Abstract
D-type cyclins play a pivotal role in G1-S progression of the cell cycle, and their expression is frequently deregulated in cancer. Cyclin D1 has a half-life of only ∼30 min as a result of its ubiquitylation and proteasomal degradation, with various F-box proteins, including Fbxo4, Fbxw8, Skp2, and Fbxo31, having been found to contribute to its ubiquitylation. We have now generated Fbxo4-deficient mice and found no abnormalities in these animals. Cyclin D1 accumulation was thus not observed in Fbxo4−/− mouse tissues. The half-life of cyclin D1 in mouse embryonic fibroblasts (MEFs) prepared from Fbxo4−/−, Fbxw8−/−, and Fbxo4−/−; Fbxw8−/− mice also did not differ from that in wild-type MEFs. Additional depletion of Skp2 and Fbxo31 in Fbxo4−/−; Fbxw8−/− MEFs by RNA interference did not affect cyclin D1 stability. Although Fbxo31 depletion in MEFs increased cyclin D1 abundance, this effect appeared attributable to upregulation of cyclin D1 mRNA. Furthermore, abrogation of the function of the Skp1–Cul1–F-box protein (SCF) complex or the anaphase-promoting complex/cyclosome (APC/C) complexes did not alter the half-life of cyclin D1, whereas cyclin D1 degradation was dependent largely on proteasome activity. Our genetic analyses thus do not support a role for any of the four F-box proteins examined in cyclin D1 degradation during normal cell cycle progression. They suggest the existence of other ubiquitin ligases that target cyclin D1 for proteolysis.
INTRODUCTION
Progression of the cell cycle in eukaryotic cells depends on the activity of a series of protein complexes composed of cyclins and cyclin-dependent kinases (CDKs). Transition from G1 phase of the cell cycle to S phase is promoted by D-type cyclins (cyclins D1, D2, and D3), which were discovered as factors whose expression is increased by growth signals and are therefore considered mediators of signaling that links extracellular stimuli to the cell cycle machinery (4, 22, 24, 43). These proteins form complexes with CDK4 or CDK6 that phosphorylate and inactivate the product (pRb) of the retinoblastoma tumor suppressor gene and the pRb-related proteins p107 and p130 (20, 21, 23). This process is thought to be indispensable for progression of the cell cycle from G1 to S phase, given that cells deficient in D-type cyclins or in both CDK4 and CDK6 show marked defects in such cell cycle progression when stimulated with serum (16, 19).
Consistent with their functions, D-type cyclins, especially cyclin D1, are overexpressed in various types of malignant tumors, such as malignant lymphoma (24, 43), breast cancer (8), and esophageal cancer (36). Such overexpression of cyclin D1 is thought to result from upregulation of gene transcription or mRNA translation or from impaired protein degradation. Whereas transcriptional regulation of the cyclin D1 gene has been extensively studied and is well understood (15), the mechanism of cyclin D1 degradation has remained unclear. Cyclin D1 has been shown to be phosphorylated at Thr286 by glycogen synthase kinase 3β (GSK3β) and to undergo subsequent export from the nucleus and ubiquitylation-dependent proteolysis (2, 9, 10).
The proteolysis of core components of the cell cycle machinery depends largely on two RING finger-type ubiquitin ligases, the Skp1–Cul1–F-box protein (SCF) complex and the anaphase-promoting complex/cyclosome (APC/C), whose dysregulation often results in cancer development (28). The SCF complex is thought to function mainly from G1 phase to M phase. The ubiquitylation of cyclin D1 after the onset of S phase has therefore been proposed to be dependent on the SCF complex. The SCF complex consists of the RING finger protein Rbx1 (also known as Roc1 or Hrt1) in addition to the adaptor protein Skp1, the scaffold protein Cul1, and an F-box protein, the latter of which serves as the substrate recognition subunit of the SCF complex. About 70 members of the F-box protein family have been identified in mammals (3), among which four F-box proteins (Fbxo4, Fbxw8, Skp2, and Fbxo31) have independently been shown to contribute to cyclin D1 ubiquitylation (5, 18, 30, 35, 44). In addition, glucose deprivation was shown to induce cyclin D1 degradation in certain cultured cells, with an SCF complex containing the F-box protein β-TrCP having been found to be responsible for this type of proteolysis (41). However, it remains unclear which F-box protein plays the key role in regulation of cyclin D1 stability.
We have now adopted a genetic approach to test which of four F-box proteins is most relevant for cyclin D1 degradation. We generated Fbxo4−/−, Fbxw8−/−, and Fbxo4−/−; Fbxw8−/− mice and found that the stability of cyclin D1 was unchanged in cells from these animals. Additional depletion of Skp2 and Fbxo31 in Fbxo4−/−; Fbxw8−/− mouse embryonic fibroblasts (MEFs) did not affect the half-life of cyclin D1. Furthermore, abrogation of the function of SCF or APC/C complexes by expression of a dominant negative Cul1 mutant or depletion of APC2, respectively, had no effect on the half-life of cyclin D1. Collectively, our results suggest that the four F-box proteins examined are dispensable for cyclin D1 degradation, at least during normal progression of the cell cycle.
MATERIALS AND METHODS
Generation of Fbxo4−/− mice.
Cloned DNA corresponding to the Fbxo4 locus was amplified from the genome of E14 mouse embryonic stem (ES) cells with the use of LA-Taq polymerase (Takara, Shiga, Japan). The targeting vector was constructed by replacement of an 8.2-kb fragment of genomic DNA spanning exons 1 to 5 of Fbxo4 with IRES-lacZ and PGK-neo-poly(A)-loxP cassettes (6). The vector thus contained 8.2- and 1.2-kb regions of homology located 5′ and 3′, respectively, relative to IRES-lacZ and the neomycin resistance gene (neo). A diphtheria toxin A (DT-A) gene cassette was ligated at the 3′ end of the targeting construct. Maintenance, transfection, and selection of ES cells were performed as described previously (25). The recombination event was confirmed by Southern blot analysis with a 0.5-kb fragment of genomic DNA that flanks the 3′ homology region as the probe (see Fig. 1B and C). The expected sizes of hybridizing fragments after digestion of genomic DNA with PstI are 5.1 and 4.1 kb for the wild-type and mutant Fbxo4 alleles, respectively. Mutant ES cells were microinjected into C57BL/6 blastocysts, and resulting male chimeras were mated with female C57BL/6 mice. The germ line transmission of the mutant allele was confirmed by Southern blot analysis. Heterozygous offspring were intercrossed to produce homozygous mutant animals and their littermate controls. Total RNA was extracted from MEFs for reverse transcription (RT) and PCR analysis with primers (forward and reverse, respectively) for Fbxo4 (5′-GATGTCTTCTGAGGACCTTTGCC-3′ and 5′-CCTGAAGATCCAAAAGCTGGGTC-3′) or hypoxanthine guanine phosphoribosyl transferase (HPRT) (5′-GCCTAAGATGAGCGCAAGTTG-3′ and 5′-TACTAGGCAGATGGCCACAGG-3′). All mouse experiments were approved by the animal ethics committee of Kyushu University.
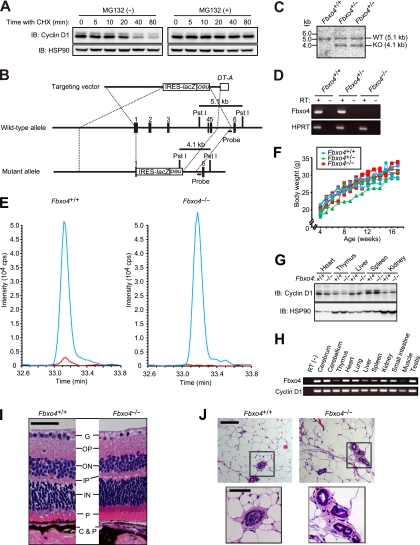
FIG 1.
Generation and phenotype of Fbxo4−/− mice. (A) Immunoblot (IB) analysis of cyclin D1 and HSP90 (loading control) in NIH 3T3 cells incubated with cycloheximide (CHX; 25 μg/ml) for the indicated times in the absence or presence of the proteasome inhibitor MG132 (10 μM). (B) Schematic representations of the wild-type mouse Fbxo4 locus, the targeting vector, and the mutant allele after homologous recombination. Exons are denoted by black boxes, and the positions of IRES-lacZ, PGK-neo-poly(A)-loxP (neo), and DT-A gene cassettes as well as PstI restriction sites are indicated. The expected sizes of DNA fragments that hybridize with the indicated probe in Southern blot analysis are shown. (C) Southern blot analysis of PstI-digested DNA from ES cells of the indicated Fbxo4 genotypes. The sizes of the hybridizing fragments corresponding to the wild-type (WT) and mutant (KO) alleles are indicated. (D) RT-PCR analysis of Fbxo4 and HPRT (control) mRNAs in Fbxo4+/+, Fbxo4+/−, and Fbxo4−/− MEFs. Reactions were performed with or without RT as indicated. (E) Extracted ion chromatograms for the Fbxo4 peptide in SRM analysis. The blue line indicates the signal for the internal standard (labeled with mTRAQ Δ4), and the red line indicates the signal for the endogenous Fbxo4 peptide (labeled with mTRAQ Δ0) for samples prepared from Fbxo4+/+ and Fbxo4−/− MEFs. (F) Representative growth curves for Fbxo4+/+, Fbxo4+/−, and Fbxo4−/− mice. (G) Immunoblot analysis of cyclin D1 and HSP90 in various tissues from 4-week-old Fbxo4+/+ or Fbxo4−/− mice. (H) RT-PCR analysis of Fbxo4 and cyclin D1 mRNAs in the indicated tissues of 12-week-old wild-type mice. (I) Histological analysis (hematoxylin-eosin staining) of the retina from 13-week-old Fbxo4−/− and wild-type littermates. G, ganglion cell layer; OP, outer plexiform layer; ON, outer nuclear layer; IP, inner plexiform layer; IN, inner nuclear layer; P, photoreceptor cell layer; C&P, choroid and pigment cells. Scale bar, 50 μm. (J) Histological analysis (hematoxylin-eosin staining) of the mammary gland from 20-week-old nulliparous Fbxo4−/− and wild-type littermates. The boxed areas in the upper panels are shown at a higher magnification in the lower panels; scale bars, 100 and 50 μm, respectively.
Histological analysis.
Tissue was fixed with 4 paraformaldehyde in phosphate-buffered saline, embedded in paraffin, and sectioned at a thickness of 3 μm. Hematoxylin-eosin staining was performed as described previously (26).
Construction of expression vectors and their introduction into cultured cells.
Mouse cDNAs for Fbxo4, Fbxw8, Skp2, Fbxo31, and Cdh1 were cloned into pENTR (Invitrogen, Carlsbad, CA) and then transferred to pMX-puro-FLAG vectors. Mouse cDNA for β-TrCP1 was directly cloned into pMX-puro-FLAG vectors. Mouse cDNA for CDK4 was directly cloned into pMX-puro-FLAG vectors. Mouse cDNA encoding Cul1 with a deletion of the COOH-terminal domain was cloned into pMX-puro-HA. The resulting plasmids were introduced into Plat-E cells by transfection with the Fu-Gene reagent (Roche, Indianapolis, IN) for the generation of recombinant retroviruses. NIH 3T3 cells were infected with the resulting retroviruses and subjected to selection in medium containing puromycin (2 μg/ml).
Preparation of MEFs and cell culture.
Primary MEFs were prepared from E13.5 embryos and cultured as previously described (25). For analysis of synchronized cells, confluent MEFs were rendered quiescent by culture for 48 h in medium supplemented with 0.1 fetal bovine serum (FBS). The cells were then cultured in medium containing 10 FBS to induce reentry into the cell cycle. For analysis of asynchronous cells, MEFs (1 × 106) plated in 10-cm culture dishes were cultured for 24 h. For growth rate analysis, MEFs (1 × 105) plated in duplicate 6-cm culture dishes were counted every day and supplied with fresh medium every 2 days.
SRM.
Mouse cDNA for Fbxo4 was subcloned into pET-30a (Novagen, Madison, WI). Fbxo4 with an NH2-terminal hexahistidine (His6) tag was then expressed in 5′-GGACCCGAGAAGACCTCCTT-3′ and 5′-GCACATCACTCAGAATTTCAATGG-3′; cyclin D1, 5′-TTCCCTTGACTGCCGAGAAG-3′ and 5′-TGAGCTTGTTCACCAGAAGCA-3′; Fbxo4, 5′-GATGTCTTCTGAGGACCTTTGCC-3′ and 5′-CCTGAAGATCCAAAAGCTGGGTC-3′; Fbxw8, 5′-TATCTGAACAGAAGAGAGCTGGG-3′ and 5′-GCAGGTGTAATCGGAGAATCTGC-3′; Skp2, 5′-GCTTAGTCGGGAGAACTTTCCAG-3′ and 5′-GGCGGTACCACCTCTTGCAAACG-3′; Fbxo31, 5′-ATGGCGTTTGTGAGAACCTGC-3′ and 5′-TGCCACAGCCCCAAAATGT-3′; and APC2, 5′-CCCAGGCTGACCAGAAGGA-3′ and 5′-TTAACATGGCCTGGATGTATGC-3′.
RNA interference.
Construction of short hairpin RNA (shRNA) vectors and RNA interference (RNAi) were performed as described previously (14). The sequences targeted to mouse Fbxo4, Fbxw8, Skp2, Fbxo31, and APC2 mRNAs were 5′-GCACTTGTCTTGATTACATGG-3′ (Fbxo4), 5′-GCTCATCCGCGACTTGAATGA-3′ (Fbxw8), 5′-CTTAGTCGGGAGAACTTTCC-3′ (Skp2-1), 5′-GCAGGCATGAGGTTAGTTTCA-3′ (Skp2-2), 5′-CACATCCAGATTGTGAAGA-3′ (Fbxo31), and 5′-GCAGTGGTCTTGCTGTATTTC-3′ (APC2). A vector for an shRNA targeted to enhanced green fluorescent protein (EGFP) mRNA was used as a control; the targeting sequence was 5′-CACCCTGAAATTCATCTGCAC-3′.
In vitro CDK4 kinase assay.
MEFs (4 × 106) were plated in 10-cm dishes and cultured for 24 h in complete medium, for 48 h in medium containing 0.1 FBS, and then for the indicated times in medium containing 10 FBS. The cells were then harvested, and cell lysates (300 μg of protein) were incubated with antibodies to CDK4 (C-22-AC; Santa Cruz Biotechnology) for 1 h at 4°C. The resulting immunoprecipitates were washed three times with wash buffer (10 mM HEPES-NaOH [pH 7.5], 150 mM NaCl, 0.1 Triton X-100) and twice with A buffer (60 mM HEPES-NaOH [pH 7.5], 5 mM MgCl2) and were then incubated at 30°C for 30 min in 20 μl of a reaction buffer containing 50 mM HEPES-NaOH (pH 7.5), 12.5 mM MgCl2, 1 mM EGTA, 4.5 mM 2-mercaptoethanol, 50 μM ATP, 222 kBq of [γ-32P]ATP, and 2 μg of Rb-C fusion protein (Santa Cruz Biotechnology). The reaction mixtures were then subjected to SDS-polyacrylamide gel electrophoresis (PAGE) followed by autoradiography with a BAS-2500 image analyzer (FujiFilm, Tokyo, Japan).
Pulse-chase analysis.
MEFs (1 × 106) were plated in 10-cm dishes, cultured for 24 h, washed with phosphate-buffered saline, and cultured for 30 min in methionine- and cysteine-free Dulbecco's modified Eagle's medium (Sigma) supplemented with 10 dialyzed FBS and 2 mM l-glutamine. The cells were then labeled for 1 h at 37°C with 5 ml of the same medium containing [35S]methionine and [35S]cysteine (total of 3.7 MBq/ml). After removal of the radioactive medium, the cells were cultured for the indicated times in medium containing 10 FBS and were then subjected to immunoprecipitation with antibodies to cyclin D1 (72-13G-AC; Santa Cruz Biotechnology). The immunoprecipitates were subjected to SDS-PAGE followed by autoradiography with a BAS-2500 image analyzer.
Statistical analysis.
Quantitative data are presented as means ± standard deviations (SD) unless indicated otherwise and were analyzed by Student's unpaired t test. A P value of <0.05 was considered statistically significant.
RESULTS
Fbxo4 is dispensable for cyclin D1 degradation.
We first measured the stability of cyclin D1 in NIH 3T3 cells cultured with the protein synthesis inhibitor cycloheximide and in the absence or presence of the proteasome inhibitor MG132. Consistent with previous observations (9, 10), cyclin D1 was rapidly degraded, with a half-life of <30 min, in the absence of MG132, whereas this rapid turnover was almost completely abolished by the proteasome inhibitor (Fig. 1A). These results suggested that the stability of cyclin D1 is dependent largely on proteasomal degradation.
To investigate the requirement for Fbxo4 in ubiquitin-dependent degradation of cyclin D1, we generated mice deficient in this protein. The Fbxo4 gene was disrupted in mouse ES cells by replacement of a region spanning exons 1 to 5 with IRES-lacZ and PGK-neo-poly(A)-loxP cassettes (Fig. 1B). We verified the homologous recombination event by Southern blot analysis of ES cells (Fig. 1C), and we confirmed that Fbxo4 mRNA was not detectable in Fbxo4−/− mice by RT-PCR analysis (Fig. 1D). We further confirmed the absence of the Fbxo4 protein in Fbxo4−/− mice with the use of mass spectrometry-based SRM. SRM analysis of Fbxo4+/+ and Fbxo4−/− MEFs thus revealed that the selected Fbxo4 peptide was undetectable in the Fbxo4−/− cells (Fig. 1E).
Mice homozygous for the Fbxo4 mutant allele were born at the expected Mendelian ratio. The mutant mice were healthy, fertile, and phenotypically indistinguishable from wild-type littermates, and no neoplasms were apparent in the mutant animals up to 1 year of age. The rate of increase in body weight thus did not differ among Fbxo4+/+, Fbxo4+/−, and Fbxo4−/− mice (Fig. 1F). Immunoblot analysis revealed that the abundance of cyclin D1 did not differ between Fbxo4+/+ and Fbxo4−/− mice in any of the tissues examined (Fig. 1G). RT-PCR analysis showed that Fbxo4 and cyclin D1 mRNAs are distributed ubiquitously in wild-type mice at 12 weeks of age (Fig. 1H), suggesting that the lack of apparent abnormalities in Fbxo4−/− mice is not attributable to spatially or temporally restricted expression of Fbxo4. Given that loss of cyclin D1 results in hypoplasia of the retina and mammary gland (37), we examined the microscopic structure of these tissues. No abnormality was found in cell density or organization in these tissues or in any others examined (Fig. 1I and J; data not shown).
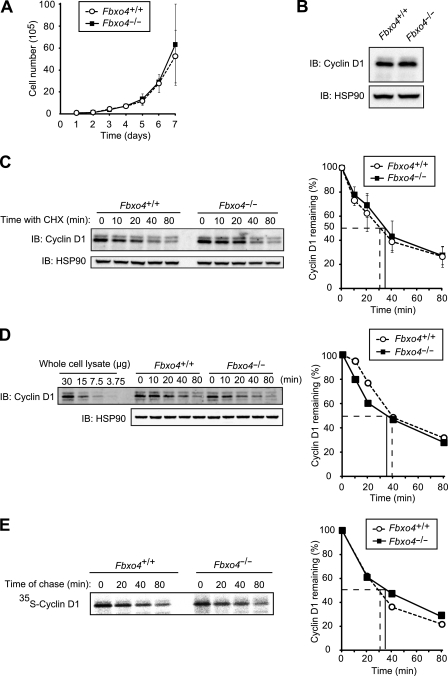
To investigate further the effects of Fbxo4 deficiency, we examined cellular functions in Fbxo4+/+ and Fbxo4−/− MEFs. There was no difference in cell proliferation rates between Fbxo4+/+ and Fbxo4−/− MEFs (Fig. 2A), and no accumulation of cyclin D1 in Fbxo4−/− MEFs was observed (Fig. 2B). Cycloheximide chase analysis revealed that the half-life of cyclin D1 in Fbxo4−/− MEFs was almost identical to that in Fbxo4+/+ MEFs (Fig. 2C). We also measured the half-life of cyclin D1 by fluorescence-based quantitative immunoblot analysis, in which the dynamic range is broader and the quantification more accurate than ECL-based analysis. Again, the half-life of cyclin D1 did not differ between Fbxo4+/+ and Fbxo4−/− MEFs with this approach (Fig. 2D). Given that cycloheximide chase analysis can give rise to secondary effects due to general inhibition of protein synthesis, we also measured the half-life of cyclin D1 by metabolic pulse-chase analysis. The half-life of cyclin D1 determined by pulse-chase analysis was almost identical to that determined by cycloheximide chase analysis, and the stability of cyclin D1 also did not differ between Fbxo4+/+ and Fbxo4−/− MEFs with this approach (Fig. 2E), suggesting that the half-life of cyclin D1 determined with the cycloheximide chase assay represents the physiological turnover rate of the protein. We therefore adopted cycloheximide chase analysis for subsequent evaluation of the stability of cyclin D1.
FIG 2.
Cyclin D1 does not accumulate in Fbxo4−/− MEFs. (A) Growth curves for Fbxo4+/+ and Fbxo4−/− MEFs. Data are means ± SD of values obtained from three MEF preparations of each genotype. (B) Immunoblot analysis of cyclin D1 in MEFs derived from Fbxo4+/+ and Fbxo4−/− mice. (C) Immunoblot analysis of cyclin D1 (left) and its quantification (right) for Fbxo4+/+ and Fbxo4−/− MEFs incubated with cycloheximide (25 μg/ml) for the indicated times. The half-life of cyclin D1 in cells of each genotype is indicated. Quantitative data are means ± SD of values obtained from four MEF preparations of each genotype. (D) Quantitative immunoblot analysis of cyclin D1 with fluorescent secondary antibodies (left) and its quantification (right) for Fbxo4+/+ and Fbxo4−/− MEFs incubated with cycloheximide for the indicated times. The indicated amounts of whole-cell lysate were analyzed as a standard. (E) Pulse-chase analysis of cyclin D1 (left) and its quantification (right) for Fbxo4+/+ and Fbxo4−/− MEFs. Cells were metabolically labeled with [35S]methionine and [35S]cysteine for 1 h and then chased with nonradioactive methionine and cysteine for the indicated times. Cell lysates were then prepared and subjected to immunoprecipitation with antibodies to cyclin D1, and the resulting precipitates were subjected to SDS-PAGE and autoradiography.
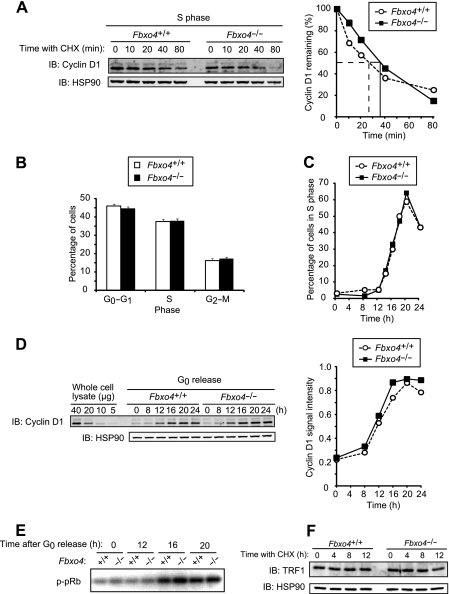
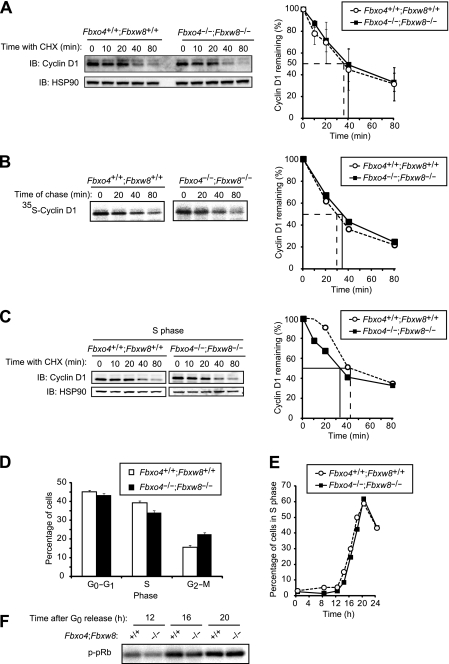
Given that cyclin D1 is degraded mainly in S phase of the cell cycle (12), we examined cyclin D1 stability in S phase. The degradation rates of cyclin D1 in S phase did not differ between Fbxo4+/+ and Fbxo4−/− MEFs (Fig. 3A). We next examined whether Fbxo4 deficiency affects cell cycle distribution. The percentages of MEFs in G0-G1, S, and G2-M phases of the cell cycle were not affected by the loss of Fbxo4 (Fig. 3B). We also examined progression of the cell cycle from G0 phase to S phase. MEFs that had been restrained in G0 phase by serum deprivation were released by serum replenishment, and the percentage of cells in S phase and cyclin D1 abundance were monitored, the latter by quantitative immunoblot analysis. Neither the progression of cells into S phase nor cyclin D1 expression differed between the two Fbxo4 genotypes (Fig. 3C and D). Kinase activity associated with the cyclin D1-CDK4 complex during cell cycle progression was also not affected by Fbxo4 deficiency (Fig. 3E). These results thus suggested that Fbxo4 is dispensable for cyclin D1 degradation.
FIG 3.
Depletion of Fbxo4 does not affect cyclin D1 function. (A) Immunoblot analysis of cyclin D1 (left) and its quantification (right) for Fbxo4+/+ and Fbxo4−/− MEFs incubated with cycloheximide in S phase. Cells were released from G0 phase and cultured for 20 h (at which time most cells were in S phase) before incubation with cycloheximide (25 μg/ml) for the indicated times. (B) Cell cycle distribution of asynchronous Fbxo4+/+ and Fbxo4−/− MEFs as determined by flow cytometry. Data are means ± SD of results from three independent experiments. (C) Kinetics of the reentry of Fbxo4+/+ and Fbxo4−/− MEFs into the cell cycle from G0 phase. Data are representative of three independent experiments. (D) Fluorescence-based quantitative immunoblot analysis of cyclin D1 (left) and its quantification (right) for Fbxo4+/+ and Fbxo4−/− MEFs released from G0 phase for the indicated times. The indicated amounts of whole-cell lysate were analyzed as a standard. A signal intensity of 1.0 corresponds to that for 40 μg of cell lysate. (E) In vitro CDK4 kinase assay for Fbxo4+/+ and Fbxo4−/− MEFs released from G0 phase for the indicated times. Cell lysates were subjected to immunoprecipitation with antibodies to CDK4, and the resulting precipitates were assayed for CDK4 kinase activity with a glutathione S-transferase (GST)-pRb fusion protein as substrate. (F) Immunoblot analysis of TRF1 in Fbxo4+/+ and Fbxo4−/− MEFs incubated with cycloheximide for the indicated times.
We also attempted to measure the half-life of TRF1, another potential target of Fbxo4 (17), in the absence or presence of Fbxo4, but we were unable to do so, because it was found to be a highly stable protein with a half-life of >12 h. Loss of Fbxo4 did not affect the steady-state abundance of TRF1 (Fig. 3F), however, suggesting that the physiological relevance of Fbxo4 to TRF1 stability is at most minimal, at least under the conditions of our analysis.
Cyclin D1 degradation occurs normally in MEFs from mice lacking both Fbxo4 and Fbxw8.
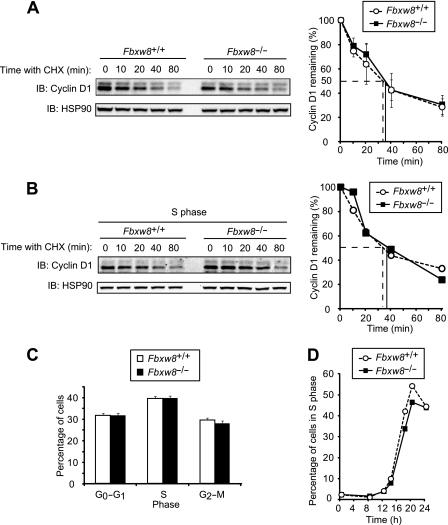
Mice lacking Fbxw8 exhibit a partially embryonic lethal phenotype as a result of a developmental defect of the placenta (39). We prepared MEFs from Fbxw8+/+ and Fbxw8−/− embryos and examined whether the degradation of cyclin D1 is impaired in the Fbxw8−/− cells. Cycloheximide chase analysis revealed that the half-life of cyclin D1 in Fbxw8−/− MEFs was indistinguishable from that in Fbxw8+/+ MEFs (Fig. 4A). The degradation rate of cyclin D1 in S phase was also not affected by Fbxw8 deficiency (Fig. 4B). The percentages of MEFs in G0-G1, S, or G2-M phases of the cell cycle (Fig. 4C) as well as the progression of cells into S phase from G0 (Fig. 4D) also did not differ between the two genotypes. These results thus suggested that Fbxw8 is also dispensable for cyclin D1 degradation.
FIG 4.
Cyclin D1 does not accumulate in Fbxw8−/− MEFs. (A) Immunoblot analysis of cyclin D1 (left) and its quantification (right) for MEFs from Fbxw8+/+ and Fbxw8−/− mice incubated with cycloheximide (25 μg/ml) for the indicated times. Quantitative data are means ± SD of values obtained from four MEF preparations of each genotype. (B) Immunoblot analysis of cyclin D1 (left) and its quantification (right) for Fbxw8+/+ and Fbxw8−/− MEFs incubated with cycloheximide for the indicated times in S phase. (C) Cell cycle distribution of asynchronous Fbxw8+/+ and Fbxw8−/− MEFs. Data are means ± SD of results from four independent experiments. (D) Kinetics of the reentry of Fbxw8+/+ and Fbxw8−/− MEFs into the cell cycle from G0 phase. Data are representative of results from three independent experiments.
A possible explanation for the apparently unaltered degradation of cyclin D1 in Fbxo4−/− and Fbxw8−/− mice was that Fbxo4 and Fbxw8 reciprocally compensate for each other's deficiency. To test this possibility, we first depleted NIH 3T3 cells of Fbxo4 or Fbxw8 by RNAi. The half-life of cyclin D1 was not affected by RNAi-mediated acute depletion of either Fbxo4 (Fig. 5A and B) or Fbxw8 (Fig. 5C and D). We next examined the abundance of Fbxw8 or Fbxo4 mRNA in Fbxo4−/− and Fbxw8−/− MEFs, respectively, but we did not detect a compensatory increase in the amount of either mRNA (Fig. 5E). We then generated mice lacking both Fbxo4 and Fbxw8 (Fbxo4−/−; Fbxw8−/− mice). Cycloheximide chase analysis (Fig. 6A) as well as pulse-chase analysis (Fig. 6B) revealed that the half-life of cyclin D1 in Fbxo4−/−; Fbxw8−/− MEFs was almost identical to that in Fbxo4+/+; Fbxw8+/+ cells. The degradation rate of cyclin D1 in S phase was also similar in Fbxo4−/−; Fbxw8−/− and Fbxo4+/+; Fbxw8+/+ cells (Fig. 6C). Furthermore, both the percentages of cells in G0-G1, S, or G2-M phases of the cell cycle (Fig. 6D) as well as the progression of cells into S phase from G0 (Fig. 6E) did not differ between Fbxo4−/−; Fbxw8−/− and Fbxo4+/+; Fbxw8+/+ MEFs. The kinase activity associated with cyclin D1-CDK4 in Fbxo4−/−; Fbxw8−/− MEFs was also almost identical to that in wild-type MEFs (Fig. 6F). These results thus suggested that neither Fbxo4 nor Fbxw8 is essential for cyclin D1 degradation, at least in MEFs.
FIG 5.
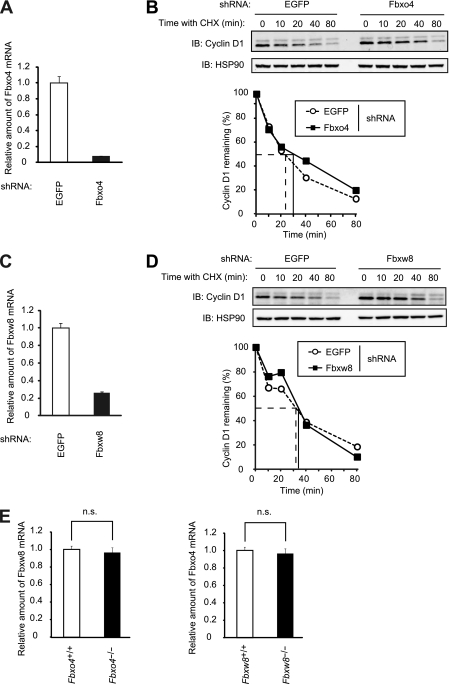
Acute depletion of Fbxo4 or Fbxw8 does not affect the half-life of cyclin D1. (A) Quantitative RT-PCR analysis of Fbxo4 mRNA in NIH 3T3 cells infected with a retroviral vector encoding control (EGFP) or Fbxo4 shRNAs. Data are means ± SD of triplicates from a representative experiment. (B) Immunoblot analysis of cyclin D1 (top) and its quantification (bottom) for NIH 3T3 cells infected as described for panel A and incubated with cycloheximide (25 μg/ml) for the indicated times. (C) Quantitative RT-PCR analysis of Fbxw8 mRNA in NIH 3T3 cells infected with a retroviral vector encoding control (EGFP) or Fbxw8 shRNAs. Data are means ± SD of triplicates from a representative experiment. (D) Immunoblot analysis of cyclin D1 (top) and its quantification (bottom) for NIH 3T3 cells infected as described for panel C and incubated with cycloheximide for the indicated times. (E) Quantitative RT-PCR analysis of Fbxw8 mRNA in Fbxo4+/+ and Fbxo4−/− MEFs and of Fbxo4 mRNA in Fbxw8+/+ and Fbxw8−/− MEFs. Data are means ± SD of triplicates from a representative experiment. n.s., not significant (Student's unpaired t test).
FIG 6.
Cyclin D1 does not accumulate in Fbxo4−/−; Fbxw8−/− MEFs. (A) Immunoblot analysis of cyclin D1 (left) and its quantification (right) for Fbxo4+/+; Fbxw8+/+ and Fbxo4−/−; Fbxw8−/− MEFs incubated with cycloheximide (25 μg/ml) for the indicated times. Quantitative data are means ± SD of values obtained from five MEF preparations of each genotype. (B) Pulse-chase analysis of cyclin D1 (left) and its quantification (right) for Fbxo4+/+; Fbxw8+/+ and Fbxo4−/−; Fbxw8−/− MEFs. (C) Immunoblot analysis of cyclin D1 (left) and its quantification (right) for Fbxo4+/+; Fbxw8+/+ and Fbxo4−/−; Fbxw8−/− MEFs incubated with cycloheximide for the indicated times in S phase. (D) Cell cycle distribution of asynchronous Fbxo4+/+; Fbxw8+/+ and Fbxo4−/−; Fbxw8−/− MEFs. Data are means ± SD from four independent experiments. (E) Kinetics of the reentry of Fbxo4+/+; Fbxw8+/+ and Fbxo4−/−; Fbxw8−/− MEFs into the cell cycle from G0 phase. Data are representative of three independent experiments. (F) In vitro CDK4 kinase assay for Fbxo4+/+; Fbxw8+/+ and Fbxo4−/−; Fbxw8−/− MEFs released from G0 phase for the indicated times.
Ablation of four F-box proteins does not affect cyclin D1 degradation.
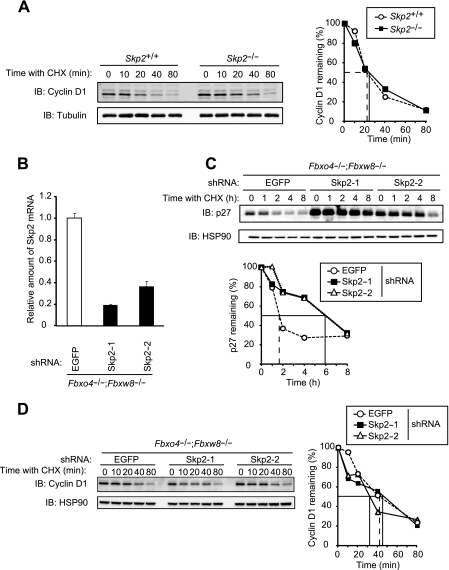
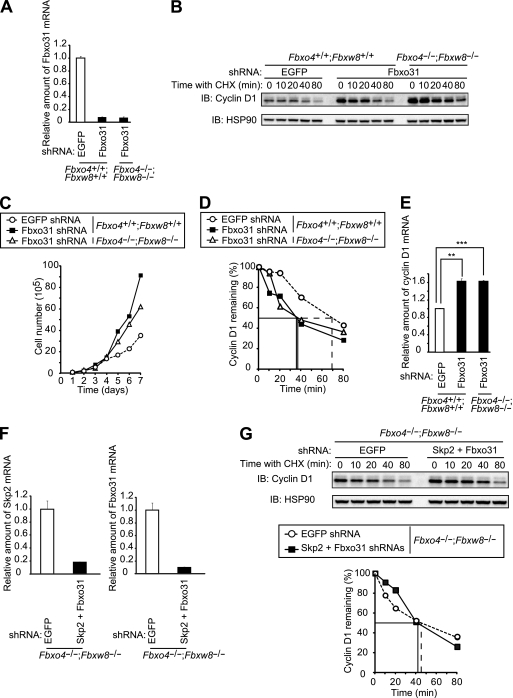
Skp2 has been implicated in cyclin D1 degradation (44). To examine such a role for Skp2, we first compared the stability of cyclin D1 between Skp2+/+ and Skp2−/− MEFs. No difference between the two genotypes was detected (Fig. 7A). We further examined whether Skp2 might compensate for the lack of Fbxo4 and Fbxw8 in mediation of cyclin D1 degradation in Fbxo4−/−; Fbxw8−/− MEFs. Depletion of Skp2 by RNAi resulted in a marked increase in both the steady-state level and stability of p27, the major substrate of Skp2 (27), in Fbxo4−/−; Fbxw8−/− MEFs, validating the attenuation of Skp2 function (Fig. 7B and C). Such depletion of Skp2 did not affect the turnover rate of cyclin D1, however, in Fbxo4+/+; Fbxw8+/+ or Fbxo4−/−; Fbxw8−/− MEFs (Fig. 7D and data not shown). In contrast, similar depletion of Fbxo31 (Fig. 8A) resulted in a marked increase in the abundance of cyclin D1 (Fig. 8B) that was accompanied by enhanced cell proliferation (Fig. 8C) in both Fbxo4+/+; Fbxw8+/+ and Fbxo4−/−; Fbxw8−/− MEFs. However, the turnover rate of cyclin D1 was actually increased by depletion of Fbxo31 in both Fbxo4+/+; Fbxw8+/+ and Fbxo4−/−; Fbxw8−/− MEFs (Fig. 8D), suggesting that the increase in the amount of cyclin D1 induced by Fbxo31 depletion is not attributable to impaired degradation of this protein. Quantitative RT-PCR analysis revealed that the abundance of cyclin D1 mRNA was significantly increased as a result of Fbxo31 depletion (Fig. 8E). These results thus suggested that the upregulation of cyclin D1 associated with Fbxo31 deficiency is likely the result of increased transcription of the cyclin D1 gene, although the mechanism underlying this effect awaits further investigation.
FIG 7.
Cyclin D1 does not accumulate in Skp2-depleted Fbxo4−/−; Fbxw8−/− MEFs. (A) Immunoblot analysis of cyclin D1 (left) and its quantification (right) for MEFs from Skp2+/+ and Skp2−/− mice incubated with cycloheximide (25 μg/ml) for the indicated times. α-Tubulin was examined as a loading control. (B) Quantitative RT-PCR analysis of Skp2 mRNA in Fbxo4−/−; Fbxw8−/− MEFs infected with a retroviral vector encoding control (EGFP) or one of two Skp2 shRNAs. Data are means ± SD of triplicates from a representative experiment. (C) Immunoblot analysis of p27 (top) and its quantification (bottom) for MEFs infected as described for panel B and incubated with cycloheximide for the indicated times. (D) Immunoblot analysis of cyclin D1 (left) and its quantification (right) for MEFs infected as described for panel B and incubated with cycloheximide for the indicated times.
FIG 8.
Cyclin D1 is degraded in the absence of Fbxo4, Fbxw8, Skp2, and Fbxo31. (A) Quantitative RT-PCR analysis of Fbxo31 mRNA in Fbxo4+/+; Fbxw8+/+ and Fbxo4−/−; Fbxw8−/− MEFs infected with a retroviral vector encoding EGFP or Fbxo31 shRNAs. Data are means ± SD of triplicates from a representative experiment. (B) Immunoblot analysis of cyclin D1 in MEFs as described for panel A incubated with cycloheximide (25 μg/ml) for the indicated times. (C) Growth curves for MEFs as described for panel A. Data are representative of results from three independent experiments. (D) Quantification of cyclin D1 abundance in the experiment shown in panel B. (E) Quantitative RT-PCR analysis of cyclin D1 mRNA in MEFs as described for panel A. Data are means ± SD of triplicates from a representative experiment. **, P < 0.005; ***, P < 0.0005 (Student's unpaired t test). (F) Quantitative RT-PCR analysis of Skp2 and Fbxo31 mRNAs in Fbxo4−/−; Fbxw8−/− MEFs infected with retroviral vectors encoding EGFP or both Skp2 and Fbxo31 shRNAs. Data are means ± SD of triplicates from a representative experiment. (G) Immunoblot analysis of cyclin D1 (top) and its quantification (bottom) for MEFs, as described for panel F, incubated with cycloheximide for the indicated times.
Finally, we depleted Fbxo4−/−; Fbxw8−/− MEFs of Skp2 and Fbxo31 by RNAi in order to examine whether cyclin D1 degradation occurs in the absence of all four F-box proteins (Fig. 8F). Cyclin D1 was indeed degraded in MEFs deficient in all four F-box proteins as efficiently as in control Fbxo4−/−; Fbxw8−/− MEFs (Fig. 8G). We therefore concluded that none of these four F-box proteins plays a key role in the degradation of cyclin D1, at least during normal progression of the cell cycle.
Cyclin D1 degradation is independent of SCF and APC/C complexes.
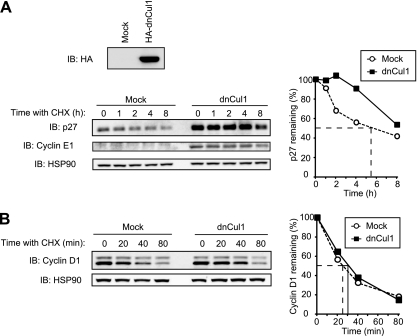
We next examined whether the stability of cyclin D1 is regulated by the SCF complex. To this end, we expressed a dominant negative mutant of Cul1 (dnCul1) that lacks the COOH-terminal domain responsible for binding to Rbx1. As a positive control, we showed that the stability of p27 (a substrate of SCFSkp2) and cyclin E1 (a substrate of SCFFbxw7) was markedly increased by the expression of dnCul1 in NIH 3T3 cells, validating that the function of these SCF complexes was indeed impaired (Fig. 9A). The half-life of cyclin D1 was not affected by dnCul1 expression, however (Fig. 9B). These results thus suggested that the SCF complex is dispensable for cyclin D1 degradation.
FIG 9.
Inhibition of Cul1 function does not affect the stability of cyclin D1. (A) Immunoblot analysis with antibodies to the hemagglutinin epitope (HA; top) or to p27 and cyclin E1 (bottom left), as well as quantification of p27 (bottom right), for NIH 3T3 cells infected with a retroviral vector for an HA-tagged dominant negative mutant (dn) of Cul1 (or subjected to mock infection) and exposed to cycloheximide (25 μg/ml) for the indicated times. (B) Immunoblot analysis of cyclin D1 (left) and its quantification (right) for cells as described for panel A exposed to cycloheximide for the indicated times.
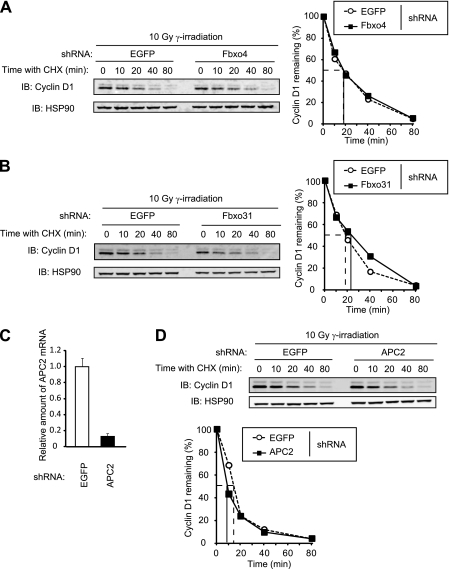
SCFFbxo4, SCFFbxo31, and APC/CCdh1 have been shown to promote cyclin D1 degradation in response to DNA damage (32, 33, 35). We therefore also examined the role of these complexes in cyclin D1 degradation triggered by DNA damage. Cycloheximide chase analysis revealed that depletion of Fbxo4, Fbxo31, or APC2 by RNAi did not affect the turnover rate of cyclin D1 after exposure of NIH 3T3 cells to gamma radiation (Fig. 10), suggesting that these proteins are dispensable for cyclin D1 degradation after DNA damage.
FIG 10.
The absence of Fbxo4, Fbxo31, or APC2 does not affect the stability of cyclin D1 after DNA damage. (A) Immunoblot analysis of cyclin D1 (left) and its quantification (right) for NIH 3T3 cells infected with a retroviral vector encoding control (EGFP) or Fbxo4 shRNAs. Cells were incubated with cycloheximide (25 μg/ml) for the indicated times beginning 1 h after gamma irradiation (10 Gy). (B) Immunoblot analysis of cyclin D1 (left) and its quantification (right) for NIH 3T3 cells infected with a retroviral vector encoding EGFP or Fbxo31 shRNAs and treated as described for panel A. (C) Quantitative RT-PCR analysis of APC2 mRNA in NIH 3T3 cells infected with a retroviral vector encoding EGFP or APC2 shRNAs. Data are means ± SD of triplicates from a representative experiment. (D) Immunoblot analysis of cyclin D1 (top) and its quantification (bottom), as described for panel C, for cells incubated with cycloheximide for the indicated times beginning 1 h after gamma irradiation.
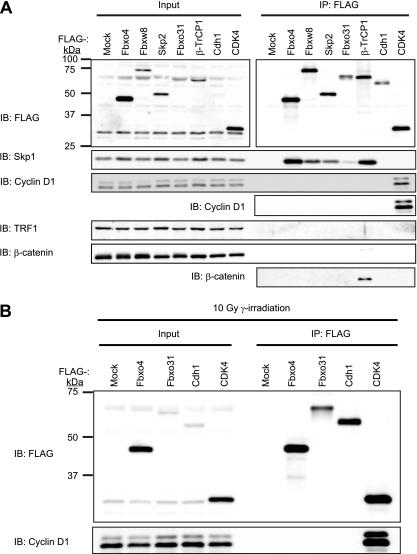
We finally examined whether endogenous cyclin D1 binds to Fbxo4, Fbxw8, Fbxo31, Skp2, β-TrCP1, or Cdh1 proteins tagged with the FLAG epitope and stably expressed in NIH 3T3 cells. All F-box proteins tested (Fbxo4, Fbxw8, Skp2, Fbxo31, and β-TrCP1) interacted with Skp1, the adaptor protein of the SCF complex; CDK4 interacted with cyclin D1; and β-TrCP1 interacted with its substrate β-catenin (Fig. 11A), validating the effectiveness of the coimmunoprecipitation experiments. In contrast, none of the F-box proteins tested nor Cdh1 interacted with endogenous cyclin D1 (Fig. 11A). The interactions between these proteins and cyclin D1 were also examined after DNA damage. Even after exposure of the cells to gamma radiation, Fbxo4, Fbxo31, and Cdh1 did not interact with endogenous cyclin D1 (Fig. 11B). These results thus supported our conclusion that none of the ubiquitin ligases examined contributes to cyclin D1 ubiquitylation, at least under our experimental conditions.
FIG 11.
Interactions between ubiquitin ligases and cyclin D1. (A) Lysates of NIH 3T3 cells infected with a retroviral vector encoding FLAG-tagged Fbxo4, Fbxw8, Skp2, Fbxo31, β-TrCP1, Cdh1, or CDK4 or subjected to mock infection were subjected to immunoprecipitation (IP) with antibodies to FLAG, and the resulting precipitates as well as the original lysates (Input) were subjected to immunoblot analysis with antibodies to FLAG or to the indicated proteins. (B) NIH 3T3 cells infected with a retroviral vector encoding FLAG-tagged Fbxo4, Fbxo31, Cdh1, or CDK4 were subjected to gamma irradiation (10 Gy). One hour after irradiation, the cells were lysed and subjected to immunoprecipitation with antibodies to FLAG, and the resulting precipitates as well as the original lysates were subjected to immunoblot analysis with antibodies to FLAG or to cyclin D1.
DISCUSSION
The half-life of cyclin D1 is <30 min even in the absence of DNA damage or glucose deficiency, and the degradation of this protein is fully dependent on the proteasome. We have now focused on cyclin D1 degradation during normal progression of the cell cycle and have investigated which ubiquitin ligase plays the prominent role in this process. We did not directly address the role of SCFβ-TrCP in the present study, given that this complex seems to participate in cyclin D1 degradation under specific conditions, such as glucose deprivation (41), but the half-life of cyclin D1 in MEFs that lack both β-TrCP1 and β-TrCP2 (also known as Fbxw11 or HOS) is similar to that in wild-type MEFs (our unpublished results). We have now shown that four SCF-type ubiquitin ligases (SCFFbxo4, SCFFbxw8, SCFSkp2, and SCFFbxo31), all of which have previously been found to participate in ubiquitylation of cyclin D1, are dispensable for cyclin D1 degradation during normal progression of the cell cycle.
Whereas the accumulation of cyclin D1 is essential for mitogen-dependent cell cycle progression, the biological relevance and molecular mechanism of cyclin D1 degradation have remained unclear. Timely proteolysis of cyclin D1 has been thought to be required for normal progression of the cell cycle, with deregulated expression of cyclin D1 having been shown to result in cancer development (40). GSK3β-dependent phosphorylation of cyclin D1 on Thr286 triggers the CRM1-mediated nuclear export of this protein followed by its ubiquitylation and proteasomal degradation. Replacement of Thr286 with Ala (T286A) resulted in a marked increase in the stability of cyclin D1 (9, 10). Forced expression of the T286A mutant of cyclin D1 also promoted anchorage-independent growth of NIH 3T3 cells in soft agar (2). SCFFbxo4 was identified as a ubiquitin ligase that is responsible for the degradation of Thr286-phosphorylated cyclin D1 after its translocation to the cytoplasm. Mutations of FBXO4 that impair the ability of the encoded protein to mediate cyclin D1 ubiquitylation have been detected in 14 of primary esophageal cancers (5). Impairment of SCFFbxo4 function by RNAi-mediated depletion of Fbxo4 in some cell lines attenuated cyclin D1 ubiquitylation, resulting in cyclin D1 accumulation and promotion of cell cycle progression (18). However, our genetic analysis did not find such a change in cyclin D1 stability or expression level in Fbxo4−/− mice. Furthermore, cell cycle progression seemed intact and no predisposition to cancer was apparent (data not shown) in these animals.
Degradation of cyclin D1 also appears to be essential for normal replication of DNA, given that acute overexpression of cyclin D1 represses DNA replication as a result of its interaction with proliferating cell nuclear antigen (PCNA) and CDK2 (11). Fbxw8 was shown to play a pivotal role in cancer cell proliferation by promoting cyclin D1 degradation in S phase (30). However, it has been unclear whether Fbxw8 is necessary for cell cycle progression in normal cells. We previously found that Fbxw8 was not expressed at a detectable level in any of the adult mouse tissues examined with the exception of the placenta (39). Moreover, we have now shown that Fbxw8 is dispensable for cyclin D1 degradation in MEFs. Together, these observations suggest that Fbxw8 might be required for cyclin D1 degradation only in certain cancer cell lines. Unlike mice deficient in Fbxo4, Fbxw8-deficient mice exhibit pre- and postnatal growth retardation resulting from a defect in placental development (39). The present study shows that Fbxw8 is dispensable for the degradation of cyclin D1, suggesting that the phenotype of Fbxw8-deficient mice is attributable to the accumulation of substrates other than cyclin D1. Given that IRS-1, a key component of signaling pathways activated by the insulin and insulin-like growth factor 1 (IGF-1) receptors, has been implicated as a target of Fbxw8 (42), the accumulation of IRS-1 in the placenta might be responsible for the growth retardation observed in mice deficient in Fbxw8.
Whereas Skp2 was previously shown to contribute to cyclin D1 ubiquitylation (44), a direct association between cyclin D1 and Skp2 was not demonstrated. Rather, Skp2 might interact indirectly with cyclin D1 through p27, a well-established substrate of Skp2 that interacts with cyclin D1 (7). It has therefore remained unclear whether cyclin D1 is a bona fide substrate of Skp2. We have now shown that Skp2 is dispensable for cyclin D1 degradation.
DNA damage induces the ubiquitin-dependent degradation of cyclin D1 and consequent arrest of the cell cycle at G1 phase (31). A recent study showed that Fbxo31 mediates cyclin D1 degradation in response to DNA damage, whereas it does not contribute to cyclin D1 degradation during the normal cell cycle (35). Our results now also indicate that Fbxo31 is not responsible for cyclin D1 degradation during the normal cell cycle. However, we observed that depletion of Fbxo31 resulted in an accumulation of cyclin D1 that appeared to be attributable to an increase in mRNA abundance. This observation suggests the possibility that Fbxo31 targets a protein (or proteins) that controls transcription of the cyclin D1 gene. Our results further indicated that Fbxo31 is not responsible for cyclin D1 degradation after DNA damage. SCFFbxo4 and APC/CCdh1 were also identified as ubiquitin ligases responsible for degradation of cyclin D1 after DNA damage (1, 33). APC/CCdh1 is activated in S or G2 phase in response to genotoxic stress, although APC/C is thought to operate predominantly from M phase to G1 phase during normal progression of the cell cycle. Our results do not support a role for Fbxo4 or Cdh1 in cyclin D1 degradation after DNA damage. However, given that our data were based only on assay of in vivo binding and cycloheximide chase analysis in mouse fibroblasts, the role of these proteins in degradation of cyclin D1 in other situations remains to be examined.
Why do our results differ from those of previous studies? In our study, we focused mainly on cyclin D1 degradation that occurs during normal progression of the cell cycle. It is thus possible that Fbxo4, Fbxw8, Skp2, and Fbxo31, in addition to β-TrCP, contribute to cyclin D1 degradation under specific conditions, such as in cancer or in cells deprived of glucose, but not during regular progression of the cell cycle. Our data suggest the existence of other ubiquitin ligases that mediate cyclin D1 ubiquitylation during normal cell cycle progression. Our results also suggest that SCF and APC/C complexes do not contribute to control of cyclin D1 stability. Degradation of cyclin D1 is dependent on its phosphorylation at Thr286, and all four SCF-type ubiquitin ligases examined in the present study were previously shown to mediate cyclin D1 ubiquitylation in a manner dependent on such phosphorylation. A splicing variant of cyclin D1 (cyclin D1b) that lacks the amino acid sequence encoded by exon 5, including Thr286, was found not to differ in stability from canonical cyclin D1 (38), suggesting the existence of a mechanism for cyclin D1 degradation that is not dependent on Thr286 phosphorylation. Furthermore, another pathway for cyclin D1 degradation that is independent of ubiquitylation and Thr286 phosphorylation has been found to operate (29). Collectively, these observations suggest the existence of multiple pathways for the regulation of cyclin D1 stability, and these pathways might contribute differentially to cyclin D1 degradation in a context-dependent manner. Future studies are required to identify bona fide ubiquitin ligases that contribute to cyclin D1 degradation during the normal cell cycle.
ACKNOWLEDGMENTS
This study was supported in part by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by the Global Center of Excellence Program of the Japanese government.
We thank N. Ishida for technical advice, A. Hatano, S. Kotoshiba, and other laboratory members for technical assistance, and A. Ohta for help in preparation of the manuscript.
We declare no conflicts of interest.
Footnotes
Published ahead of print 28 November 2011
REFERENCES
- 1. Agami R, Bernards R. 2000. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell 102:55–66 [DOI] [PubMed] [Google Scholar]
- 2. Alt JR, Cleveland JL, Hannink M, Diehl JA. 2000. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 14:3102–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ang XL, Wade Harper J. 2005. SCF-mediated protein degradation and cell cycle control. Oncogene 24:2860–2870 [DOI] [PubMed] [Google Scholar]
- 4. Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. 1993. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 7:812–821 [DOI] [PubMed] [Google Scholar]
- 5. Barbash O, et al. 2008. Mutations in Fbx4 inhibit dimerization of the SCFFbx4 ligase and contribute to cyclin D1 overexpression in human cancer. Cancer Cell 14:68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bessho Y, et al. 2001. Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 15:2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blain SW. 2008. Switching cyclin D-Cdk4 kinase activity on and off. Cell Cycle 7:892–898 [DOI] [PubMed] [Google Scholar]
- 8. Buckley MF, et al. 1993. Expression and amplification of cyclin genes in human breast cancer. Oncogene 8:2127–2133 [PubMed] [Google Scholar]
- 9. Diehl JA, Cheng M, Roussel MF, Sherr CJ. 1998. Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12:3499–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Diehl JA, Zindy F, Sherr CJ. 1997. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 11:957–972 [DOI] [PubMed] [Google Scholar]
- 11. Fukami-Kobayashi J, Mitsui Y. 1999. Cyclin D1 inhibits cell proliferation through binding to PCNA and cdk2. Exp. Cell Res. 246:338–347 [DOI] [PubMed] [Google Scholar]
- 12. Guo Y, et al. 2005. Phosphorylation of cyclin D1 at Thr 286 during S phase leads to its proteasomal degradation and allows efficient DNA synthesis. Oncogene 24:2599–2612 [DOI] [PubMed] [Google Scholar]
- 13. Ishida N, et al. 2002. Phosphorylation of p27Kip1 on serine 10 is required for its binding to CRM1 and nuclear export. J. Biol. Chem. 277:14355–14358 [DOI] [PubMed] [Google Scholar]
- 14. Kamura T, et al. 2004. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27Kip1 at G1 phase. Nat. Cell Biol. 6:1229–1235 [DOI] [PubMed] [Google Scholar]
- 15. Kim JK, Diehl JA. 2009. Nuclear cyclin D1: an oncogenic driver in human cancer. J. Cell. Physiol. 220:292–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kozar K, et al. 2004. Mouse development and cell proliferation in the absence of D-cyclins. Cell 118:477–491 [DOI] [PubMed] [Google Scholar]
- 17. Lee TH, Perrem K, Harper JW, Lu KP, Zhou XZ. 2006. The F-box protein FBX4 targets PIN2/TRF1 for ubiquitin-mediated degradation and regulates telomere maintenance. J. Biol. Chem. 281:759–768 [DOI] [PubMed] [Google Scholar]
- 18. Lin DI, et al. 2006. Phosphorylation-dependent ubiquitination of cyclin D1 by the SCFFBX4-αB crystallin complex. Mol. Cell 24:355–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Malumbres M, et al. 2004. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell 118:493–504 [DOI] [PubMed] [Google Scholar]
- 20. Matsushime H, et al. 1992. Identification and properties of an atypical catalytic subunit (p34PSK-J3/cdk4) for mammalian D type G1 cyclins. Cell 71:323–334 [DOI] [PubMed] [Google Scholar]
- 21. Matsushime H, et al. 1994. D-type cyclin-dependent kinase activity in mammalian cells. Mol. Cell. Biol. 14:2066–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matsushime H, Roussel MF, Ashmun RA, Sherr CJ. 1991. Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell 65:701–713 [DOI] [PubMed] [Google Scholar]
- 23. Meyerson M, Harlow E. 1994. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol. Cell. Biol. 14:2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Motokura T, et al. 1991. A novel cyclin encoded by a bcl1-linked candidate oncogene. Nature 350:512–515 [DOI] [PubMed] [Google Scholar]
- 25. Nakayama K, et al. 1996. Mice lacking p27Kip1 display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell 85:707–720 [DOI] [PubMed] [Google Scholar]
- 26. Nakayama K, et al. 2000. Targeted disruption of Skp2 results in accumulation of cyclin E and p27Kip1, polyploidy and centrosome overduplication. EMBO J. 19:2069–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nakayama K, et al. 2004. Skp2-mediated degradation of p27 regulates progression into mitosis. Dev. Cell 6:661–672 [DOI] [PubMed] [Google Scholar]
- 28. Nakayama KI, Nakayama K. 2006. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer 6:369–381 [DOI] [PubMed] [Google Scholar]
- 29. Newman RM, et al. 2004. Antizyme targets cyclin D1 for degradation. A novel mechanism for cell growth repression. J. Biol. Chem. 279:41504–41511 [DOI] [PubMed] [Google Scholar]
- 30. Okabe H, et al. 2006. A critical role for FBXW8 and MAPK in cyclin D1 degradation and cancer cell proliferation. PLoS One 1:e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pagano M, Theodoras AM, Tam SW, Draetta GF. 1994. Cyclin D1-mediated inhibition of repair and replicative DNA synthesis in human fibroblasts. Genes Dev. 8:1627–1639 [DOI] [PubMed] [Google Scholar]
- 32. Pawar SA, et al. 2010. C/EBPδ targets cyclin D1 for proteasome-mediated degradation via induction of CDC27/APC3 expression. Proc. Natl. Acad. Sci. U. S. A. 107:9210–9215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pontano LL, et al. 2008. Genotoxic stress-induced cyclin D1 phosphorylation and proteolysis are required for genomic stability. Mol. Cell. Biol. 28:7245–7258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rappsilber J, Ishihama Y, Mann M. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75:663–670 [DOI] [PubMed] [Google Scholar]
- 35. Santra MK, Wajapeyee N, Green MR. 2009. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature 459:722–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shinozaki H, et al. 1996. Cyclin D1 amplification as a new predictive classification for squamous cell carcinoma of the esophagus, adding gene information. Clin. Cancer Res. 2:1155–1161 [PubMed] [Google Scholar]
- 37. Sicinski P, et al. 1995. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell 82:621–630 [DOI] [PubMed] [Google Scholar]
- 38. Solomon DA, et al. 2003. Cyclin D1 splice variants. Differential effects on localization, RB phosphorylation, and cellular transformation. J. Biol. Chem. 278:30339–30347 [DOI] [PubMed] [Google Scholar]
- 39. Tsunematsu R, et al. 2006. Fbxw8 is essential for Cul1-Cul7 complex formation and for placental development. Mol. Cell. Biol. 26:6157–6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang TC, et al. 1994. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature 369:669–671 [DOI] [PubMed] [Google Scholar]
- 41. Wei S, et al. 2008. A novel mechanism by which thiazolidinediones facilitate the proteasomal degradation of cyclin D1 in cancer cells. J. Biol. Chem. 283:26759–26770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu X, et al. 2008. The CUL7 E3 ubiquitin ligase targets insulin receptor substrate 1 for ubiquitin-dependent degradation. Mol. Cell 30:403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang WI, Zukerberg LR, Motokura T, Arnold A, Harris NL. 1994. Cyclin D1 (Bcl-1, PRAD1) protein expression in low-grade B-cell lymphomas and reactive hyperplasia. Am. J. Pathol. 145:86–96 [PMC free article] [PubMed] [Google Scholar]
- 44. Yu ZK, Gervais JL, Zhang H. 1998. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21CIP1/WAF1 and cyclin D proteins. Proc. Natl. Acad. Sci. U. S. A. 95:11324–11329 [DOI] [PMC free article] [PubMed] [Google Scholar]