict laforin's localization to the cytoplasmic portion (Fig. 2E). Since glucosamine and forskolin are also known to modulate the glycosylation of cellular proteins via the HBP (21), a glucose dependent process, we also checked whether protein glycosylation could play a role in laforin's nuclear translocation. For this, COS-7 cells transiently expressing laforin were treated (24 h) with DON or BADGP, two potent inhibitors of the glycosylation process (25). While DON did reduce glycosylation of cellular proteins, it did not induce nuclear translocation of laforin (Fig. 2F). A similar observation was made when BADGP was used (data not shown), suggesting that glycosylation may not play a major role in laforin's nuclear translocation. We finally checked whether the observed effects of the nonmetabolizable glucose analogues 2-DOG and 3-OMG on laforin's subcellular localization could be due to their effects on cellular glycogen content. Treatment with 2-DOG, but not 3-OMG, indeed led to increased glycogen content, even when cells were deprived of normal glucose, and this increased glycogen content correlated with laforin localization within the cytoplasm (Fig. 2G). Taken together, these results suggest that cellular glycogen is a major determinant for laforin's subcellular localization.
Laforin's subcellular localization is dependent on its ability to bind to glycogen.
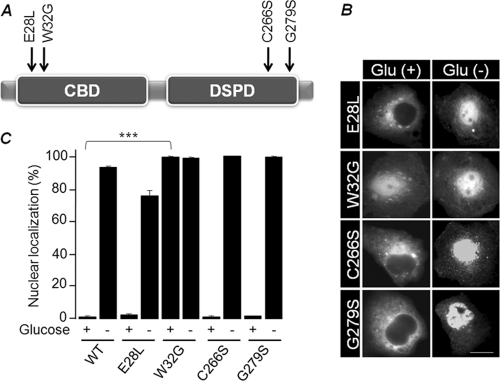
Laforin is known to bind to a glycogen complex through its amino-terminal CBD (5, 18, 49), and some LD-associated mutations affect laforin's ability to bind to glycogen and/or its subcellular localization (41). We were therefore interested in whether laforin's glycogen-binding property is critical for its glycogen-dependent subcellular localization. For this, we selected four missense mutants of laforin: two affecting the CBD (E28L and W32G) and two affecting the phosphatase domain (C266S and G279S) (Fig. 3A). Intriguingly, laforin mutants E28L, C266S, and G279S (all three are inactive phosphatases [41]) were able to translocate to the nucleus upon glucose deprivation (Fig. 3B and C). On the other hand, mutant W32G, which is an inactive phosphatase and does not bind to glycogen in vitro (14, 18, 27, 49), was localized predominantly in the nucleus regardless of glucose availability (Fig. 3B and C). Therefore, the carbohydrate-binding ability, and not the phosphatase activity, could be the critical factor that determines laforin's subcellular localization.
Fig 3.
Subcellular localization of laforin mutants. (A) Schematic diagram of the domain organization of laforin and the positions of missense mutations (CBD and dual-specificity phosphatase domain [DSPD]). (B) Representative images of the subcellular localization of laforin mutants in the presence/absence of glucose. Bar, 10 μm. (C) Bar diagram showing nuclear localization of laforin mutants in the presence/absence of glucose. ***, P < 0.0005.
Intracellular glucose availability regulates malin-dependent degradation of laforin.
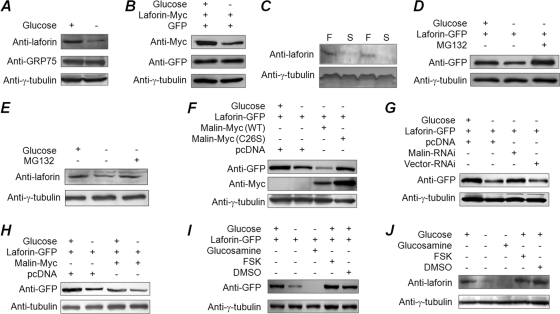
We consistently found the cellular level of transiently expressed laforin to be lower when cells where grown in glucose-free medium. Since laforin is one of the substrates of the malin E3 ubiquitin ligase, we tested whether intracellular glucose availability modulated laforin's affinity toward malin and its degradation. There was a significant reduction in the cellular level of laforin upon glucose deprivation (24 h) (Fig. 4A and B). Endogenous laforin was also found at reduced levels in the livers mice that were starved for 24 h compared to mice fed ad libitum (Fig. 4C). An increase in laforin level was observed when glucose-starved cells were treated with the proteasomal blocker MG132 (for the last 16 h of the 24-h glucose deprivation) (Fig. 4D and E), suggesting that laforin is degraded through the proteasome when cells are starved of glucose. Similarly, the laforin level was lower in glucose-deprived cells that coexpressed wild-type malin than in the control set, which coexpressed the catalytically inactive mutant malin C26S (Fig. 4F), and the laforin level increased when endogenous malin was partially knocked down (Fig. 4G). Malin enhanced laforin degradation during glucose deprivation at a higher rate than under glucose-fed conditions (Fig. 4H). Thus, malin appears to promote the degradation of laforin in a glucose-dependent manner.
Fig 4.
Intracellular glucose availability regulates malin-dependent degradation of laforin. (A and B) Lysates of cells grown in the presence or absence of glucose were immunoblotted with an antibody to detect endogenous laforin, tubulin or GRP75 (A) or transiently expressed laforin (B). (C) Endogenous laforin level in liver tissue lysates (70 μg of protein/lane) from mice that were either fed ad libitum (F) or starved for 24 h (S). (D and E) Cellular levels of the overexpressed laforin (D) or its endogenous form (E) in cells that were grown in the presence/absence of glucose and/or MG132, as indicated. (F and G) Cells coexpressing GFP-tagged laforin with wild-type malin or its mutant C26S (F) or a knockdown construct for malin (G) were evaluated by immunoblotting. (H) Cells expressing the indicated constructs in the presence or absence of glucose were processed for immunoblotting to establish the relative levels of GFP-tagged laforin. (I and J) Cells were exposed to pharmacological agents as indicated, and the cell lysates were processed for immunoblotting to detect the level of transiently expressed GFP-tagged laforin (I) or endogenous laforin (J).
Since laforin's nuclear localization is dependent on the cellular glycogen content rather than on the free glucose level, we next tested whether laforin's stability was also dependent on cellular glycogen content. For this, cells were treated with either glucosamine or forskolin in the absence or presence of glucose, respectively (24 h), and the cellular levels of endogenous or transiently expressed laforin were evaluated. Surprisingly, glucosamine treatment led to a further reduction in the level of laforin (Fig. 4I and J), although the cellular glycogen content was comparable to that of glucose-fed cells (Fig. 2E). On the other hand, forskolin treatment did not significantly alter the laforin level (Fig. 4I and J), although the cellular glycogen content was significantly lower than in glucose-fed cells (Fig. 2E). These results strongly suggest that cellular glucose availability and not the glycogen content determines laforin's stability.
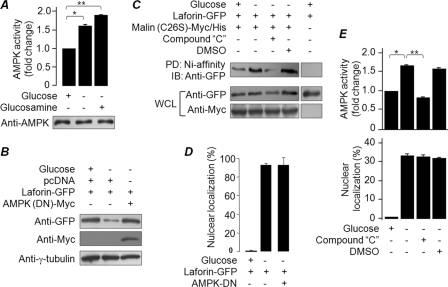
AMPK activity is required for degradation of laforin under glucose deprivation.
AMPK is an energy sensor kinase, since a drop in the intracellular ATP level (for example, during glucose deprivation) rapidly activates AMPK (22). Since the laforin-malin interaction is dependent on AMPK (37, 42), we next tested whether the glucose-dependent degradation of laforin by malin could be mediated by AMPK. Glucose deprivation or glucosamine treatment led to a significant increase in AMPK activity without a change in its level (Fig. 5A). Glucosamine treatment is known to enhance the AMPK activity by reducing the cellular ATP level (24, 29), and similar observations were made in the present study as well (Fig. 2C), suggesting that the increased AMPK activity could be one of the key regulators of laforin's stability during glucose deprivation as well as during glucosamine treatment. Indeed, we found that coexpression of a dominant negative form of AMPK (DN-AMPK) in glucose-free medium resulted in an increase in the cellular level of laforin (Fig. 5B). We next checked whether AMPK regulates the affinity between laforin and malin when cells are deprived of glucose. For this, laforin was expressed with a His-tagged, catalytically inactive malin mutant (C26S), and the cells were either treated with an inhibitor for AMPK (compound C; 12 h) or with the vehicle (dimethyl sulfoxide; 12 h), and the His-tagged malin was pulled down using Ni-affinity resins. The C26S mutant malin was used in the pulldown assay, because the signal intensity of the pulled-down product would represent the amount of protein available for the interaction and not the amount that escaped malin-mediated degradation (13). The mutant malin exhibited a robust interaction with laforin when the cells were deprived of glucose, and this interaction was compromised when the cells were treated with the AMPK inhibitor (Fig. 5C). However, AMPK blockade (treatment with compound C or coexpression of AMPK-DN) did not prevent laforin's nuclear translocation when cells were deprived of glucose (Fig. 5A, D, and E). Thus, AMPK appears to regulate the stability and not the subcellular localization of laforin.
Fig 5.
AMPK activity is required for the degradation of laforin under glucose deprivation. (A) Bar diagram showing the fold difference in AMPK activity in cells treated with the indicated compounds. The immunoblot (lower panel) shows AMPK levels in the cell lysates. (B) Cells expressing AMPK-DN and/or GFP-tagged laforin were maintained in medium containing or deprived of glucose and processed for immunoblotting, as indicated. (C) Cells transfected with constructs that code for GFP-tagged laforin and Myc/His-tagged mutant malin (C26S) were processed for a Ni affinity-based pulldown assay. The pulled-down (PD) and whole-cell lysates (WCL) were immunoblotted with the indicated antibodies. (D) Bar diagram showing the effects of DN-AMPKon laforin nuclear localization under glucose deprivation conditions. (E) Bar diagram showing the difference in AMPK activity (upper panel) or the frequency of cells with nuclear localization of laforin (lower panel) when grown in the presence or absence of glucose or compound C, as indicated. **, P < 0.005; *, P < 0.05.
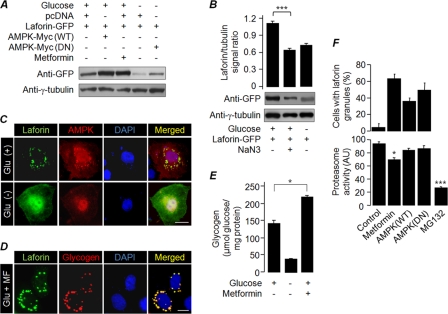
Since overexpression of AMPK-DN resulted in an increase in the level of laforin in cells that were starved of glucose, we next checked whether overexpression of AMPK or enhancement of its activity in the presence of glucose would bring down the cellular level of laforin. Intriguingly, activation of AMPK, either by its overexpression or by treating the cells with metformin, a drug that activates AMPK independently of cellular ATP/AMP levels (23), resulted in a significant increase in the level of laforin in cells that were grown in glucose-containing medium (Fig. 6A). This could possibly mean that activation of AMPK is necessary but not sufficient to degrade laforin and that AMPK might require other factors for its action on laforin. The energy-sensing property of AMPK raises the possibility that the cellular ATP level could be a cofactor that contributes to laforin's stability, since the ATP level goes down when cells are deprived of glucose. To test this possibility, cells that transiently expressed laforin were treated with sodium azide (12 h), a compound that reduces the cellular ATP content even in the presence of glucose (Fig. 2C), and the cellular level of laforin was evaluated. Sodium azide treatment resulted in a significant reduction in the cellular laforin level, even in the presence of glucose (Fig. 6B). However, the same treatment did not alter laforin's subcellular localization (Fig. 2B), suggesting that the cellular ATP level plays a critical role in determining laforin stability in addition to regulating the activity of AMPK. We next evaluated the subcellular distribution of laforin upon coexpression of AMPK or by treating the transfected cells with metformin (24 h). Intriguingly, both conditions resulted in the recruitment of laforin into perinuclear granule-like structures in transfected cells (in ∼40% cells upon AMPK coexpression and in ∼72% cells treated with metformin) that also stained positive for glycogen (Fig. 6C and D). However, such a staining pattern was not observed when cells were maintained in glucose-free medium (Fig. 6C, lower panel), suggesting that glucose deprivation may have resulted in the utilization of the glycogen reserve, thereby abolishing the granule-like staining pattern. Overexpression of AMPK is known to localize this protein onto cytoplasmic glycogen granules (35), suggesting that the activation of AMPK results in an increase in intracellular glycogen content. Indeed, metformin-treated cells grown in glucose-containing medium showed increased intracellular glycogen content compared to untreated cells (Fig. 6E). There was also a moderate yet significant reduction in proteasomal activity upon metformin treatment (Fig. 6F, lower panel). However, these perinuclear granules are unlikely to be “aggresomes,” since coexpression of AMPK (DN or the wild-type) did result in similar recruitment of laforin without compromising proteasomal activity (Fig. 6F). Thus, sequestration of laforin to glycogen particles upon AMPK activation in the presence of glucose could be one of the reasons for its elevated levels in metformin-treated cells. Nonetheless, the present results are compelling enough to suggest that while the subcellular localization of laforin is dependent on the cellular glycogen content, the stability of laforin is dependent on the cellular ATP level and the AMPK activity.
Fig 6.
AMPK is necessary but not sufficient to degrade laforin. (A) Cells expressing laforin-GFP, wild-type AMPK (AMPK-WT), or AMPK-DN were treated the indicated compounds and processed for immunoblotting. (B) Cells expressing GFP-tagged laforin were grown in media containing the indicated compounds and processed for immunoblotting. The signal intensities (laforin/tubulin ratio) were measured and plotted as a bar diagram to reveal the fold difference. (C and D) Representative images of the subcellular localization of laforin and wild-type AMPK when coexpressed in COS-7 cells grown in the presence/absence of glucose (C) or the recruitment of overexpressed laforin to the glycogen particle when Neuro2A cells were grown in a medium containing glucose and metformin (Glu+MF) (D). Bar, 10 μm (C and D). (E) Bar diagram showing intracellular glycogen content in COS-7 cells grown in the presence or absence of glucose and/or metformin (MF). (F) Bar diagram (upper panel) showing nuclear localization of laforin in cells treated with metformin or when coexpressed with the wild-type (WT) AMPK or its DN form. The lower panel shows proteasome activity in the lysates of cells under various conditions. Cells treated with MG132 and metformin were transfected with an empty vector. * and ***, P < 0.05 and 0.0005, respectively.
The laforin-malin complex regulates cellular glucose uptake by modulating subcellular localization of glucose transporters.
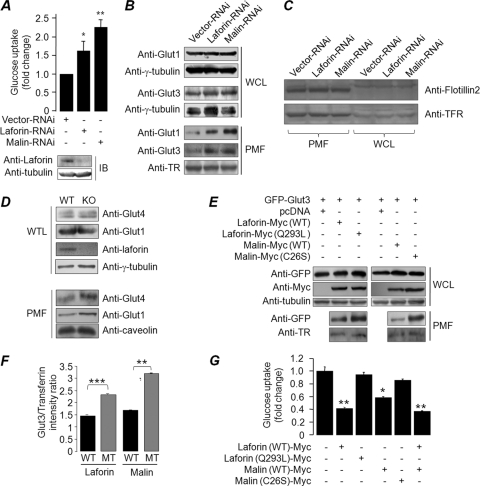
Based on our observations that glycogen metabolites regulate the stability and subcellular localization laforin and previous reports on the role of the laforin-malin complex in glycogen metabolism, we reasoned that the laforin-malin complex might regulate cellular glucose uptake, the first step of the glycogen metabolic pathway. For testing this hypothesis, we used Neuro2a cells, as this cell line is known to store much less glycogen (48), and therefore differences in the glycogen level could be easily measured. The expression of laforin or malin was partially suppressed by the RNAi approach, and cellular glucose uptake was measured using the fluorescent d-glucose analogue 2-NBDG. Partial knockdown of laforin or malin resulted in a significant increase in basal glucose uptake compared to the control set (Fig. 7A). Similar observations were made in COS-7 cells, although the difference was minimal compared to that in Neuro2A cells (data not shown). To elucidate the mechanism by which cells enhance glucose uptake upon the loss of laforin or malin, we first evaluated the relative levels of endogenous glucose transporters Glut1 and Glut3 in the membrane fraction of COS-7 and Neuor2A cells. Glut3 is expressed specifically in the neuronal cells, while Glut1 is expressed in most tissues, although both transporters are involved in basal glucose uptake (31). Loss of laforin or malin did not alter the cellular level of either Glut 1 (COS-7 cells) or Glut3 (Neuro2A cells) (Fig. 7B). However, their loss resulted in a significant increase in the level of Glut1 and Glut3 in the plasma membrane fraction (Fig. 7B). No such difference, however, was noted for the transferrin receptor, a protein that also localizes in the plasma membrane, suggesting that the difference in the signal intensity observed for Glut1 and Glut3 is specific to these proteins and not due to the difference in the protein content of the membrane fraction per se. The enrichment of plasma membrane proteins in the fractionated samples was established by probing fractions with antibodies for the transferrin receptor or flotillin 2 (Fig. 7C). The Glut1 and Glut4 proteins were found to be enriched in the plasma membrane fraction of the soleus skeletal muscle tissue of laforin-deficient mice (Fig. 7D), suggesting that such targeting may not be restricted to cell lines. Thus, laforin and malin appear to negatively regulate the targeting of glucose transporters to the plasma membrane, and defects in this process could result in increased glucose uptake in the absence of laforin or malin.
Fig 7.
The laforin-malin complex regulates glucose homeostasis. (A) Neuro2A cells were transfected with the indicated RNAi construct, and the difference in the uptake of labeled glucose analogue 2-NBDG was measured and plotted. (B) COS-7 and Neuro2A cells were transiently transfected with the indicated RNAi construct, and the level of endogenous glucose transporter Glut1 (COS-7) or Glut3 (Neuro2A) was evaluated in the whole-cell lysate (WCL) and in the plasma membrane fraction (PMF). The levels of tubulin and transferrin receptor (TFR) served as loading controls. (C) Equal quantities (10 μg/lane) of protein samples representing the WCL and PMF of Neuro2A cells shown in panel B were loaded in the same gel, resolved, and immunoblotted with antibodies for the indicated membrane proteins to reveal the relative enrichment of membrane proteins in the PMF. (D) Immunoblot showing levels of Glut1 and Glut4 proteins in whole-tissue lysate (WTL) and the PMF of soleus skeletal muscle tissue of laforin knockout mice (KO) and their wild-type littermates (WT). Tubulin and caveolin served as loading controls for WTL and PMF, respectively. (E) Neuro2A cells were transfected with a combination of constructs that code for GFP-Glut3 or the WT or the mutants of laforin or malin, and the relative levels of Glut3 in the WCL or in the PMF were evaluated by immunoblotting. (F) Signal intensities (Glut3/transferrin receptor ratio) of the immunoblot for the PMF shown in panel E (lower panel) were measured and plotted to reveal the fold difference. (G) Bar diagram showing glucose uptake in Neuro2A cells transiently expressing the wild-type or the mutant form of laforin or malin as indicated. Values were normalized over protein content and are presented as the fold change compared to pcDNA-transfected cells. *, **, and ***, P < 0.05, 0.005, and 0.0005, respectively.
Glucose transporters are normally localized in the vesicular compartments of the cytoplasm and are targeted to the plasma membrane, allowing glucose import upon appropriate signaling (4, 28). Therefore, we wanted to test whether laforin/malin regulate the intracellular trafficking of glucose transporters. For this we chose Neuro2A cells, wherein the overexpressed Glut3 (but not the endogenous form) resides primarily at the plasma membrane (40). When Glut3 was coexpressed with laforin or malin, a significant amount of overexpressed Glut3 was retained in the cytoplasmic compartment in around 70% of the cells (data not shown). To further validate this observation, we fractioned the plasma membranes and evaluated the Glut3 by immunoblotting. While the level of Glut3 in the whole-cell lysate did not show any change, there was a significant reduction in the level of Glut3 in the plasma membrane fraction of cells that coexpressed the wild-type form of laforin or malin compared to the sets that coexpressed the catalytically inactive mutant forms of laforin or malin (Fig. 7E and F). Corroborating this observation, overexpression of wild-type laforin and malin, but not their mutants, significantly reduced the cellular glucose uptake (Fig. 7G). Taken together, our results suggest that laforin and malin are important for the cytoplasmic retention of glucose transporters and that the functional loss of laforin or malin results in the targeting of glucose transporters to the plasma membrane and excessive glucose uptake.
Laforin and malin regulate glycogen synthesis but not its breakdown.
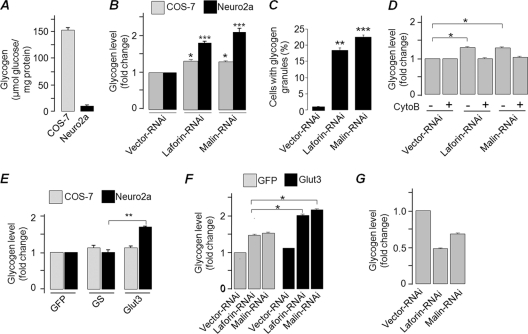
Since loss of laforin or malin resulted in the excessive uptake of glucose, we wanted to check whether loss of these proteins and the plasma membrane localization of glucose transporter would induce glycogen accumulation. For this, we selected two distinct cell types: (i) Neuro2A, a cell line of neuronal origin that stores a very small amount of glycogen, and (ii) COS-7, a nonneuronal cell type that stores ∼100-fold-higher levels of glycogen than Neuro2A cells (Fig. 8A). Transient knockdown of laforin or malin resulted in a significant increase in the intracellular glycogen level in both Neuro2A and COS-7 cells (Fig. 8B). Corroborating this observation, nearly 20% of the cells that were transfected with the laforin or malin RNAi construct showed glycogen granules when stained with an antibody against glycogen, while the control set did not show any such staining (Fig. 8C). To further establish that the enhanced glycogen content was indeed due to increased glucose uptake, we treated cells with an inhibitor of glucose uptake (cytochalasin B) and measured their glycogen content. The cellular glycogen contents of cells lacking laforin or malin did not differ significantly from cells that were transfected with control vector (Fig. 8D), suggesting that loss of laforin or malin promotes intracellular glycogen accumulation via increased glucose uptake. We tested this possibility by overexpressing Glut3 in Neuro2A and COS-7 cells. Overexpressed Glut3 translocates to the plasma membrane in Neuro2A cells but not in COS-7 cells (data not shown). We therefore measured the glycogen contents of Neuro2A and COS-7 cells overexpressing Glut3, glycogen synthase, or GFP (the latter two being controls). Overexpression of Glut3 resulted in a significant increase in the cellular glycogen content, but only in Neuro2A cells (Fig. 8E). However, knockdown of laforin or malin caused a modest but significant increase in the glycogen content, even in COS-7 cells that overexpressed Glut3 (Fig. 8F). Taken together, our results suggest that laforin and malin are critical regulators of glucose transport, that they regulate this process by modulating the subcellular localization of glucose transporters, and that the targeting of glucose transporters to the plasma membrane increases cellular glycogen content.
Fig 8.
Laforin and malin regulate glycogen synthesis but not its breakdown. (A) Bar diagram showing glycogen content in COS-7 and Neuro2A cells grown in a medium with 25 mM glucose. (B) Bar diagram showing relative glycogen content in Neuro2A or COS-7 cells transiently transfected with the indicated RNAi construct. The cellular glycogen level was estimated at 36 h posttransfection. (C) Neuro2A cells transfected with the indicated RNAi constructs were scored for the presence of glycogen granules, as visualized with antiglycogen antibody, and plotted. (D) Bar diagram showing glycogen content of COS-7 cells transiently transfected with the indicated RNAi construct and treated/not treated with cytochalasin B (CytoB). (E) Bar diagram showing the fold difference in the glycogen content of COS-7 and Neuro2A cells transiently expressing GFP, glycogen synthase (GS), or Glut3, as indicated. (F) Bar diagram showing the fold change in the glycogen content of COS-7 cells cotransfected with GFP, Glut3, and the RNAi construct. (G) COS-7 cells transiently transfected with the indicated RNAi construct were transferred to glucose-free medium at 36 h posttransfection, and the cellular glycogen content was measured after 12 h of incubation and plotted as the fold change. *, **, and ***, P < 0.05, 0.005, and 0.0005, respectively.
We next wanted to check whether laforin and malin are involved in glycogen degradation, since inhibition of this process could also result in an increased glycogen level within the cell. To test this possibility, COS-7 cells were transiently transfected with a knockdown construct for laforin or malin or with the control vector, and the cells were exposed to glucose-free medium at 36 h posttransfection to degrade the glycogen reserve for the next 12 h. As shown in Fig. 8G, the cellular glycogen content was lower in cells that expressed the knockdown construct than cells that expressed the control vector, suggesting that laforin and malin are not critical for glycogen degradation. Thus, the increase in glycogen level observed in the absence of laforin or malin could be due to increased glycogen synthesis via excessive glucose uptake and not due to a defect in the glycogen degradation process.
DISCUSSION
Laforin and malin are thought to function as nonredundant partners in a functional complex (17), and several studies have shown that this is indeed the case (14, 19, 39). Here we showed that laforin and malin, through an unknown mechanism but as nonredundant partners, sense the intracellular glycogen level and negatively regulate the cellular uptake of glucose. Partial knockdown of either one resulted in excessive intracellular glycogen accumulation, primarily via enhanced cellular glucose import.
The glycogen-dependent nuclear translocation observed for laforin is similar to the that shown for muscle glycogen synthase (MGS) (8); both of them translocate to the nucleus when glycogen stores are depleted within the cell. Our results also demonstrated that carbohydrate-binding ability is required for laforin to remain in cytoplasm, and therefore the absence or a very low level of glycogen is unable to retain laforin within the cytoplasm and therefore it translocates to the nucleus. While the significance of the nuclear translocation of laforin is not obvious to us, it should be noted here that the nuclear MGS is thought to be involved in transcriptional regulation (8) and that laforin, along with malin, is known to translocate to the nucleus upon thermal stress and to regulate heat shock-induced transcription (39). Thus, laforin appears to have additional functions in the nucleus when cells are under physiological stress, including the glucose deprivation conditions tested in the present study.
Laforin is known to be degraded by malin via the ubiquitin-proteasome pathway (20); however, the physiological significance for this function is not well understood. We showed here that, under physiological conditions, laforin's stability was correlated with the cellular glycogen content. Since loss of laforin results in increased glucose uptake, it is reasonable to suggest that malin promotes the degradation of laforin, probably via increases in the cellular glucose level. This suggestion was supported by our observations that laforin displayed increased affinity toward malin when cells were deprived of glucose, and there was a significant decrease in the level of laforin when cells were starved of glucose, which could be rescued when malin was partially knocked down or when proteasomes were blocked. However, the laforin-malin affinity and the malin-mediated degradation of laforin seem to be regulated by at least by two factors: AMPK activity and cellular ATP levels. AMPK is one of the critical enzymes involved in cellular energy homeostasis and is known to be activated when cells are deprived of energy source (22). Indeed, an earlier study (42) suggested that the laforin-malin interaction is mediated by AMPK, although the functional significance of this role was not explored. We showed here that active AMPK is required for malin to promote the degradation of laforin when cells are deprived of glucose. AMPK is less active when cells are not under metabolic stress, and one of the triggers that activate AMPK is stimuli that change the AMP:ATP ratio (22). Thus, there appears to be an inverse correlation between AMPK activity and laforin level. Intriguingly, active AMPK is unable to promote the degradation of laforin when cellular ATP levels were high, suggesting that AMPK is required but not sufficient for the degradation of laforin. These data clearly indicate that the factor(s) that senses the cellular ATP level could regulate the stability of laforin with the help of AMPK. Similarly, that active AMPK (as during glucose deprivation) is unable to prevent the nuclear translocation of laforin in cells that are deprived of glycogen suggests the possible involvement of an additional regulatory factor(s) that functions independently of AMPK. Thus, ATP depletion is likely to activate AMPK as well as an unknown factor(s), which together might promote the degradation of laforin. The nuclear translocation or/and the degradation of laforin may help in the restoration of the energy level by enhancing cellular glucose uptake within the cell. Taken together, our data suggest that the laforin-malin complex is one of the critical players in cellular energy homeostasis.
Previous studies that investigated the possible role of laforin/malin in glycogen metabolism focused on the enzymes involved in glycogen synthesis or its degradation (11, 14, 42, 44–46). Studies from laforin- or malin-deficient animals have demonstrated that none of the critical enzymes show any difference in their level or activity (11, 44). We reasoned that the laforin-malin complex could indirectly regulate glucose homeostasis, and we demonstrated here that loss of laforin and malin results in excessive glucose uptake. While the manuscript was in preparation, Vernia et al. (47) reported excessive glucose uptake in the tissues of laforin-deficient mice, suggesting that loss of laforin (and possibly malin) might have similar effects at both tissue and cellular levels. We have shown here that loss of laforin or malin increases the abundance of glucose transporters in the plasma membrane and hence excessive glucose uptake. Similar observations were also made in the muscle tissue of laforin-deficient mice. Intriguingly, laforin and malin appear to regulate the translocation of the glucose transporters Glut1, Glut3, and Glut4 to the plasma membrane, although their cellular level remains unchanged. Glut1 and Glut3 represent the two major insulin-independent glucose transporters found in nonneuronal and neuronal tissues, respectively, and Glut4 is exclusively expressed in striated muscle (4). In this regard it is interesting that the plasma insulin level did not change in laforin-deficient mice (47), and we did not use insulin in our glucose uptake assays. Therefore, the laforin-malin complex possibly regulates glucose homeostasis via an insulin-independent glucose uptake mechanism, a constitutive process that accounts for most of the basal glucose uptake in tissues (4). Glucose transporters are found both at cytoplasmic vesicles and at the plasma membrane (4, 28). Under conditions of a glucose requirement, vesicles target the glucose transporters to the plasma membrane via exocytosis, and when the glucose demand is met, most of the transporters are shunted back to the cytoplasmic vesicle via endocytosis (28). What we have shown here is that coexpression of laforin or malin is able to restrict Glut3 in the cytoplasmic vesicles. While our observations are strong enough to suggest that the laforin-malin complex regulates the subcellular trafficking of glucose transporters, the specific process by which laforin and malin are able to modulate the vesicular trafficking was not addressed in the present study. Clearly, further studies are required to elucidate this mechanism.
In summary, we have shown that the laforin-malin complex regulates glucose uptake and not the degradative process of cellular glycogen. Thus, excessive substrate (cellular glucose level) appears to be the trigger for the abnormally high levels of glycogen seen in LD. Thus, laforin/malin deficiency would result in an abnormal increase in cellular glycogen that is less branched and hyperphosphoryled, leading to the genesis of Lafora bodies. Our study indicates that factors that regulate cellular glucose uptake could be potential therapeutic targets for the treatment of LD.
ACKNOWLEDGMENTS
This work was supported by a sponsored research grant from the Department of Science and Technology, Government of India to S.G. S.G. is a DAE-SRC Outstanding Research Investigator (supported by the Department of Atomic Energy, Government of India) and Joy-Gill Chair Professor. P.S. and S.S received research fellowships from the Council of Scientific and Industrial Research, Government of India.
We thank Kazuhiro Yamakawa (RIKEN Brain Science Institute, Japan) for generously providing muscle tissue samples from laforin-deficient mice, Juan P. Bolanos (Universitario de Salamanca, Spain) for expression constructs for the glucose transporter, Otto Baba (Tokyo Medical and Dental University, Japan) for the glycogen antibody, and Balaji Prakash (IIT Kanpur) for extending the use of his research facilities for some of the experiments reported in the present study. We also thank the anonymous reviewers for their comments and suggestions on an earlier version of this article.
Footnotes
Published ahead of print 28 November 2011
REFERENCES
- 1. Becker M, Newman S, Ismail-Beigi F. 1996. Stimulation of GLUT1 glucose transporter expression in response to inhibition of oxidative phosphorylation: role of reduced sulfhydryl groups. Mol. Cell. Endocrinol. 121:165–170 [DOI] [PubMed] [Google Scholar]
- 2. Bell B, Xing H, Yan K, Gautam N, Muslin AJ. 1999. KSR-1 binds to G-protein βγ subunits and inhibits βγ-induced mitogen-activated protein kinase activation. J. Biol. Chem. 274:7982–7986 [DOI] [PubMed] [Google Scholar]
- 3. Boer P, Sperling O. 2004. Modulation of glycogen phosphorylase activity affects 5-phosphoribosyl-1-pyrophosphate availability in rat hepatocyte cultures. Nucleosides Nucleotides Nucleic Acids 23:1235–1239 [DOI] [PubMed] [Google Scholar]
- 4. Carruthers A, DeZutter J, Ganguly A, Devaskar SU. 2009. Will the original glucose transporter isoform please stand up! Am. J. Physiol. 297:E836–E848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chan EM, et al. 2004. Laforin preferentially binds the neurotoxic starch-like polyglucosans, which form in its absence in progressive myoclonus epilepsy. Hum. Mol. Genet. 13:1117–1129 [DOI] [PubMed] [Google Scholar]
- 6. Chan EM, et al. 2003. Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat. Genet. 35:125–127 [DOI] [PubMed] [Google Scholar]
- 7. Cheng A, et al. 2007. A role for AGL ubiquitination in the glycogen storage disorders of Lafora and Cori's disease. Genes Dev. 21:2399–2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cid E, Cifuentes D, BaquÉ S, Ferrer JC, Guinovart JJ. 2005. Determinants of the nucleocytoplasmic shuttling of muscle glycogen synthase. FEBS J. 272:3197–3213 [DOI] [PubMed] [Google Scholar]
- 9. Coore HG, Randle PJ. 1964. Inhibition of glucose phosphorylation by mannoheptulose. Biochem. J. 91:56–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davies SP, Carling D, Hardie DG. 1989. Tissue distribution of the AMP-activated protein kinase, and lack of activation by cyclic-AMP-dependent protein kinase, studied using a specific and sensitive peptide assay. Eur. J. Biochem. 186:123–128 [DOI] [PubMed] [Google Scholar]
- 11. DePaoli-Roach AA, et al. 2010. Genetic depletion of the malin E3 ubiquitin ligase in mice leads to Lafora bodies and the accumulation of insoluble laforin. J. Biol. Chem. 285:25372–25381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dombrowski L, Roy D, Marcotte B, Marette A. 1996. A new procedure for the isolation of plasma membranes, T tubules, and internal membranes from skeletal muscle. Am. J. Physiol. 270:E667–E676 [DOI] [PubMed] [Google Scholar]
- 13. Dubey D, Ganesh S. 2008. Modulation of functional properties of laforin phosphatase by alternative splicing reveals a novel mechanism for the EPM2A gene in Lafora progressive myoclonus epilepsy. Hum. Mol. Genet. 17:3010–3020 [DOI] [PubMed] [Google Scholar]
- 14. Fernández-Sánchez ME, et al. 2003. Laforin, the dual-phosphatase responsible for Lafora disease, interacts with R5 (PTG), a regulatory subunit of protein phosphatase-1 that enhances glycogen accumulation. Hum. Mol. Genet. 12:3161–3171 [DOI] [PubMed] [Google Scholar]
- 15. Ganesh S, et al. 2000. Laforin, defective in the progressive myoclonus epilepsy of Lafora type, is a dual-specificity phosphatase associated with polyribosomes. Hum. Mol. Genet. 9:2251–2261 [DOI] [PubMed] [Google Scholar]
- 16. Ganesh S, et al. 2002. Targeted disruption of the Epm2a gene causes formation of Lafora inclusion bodies, neurodegeneration, ataxia, myoclonus epilepsy and impaired behavioral response in mice. Hum. Mol. Genet. 11:1251–1262 [DOI] [PubMed] [Google Scholar]
- 17. Ganesh S, Puri R, Singh S, Mittal S, Dubey D. 2006. Recent advances in the molecular basis of Lafora's progressive myoclonus epilepsy. J. Hum. Genet. 51:1–8 [DOI] [PubMed] [Google Scholar]
- 18. Ganesh S, et al. 2004. The carbohydrate-binding domain of Lafora disease protein targets Lafora polyglucosan bodies. Biochem. Biophys. Res. Commun. 313:1101–1109 [DOI] [PubMed] [Google Scholar]
- 19. Garyali P, et al. 2009. The laforin-malin complex suppresses the cellular toxicity of misfolded proteins by promoting their degradation through the ubiquitin-proteasome system. Hum. Mol. Genet. 18:688–700 [DOI] [PubMed] [Google Scholar]
- 20. Gentry MS, Worby CA, Dixon JE. 2005. Insights into Lafora disease: malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Proc. Natl. Acad. Sci. U. S. A. 102:8501–8506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guinez C, et al. 2008. Protein ubiquitination is modulated by O-GlcNAc glycosylation. FASEB J. 22:2901–2911 [DOI] [PubMed] [Google Scholar]
- 22. Hardie DG. 2007. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 8:774–785 [DOI] [PubMed] [Google Scholar]
- 23. Hawley SA, Gadalla AE, Olsen GS, Hardie DG. 2002. The antidiabetic drug metformin activates the AMP-activated protein kinase cascade via an adenine nucleotide-independent mechanism. Diabetes 51:2420–2425 [DOI] [PubMed] [Google Scholar]
- 24. Hresko RC, Heimberg H, Chi MM, Mueckler M. 1998. Glucosamine-induced insulin resistance in 3T3-L1 adipocytes is caused by depletion of intracellular ATP. J. Biol. Chem. 273:20658–20668 [DOI] [PubMed] [Google Scholar]
- 25. James LR, et al. 2002. Flux through the hexosamine pathway is a determinant of nuclear factor κB-dependent promoter activation. Diabetes 51:1146–1156 [DOI] [PubMed] [Google Scholar]
- 26. Johnston M. 1999. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet. 15:29–33 [DOI] [PubMed] [Google Scholar]
- 27. Liu Y, Wang Y, Wu C, Liu Y, Zheng P. 2006. Dimerization of laforin is required for its optimal phosphatase activity, regulation of GSK3β phosphorylation, and Wnt signaling. J. Biol. Chem. 28:34768–34774 [DOI] [PubMed] [Google Scholar]
- 28. Luiken JJ, et al. 2004. Regulation of cardiac long-chain fatty acid and glucose uptake by translocation of substrate transporters. Pflugers Arch. 448:1–15 [DOI] [PubMed] [Google Scholar]
- 29. Luo B, et al. 2007. Chronic hexosamine flux stimulates fatty acid oxidation by activating AMP-activated protein kinase in adipocytes. J. Biol. Chem. 282:7172–7180 [DOI] [PubMed] [Google Scholar]
- 30. Marshall S, Nadeau O, Yamasaki K. 2005. Glucosamine-induced activation of glycogen biosynthesis in isolated adipocytes. Evidence for a rapid allosteric control mechanism within the hexosamine biosynthesis pathway. J. Biol. Chem. 280:11018–11024 [DOI] [PubMed] [Google Scholar]
- 31. Medina RA, Owen GI. 2002. Glucose transporters: expression, regulation and cancer. Biol. Res. 35:9–26 [DOI] [PubMed] [Google Scholar]
- 32. Minassian BA, et al. 1998. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat. Genet. 20:171–174 [DOI] [PubMed] [Google Scholar]
- 33. Mittal S, Dubey D, Yamakawa K, Ganesh S. 2007. Lafora disease proteins malin and laforin are recruited to aggresomes in response to proteasomal impairment. Hum. Mol. Genet. 16:753–762 [DOI] [PubMed] [Google Scholar]
- 34. Munekata K, Sakamoto K. 2009. Forkhead transcription factor Foxo1 is essential for adipocyte differentiation. In Vitro Cell. Dev. Biol. Anim. 45:642–651 [DOI] [PubMed] [Google Scholar]
- 35. Polekhina G, et al. 2003. AMPK beta subunit targets metabolic stress sensing to glycogen. Curr. Biol. 13:867–871 [DOI] [PubMed] [Google Scholar]
- 36. Puri R, Suzuki T, Yamakawa K, Ganesh S. 2009. Hyperphosphorylation and aggregation of tau in laforin-deficient mice, an animal model for Lafora disease. J. Biol. Chem. 284:22657–22663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Romá-Mateo C, et al. 2011Laforin, a dual-specificity protein phosphatase involved in Lafora disease, is phosphorylated at Ser25 by AMP-activated protein kinase. Biochem. J. 439:265–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Salt IP, Johnson G, Ashcroft SJ, Hardie DG. 1998. AMP-activated protein kinase is activated by low glucose in cell lines derived from pancreatic beta cells, and may regulate insulin release. Biochem. J. 335:533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sengupta S, Badwar I, Upadhyay M, Singh S, Ganesh S. 2011. Malin and laforin are essential components of a protein complex that protects cells from the thermal stress. J. Cell Sci. 124:2277–2286 [DOI] [PubMed] [Google Scholar]
- 40. Shin BC, McKnight RA, Devaskar SU. 2004. Glucose transporter GLUT8 translocation in neurons is not insulin responsive. J. Neurosci. Res. 75:835–844 [DOI] [PubMed] [Google Scholar]
- 41. Singh S, Ganesh S. 2009. Lafora progressive myoclonus epilepsy: a meta-analysis of reported mutations in the first decade following the discovery of the EPM2A and NHLRC1 genes. Hum. Mutat. 30:715–723 [DOI] [PubMed] [Google Scholar]
- 42. Solaz-Fuster MC, et al. 2008. Regulation of glycogen synthesis by the laforin-malin complex is modulated by the AMP-activated protein kinase pathway. Hum. Mol. Genet. 17:667–678 [DOI] [PubMed] [Google Scholar]
- 43. Tagliabracci VS, et al. 2007. Laforin is a glycogen phosphatase, deficiency of which leads to elevated phosphorylation of glycogen in vivo. Proc. Natl. Acad. Sci. U. S. A. 104:19262–19266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tagliabracci VS, et al. 2008. Abnormal metabolism of glycogen phosphate as a cause for Lafora disease. J. Biol. Chem. 283:33816–33825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tagliabracci VS, et al. 2011. Phosphate incorporation during glycogen synthesis and Lafora disease. Cell. Metab. 13:274–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vernia S, et al. 2009. AMP-activated protein kinase phosphorylates R5/PTG, the glycogen targeting subunit of the R5/PTG-protein phosphatase 1 holoenzyme, and accelerates its down-regulation by the laforin-malin complex. J. Biol. Chem. 284:8247–8255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vernia S, et al. 2011. Laforin, a dual specificity phosphatase involved in Lafora disease, regulates insulin response and whole-body energy balance in mice. Hum. Mol. Genet. 20:2571–2584 [DOI] [PubMed] [Google Scholar]
- 48. Vilchez D, et al. 2007. Mechanism suppressing glycogen synthesis in neurons and its demise in progressive myoclonus epilepsy. Nat. Neurosci. 10:1407–1413 [DOI] [PubMed] [Google Scholar]
- 49. Wang J, Stuckey JA, Wishart M, Dixon JE. 2002. A unique carbohydrate binding domain targets the Lafora disease phosphatase to glycogen. J. Biol. Chem. 277:2377–2380 [DOI] [PubMed] [Google Scholar]
- 50. Wolfsdorf JI, Holm IA, Weinstein DA. 1999. Glycogen storage diseases: phenotypic, genetic, and biochemical characteristics, and therapy. Endocrinol. Metab. Clin. North. Am. 4:801–823 [DOI] [PubMed] [Google Scholar]
- 51. Yokoi S, Austin J, Witmer F, Sakai M. 1968. Studies in myoclonus epilepsy (Lafora body form). I. Isolation and preliminary characterization of Lafora bodies in two cases. Arch. Neurol. 19:15–33 [DOI] [PubMed] [Google Scholar]
- 52. Yokoi S, Nakayama H, Negishi T. 1975. Biochemical studies on tissues from a patient with Lafora disease. Clin. Chim Acta 62:415–423 [DOI] [PubMed] [Google Scholar]
- 53. Zhong D, et al. 2008. 2-Deoxyglucose induces Akt phosphorylation via a mechanism independent of LKB1/AMP-activated protein kinase signaling activation or glycolysis inhibition. Mol. Cancer Ther. 4:809–817 [DOI] [PubMed] [Google Scholar]