Abstract
The function of Notch signaling in murine neural crest-derived cell lineages in vivo was examined. Conditional gain (Wnt1Cre;RosaNotch) or loss (Wnt1Cre;RBP-Jf/f) of Notch signaling in neural crest cells (NCCs) in vivo results in craniofacial, cardiac, and trunk abnormalities. Severe craniofacial malformations are apparent in Wnt1Cre;RosaNotch embryos, while less severe skull abnormalities are evident in Wnt1Cre;RBP-Jf/f mice. Deficient cardiac neural crest migration, resulting in cardiac outflow tract malformations, occurs with increased or decreased Notch signaling in NCCs. Smooth muscle cell differentiation also is impaired in pharyngeal NCC derivatives in both Wnt1Cre;RosaNotch and Wnt1Cre;RBP-Jf/f embryos. Neurogenesis is absent and gliogenesis is increased in the dorsal root ganglia of Wnt1Cre;RosaNotch embryos, while neurogenesis is increased and gliogenesis is decreased in Wnt1Cre;RBP-Jf/f embryos. Together, these studies demonstrate essential cell-autonomous roles for appropriate levels of Notch signaling during NCC migration, proliferation, and differentiation with critical implications in craniofacial, cardiac, and neurogenic development and disease.
Keywords: Notch signaling, neural crest, cardiovascular malformation, migration, differentiation, neurocristopathy
Introduction
Neural crest cells (NCCs) are multi-potent progenitor cells that form by inductive interactions at the border of the neural plate and epidermal ectoderm (Knecht and Bronner-Fraser, 2002). After undergoing an epithelial to mesenchymal transition (EMT), NCCs delaminate from the dorsal neural tube and migrate along distinct paths laterally and ventrally in the embryo (Farlie et al., 2004). Cranial NCCs populate the pharyngeal arches and differentiate into bone, cartilage, cranial ganglia, and connective tissue of the head and face (Chai et al., 2000; Trainor, 2005). Cardiac NCCs contribute to the smooth muscle layer of pharyngeal arch arteries (PAA) 3, 4, and 6 and also to the cardiac outflow tract (OFT), where they are required for aorticopulmonary septation (Bergwerff et al., 1999; Waldo et al., 1998; Hutson and Kirby, 2007). NCCs also contribute to the thymus, thyroid, and parathyroid glands of the pharynx, as well as the enteric nervous system of the developing gut and dorsal root ganglia (Hutson and Kirby, 2007; Marmigere and Ernfors, 2007). Dysregulation of NCC development occurs in human neurocristopathy syndromes, including DiGeorge/22q11 deletion syndrome, characterized by cardiac, craniofacial, neurogenic, and thymic malformations, as well as Hirschprung’s disease, a deficiency in enteric nervous system development (Lindsay et al., 2001; Southard-Smith et al., 1998). The molecular mechanisms of NCC determination, migration, proliferation, and differentiation are not fully understood, but several signaling pathways including Wnt, BMP, Sox, Fgf, Shh, and Notch signaling, have been implicated in their development (Heeg-Truesdell and LaBonne, 2004; Sauka-Spengler and Bronner-Fraser, 2008).
The canonical Notch signaling pathway consists of five ligands (Delta ligand 1, 3, 4, Jagged1, 2) that interact with four Notch receptors (Notch1-4) (Gridley, 2003). Upon Notch ligand-receptor binding between neighboring cells, the Notch receptor is cleaved, and the resulting Notch intracellular domain (NICD) translocates to the nucleus. NICD binds to the recombination signal binding protein for immunoglobulin kappa J (RBP-J, also known as Rbpsuh) transcription factor for the activation of canonical Notch pathway responsive genes (Kopan and Ilagan, 2009; Kovall, 2008). NICD generated from any of the four Notch receptors can bind to RBP-J, and loss of RBP-J inhibits canonical Notch signaling from all four Notch receptors in mice (Kato et al., 1996; Kato et al., 1997). In vivo studies of gain or loss of Notch pathway activity have demonstrated a wide variety of developmental functions in many tissues and organ systems. Target genes of activated Notch signaling include critical regulators of cell migration, proliferation, and differentiation (Bolos et al., 2007; Iso et al., 2003).
Specific inhibition of the Notch signaling pathway in NCCs has demonstrated requirements for Notch signaling in vascular smooth muscle differentiation, gliogenesis, and enteric nerve cell maintenance, but the effects of increased Notch signaling in NCCs in vivo have not been reported previously (High et al., 2007; Okamura and Saga, 2008; Taylor et al., 2007). Here we report a comprehensive analysis of the effects of increased or decreased Notch pathway activity on NCC migration, proliferation, and differentiation in vivo. Mice were generated with over-expression (RosaNotch) or inhibition (RBP-Jf/f) of Notch signaling specifically in NCCs with the use of Wnt1Cre (Danielian et al., 1998; Han et al., 2002; Murtaugh et al., 2003). These studies demonstrate Notch pathway regulation of NCC migration, proliferation, and differentiation in multiple derivative cell lineages. In addition, manipulation of Notch signaling in vivo results in cranial, cardiac, and enteric NCC deficiency with characteristic congenital malformations. Therefore, precise levels of Notch signaling are necessary for regulation of NCC-derived cell lineages in multiple tissues and organ systems.
Results
Mice with gain or loss of Notch signaling in neural crest cell lineages do not survive after birth
The Notch signaling pathway is active in NCCs, but the precise roles of Notch signaling in murine NCC migration, proliferation, and differentiation have not been fully defined (Tsarovina et al., 2008; Wakamatsu et al., 2000; Williams et al., 1995; High et al., 2007). Wnt1Cre transgenic mice were utilized to selectively manipulate Notch signaling in NCCs and their derivatives. In these mice, Cre recombinase is expressed in pre-migratory NCCs starting at embryonic day (E)8.0-8.5 in the neural tube, and recombination is apparent in cranial, cardiac, and trunk NC derivatives (Chai et al., 2000; Danielian et al., 1998; Jiang et al., 2000). Notch pathway gain-of-function was achieved with Cre-dependent NICD (Notch1 intracellular domain) expression using the RosaNotch line (Murtaugh et al., 2003). RBP-Jf/f mice were used for loss of canonical Notch signaling from all of the Notch receptors, and specific loss of Notch1 was examined using Notch1fl/fl mice (Cheng et al., 2007; Han et al., 2002). Wnt1Cre;RosaNotch double transgenic embryos were obtained in expected Mendelian ratios through E14.5. However, resorbed embryos were apparent by E14.5, and no viable Wnt1Cre;RosaNotch embryos were recovered at E18.5 (Table 1). Wnt1Cre;RBP-Jf/f and Wnt1Cre;Notch1fl/fl mice were obtained in predicted Mendelian ratios at E18.5, but no live pups with these genotypes were recovered at post-natal day (PND) 1 (Table 1). Therefore increased or decreased Notch pathway activity in NCCs leads to developmental anomalies incompatible with life after birth.
Table 1.
Frequency of genotypes obtained and timing of lethality for mice with altered Notch pathway activation in NCC.
| Expected% | % Embryos (n/total) obtained for indicated genotype | ||||||
|---|---|---|---|---|---|---|---|
| E9.5 | E10.5 | E12.5 | E14.5 | E18.5 | PND1 | ||
| Wnt1Cre;RosaNotch | 50% | 55% (24/44) |
52% (68/130) |
64% (28/44) |
46% (42/92) |
0% (0/16) |
NAa |
| Wnt1 Cre;RBP-f/f | 25% | 13% (8/60) |
37% (21/57) |
14% (7/50) |
11% (3/38) |
22% (19/85) |
0% (0/57) |
| Wnt1Cre;Notchfl/fl | 25% | 33% (11/33) |
30% (8/27) |
25% (7/28) |
21% (18/86) |
23% (15/66) |
0% (0/29) |
NA=not applicable
Increased Notch signaling in neural crest cells results in severe craniofacial malformations, whereas loss of Notch signaling leads to reduced cranial skeletal elements
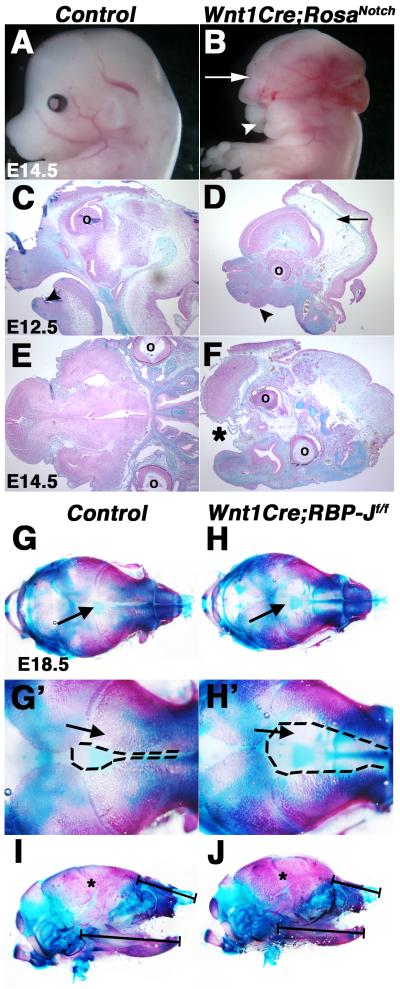
Cranial NCCs are required for craniofacial morphogenesis and contribute to the frontonasal process, palate, jaw, and skull bones, as well as cranial ganglia (Farlie et al., 2004). Craniofacial development was examined in mice with Notch pathway activation or inactivation in NCCs. Wnt1Cre;RosaNotch embryos exhibit neural tube closure defects at E10.5, along with exencephaly and a reduced jaw, resulting in micrognathia, as compared to controls at E12.5-14.5 (Fig. 1A-D; Fig. S1A-B, D-E, G-H; data not shown). Additional defects in Wnt1Cre;RosaNotch embryos include an absent skull vault, reduced frontonasal bones, and shortened nasal cartilage (Fig. 1C-F; data not shown). A severe cleft face and palate also is evident with deficient fusion of the facial midline occurring in 100% of Wnt1Cre;RosaNotch embryos (Fig. S1E, H; data not shown). Furthermore, Wnt1Cre;RosaNotch embryos exhibit reduced expression of NCC migration marker Crabp1, as well as craniofacial differentiation markers Twist1, Sox9, and Msx2, in the frontonasal region and pharyngeal arches (PA) 1 and 2, as determined by whole-mount in situ hybridization compared to controls (Fig. S2A-F, arrows; data not shown). The severe craniofacial defects and reduction of NCC marker expression observed in Wnt1Cre;RosaNotch embryos are indicative of reduced cranial NCC contributions to head structures. Therefore, ectopic expression of NICD in the cranial NC results in severe craniofacial abnormalities in NC-derived structures.
Fig. 1.
Craniofacial malformations result from activation or inactivation of Notch signaling in the neural crest. (A-F) Wnt1Cre;RosaNotch embryos (n=4) exhibit craniofacial malformations, consisting of exencephaly (B, D, arrows), micrognathia (D, arrowhead), cleft face (B, arrowhead) and palate (F, asterisk). Embryos are shown in whole mount (A,B) and in Alcian blue and nuclear fast red counter stained cranial sections (C-F). (G-H’) Alcian blue and Alizarin red stained cranial skeletal preparations of E18.5 Wnt1Cre;RBP-Jf/f embryos exhibit expanded frontal cranial sutures as a result of deficient formation of craniofacial frontal bones apparent in reduced Alizarin red-stained structures, as compared to a wildtype control littermate (arrows) (n=4). (I-J) Mandible size and nasal cartilage are reduced in Wnt1Cre;RBP-Jf/f embryos (n=6), as compared to a wildtype control littermate (n=6). ° denotes eye placement. Dotted lines (G’, H’) represent the area of proximal frontal bones. * indicate apparently normal coronal suture closure (I,J). Black brackets (I, J) denote length of mandible and nasal cartilage.
In contrast to Wnt1Cre;RosaNotch embryos, the craniofacial structures in Wnt1Cre;RBP-Jf/f mice with Notch pathway loss of function appear relatively normal throughout early embryogenesis. No defects in head or face development are apparent at E14.5 (Fig. S1C, F, I). In addition, examination of embryos at E18.5 reveals that all the major cranial and jaw bones are present (Fig. 1G-J). However, there is defective bone formation in the developing craniofacial elements in Wnt1Cre;RBP-Jf/f animals at E18.5. Alcian blue (cartilage) and Alizarin red (bone) stained skeletal preparations of E18.5 Wnt1Cre;RBP-Jf/f heads illustrate craniofacial malformations consisting of widened cranial frontal sutures and reduced frontal bone formation in Wnt1Cre;RBP-Jf/f fetuses, relative to controls (Fig. 1G-H’, arrows). However coronal suture closure is relatively unaffected (Fig. 1I-J, asterisks). In addition, the Wnt1Cre;RBP-Jf/f mandible is reduced in length (10% reduction; n=4; * P ≤ 0.05) and area (20% reduction; * P ≤ 0.01), with accompanying reduction of nasal cartilage (Fig. 1I-J, black bars). In contrast, no craniofacial abnormalities were noted in Wnt1Cre;Notch1fl/fl mice at E18.5, suggesting that multiple Notch ligands are active in these tissues. Together these results demonstrate that loss of Notch signaling in the cranial NC does not affect initial NCC contributions to the head or morphogenesis of craniofacial structures prior to E14.5. However, loss of Notch signaling with deletion of RBP-J in NCC-derived skeletal elements does result in defective frontal cranial suture closure and reduced bone formation in craniofacial skeletal structures apparent at E18.5, consistent with known roles for Notch signaling in cartilage lineage development and osteogenesis (Loomes et al., 2007; Mead and Yutzey, 2009).
Cardiac outflow tract malformations result from increased or decreased Notch signaling in cardiac neural crest
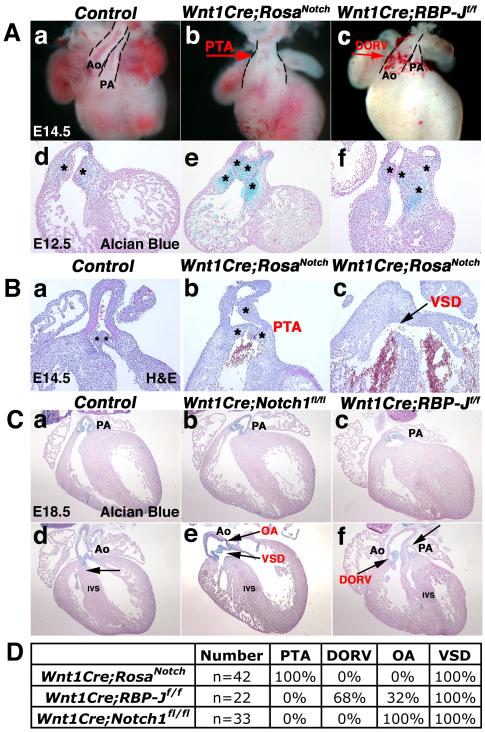
The process of conotruncal septation results in the division of the cardiac OFT into the aorta and pulmonary trunk (Kirby and Waldo, 1995; Le Lievre and Le Douarin, 1975). Cardiac OFT septation and conotruncal alignment were analyzed in mice with increased or decreased Notch signaling in NCC derivatives. As early as E12.5, defective conotruncal development is evident in mice with either gain or loss of Notch pathway function in the cardiac NC. During normal development, the OFT is septated with the pulmonary trunk aligned with the right ventricle and aortic root continuous with the left ventricle. Whole mount and sections of E12.5 and E14.5 Wnt1Cre;RosaNotch hearts illustrate a failure of the OFT to septate. The lack of septation results in a single OFT, known as persistent truncus arteriosus (PTA), with an accompanying ventricular septal defect (VSD), as compared to normal septation observed in the controls (Fig. 2Aa-b; Ba-c). In control embryos at E12.5 and E14.5, two endocardial cushions are visible in a single sagittal section. In contrast, 3-4 individual endocardial cushions are visible in comparable Wnt1Cre;RosaNotch sagittal sections of E12.5 and E14.5 embryos, indicative of abnormal OFT morphogenesis (Fig. 2Ad, e; Ba-b, asterisks). Therefore, increased Notch signaling in the cardiac NC leads to OFT malformations consistent with NC deficiency.
Fig. 2.
Neural crest-specific gain or loss of Notch signaling results in cardiac outflow tract malformations. (A) Wnt1Cre;RosaNotch hearts exhibit defective cardiac OFT septation resulting in PTA (b, red arrow; e). DORV is apparent in Wnt1Cre;RBP-Jf/f hearts (c, red arrow; f) as compared to a wildtype control littermate (a,d). Endocardial cushions are stained with Alcian blue (d-f). (B) Wnt1Cre;RosaNotch hearts exhibit PTA and an accompanied VSD (black arrow) at E14.5 apparent in H&E stained sections as compared to a Wnt1Cre-negative RosaNotch control littermate (a-c). (C) Wnt1Cre;Notch1fl/fl hearts exhibit OA and VSD (e, black arrows), while Wnt1Cre;RBP-Jf/f heart sections exhibit DORV with pulmonary stenosis (f, black arrows), as compared to E18.5 control sections (a,d). The intact IVS is indicated by an arrow in d. Valve leaflets are stained with Alcian blue (a-f). (D) Wnt1Cre;RosaNotch mice exhibit 100% penetrance of PTA and VSD at E14.5. 100% of Wnt1Cre;RBP-Jf/f embryos have OFT malformations with the majority containing DORV and the minority exhibiting OA from E14.5 until birth. Wnt1Cre;Notch1fl/fl mice exhibit 100% OA from E14.5 until birth. Dashed lines outline OFT (A). * Denotes number of heart cushions in section plane (B). Ao, aortic root; PA, pulmonary artery root; PTA, persistent truncus arteriosus; DORV, double outlet right ventricle; VSD, ventricular septal defect; OA, overriding aorta; IVS, interventricular septum. (n=8 for each genotype at each age analyzed)
Cardiac OFT development was additionally investigated in mice with NCC Notch loss-of-function. While the Wnt1Cre;RBP-Jf/f OFT septates to a greater extent than is observed in Wnt1Cre;RosaNotch embryos, OFT malalignment is apparent beginning at E12.5 (Fig. 2Ac, f). A spectrum of OFT defects is observed in Wnt1Cre;RBP-Jf/f embryos, including double outlet right ventricle (DORV) with pulmonary stenosis, in which the great vessels are transposed and align over the right ventricle (Fig. 2Ac, f; Fig. 2Cc, f). Wnt1Cre;Notch1fl/fl mice with specific loss of Notch1 in the NC exhibit less severe OFT malformations, including overriding aorta (OA) in which the aorta is aligned over the interventricular septum resulting in a membranous VSD, apparent as early as E14.5 (Fig. 2Cb,e, arrows, data not shown). These conotruncal malalignment and ventricular septal defects could be due to indirect effects of NC-deficiency on secondary heart field derivatives (Hutson and Kirby, 2007). Thus NC-related cardiac OFT defects occur in 100% of the mice with increased or decreased Notch signaling in the NC; however the severity of the malformations is dependent on the specific signaling alteration (Fig. 2D).
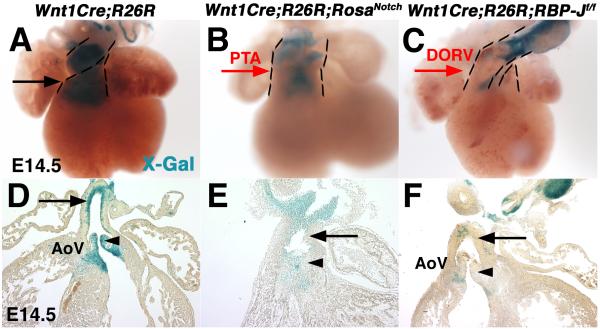
NCC contributions to the cardiac OFT were examined in mice with gain or loss of Notch signaling in NCCs. To monitor the presence of NCC derivatives in the cardiac OFT, ROSA26R (R26R) Cre-dependent LacZ reporter mice were bred into Wnt1Cre;RosaNotch and Wnt1Cre;RBP-Jf/f backgrounds (Soriano, 1999). In both Wnt1Cre;R26R;RosaNotch and Wnt1Cre;R26R;RBP-Jf/f embryos at E14.5, there is a paucity of NCCs present in the heart, apparent as a significant decrease of β-Gal positive cells in the OFT and semilunar valves, as compared to Wnt1Cre;R26R controls (Fig. 3A-F). This deficiency of NC-derived cells is consistent with cardiac OFT malformations observed in mice with increased or decreased Notch signaling in the NC.
Fig. 3.
The presence of cardiac neural crest cell derivatives is reduced in the developing OFT in mice with gain or loss of Notch signaling in the NCC associated with congenital cardiac malformations. Expression of β-gal from the R26R allele was detected by blue X-gal staining of isolated hearts. Wnt1Cre;R26R;RosaNotch (B,E) and Wnt1Cre;R26R;RBP-Jf/f (C,F) embryos have reduced presence of neural crest-derived cells in the cardiac outflow tract and aortic valves at E14.5, as compared to Wnt1Cre;R26R controls (A, D, arrows, arrowheads). Dashed lines outline the outflow tract. PTA, persistent truncus arteriosus; DORV, double outlet right ventricle; AoV, aortic valve. (n=6 for each genotype at each age analyzed)
Neural crest cell migration is aberrant with Notch pathway gain- or loss-of-function
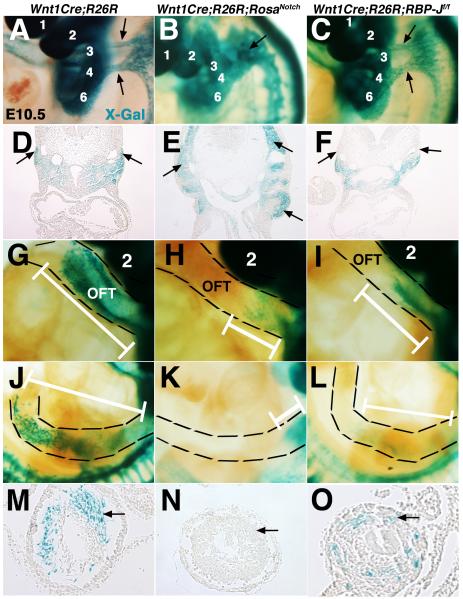
Decreased NCC-derivatives are apparent in the cardiac OFT structures at E14.5 in mice with increased or decreased Notch signaling in NCCs (Fig. 3). Embryos were examined at E10.5 in order to determine if alterations in Notch signaling levels affect NCC migration into the pharyngeal arches (PAs) and more distal cardiac and enteric structures. Cardiac NCCs migrate towards PA 3 and 4 in two distinct streams that are apparent in E10.5 Wnt1Cre;R26R lineage reporter control embryos (Fig. 4A, D). However, in Wnt1Cre;R26R;RosaNotch embryos, NCC-derivatives are present in one large solid region of β-Gal positive NCCs around the PAs (Fig. 4B, E). In addition, there is a severe deficiency of NCCs present in the OFT, and the distance of NCC migration into the OFT is decreased by 48% in Wnt1Cre;R26R;RosaNotch embryos, as compared to controls (Fig. 4G-H, Fig. S3A). In Wnt1Cre;R26R;RBP-Jf/f embryos, NCCs are present in two distinct streams of β-Gal positive cells as they migrate into PAs 3-4, similar to control embryos (Fig. 4A, C, black arrows). However, the NCC migration distance into the OFT is significantly decreased, as compared to controls (Fig. 4I, G, white bars, Fig. S3A), and later contribution to OFT structures is obviously reduced (Fig. 3C, F). Therefore, Wnt1Cre;R26R;RosaNotch embryos have aberrant migration paths of NCCs, as well as decreased contributions to the cardiac OFT. In contrast, Wnt1Cre;R26R;RBP-Jf/f embryos have apparently normal NCC migration trajectories, but also have reduced NCC contribution in the cardiac OFT. Therefore, increased or decreased Notch signaling in NCCs leads to deficient NC contribution to the developing cardiac OFT.
Fig. 4.
Cardiac and trunk neural crest cell migration is defective in mice with gain or loss of Notch signaling in neural crest derivatives. (A-F) X-Gal-stained E10.5 embryos and sections illustrate abnormal NCC migration into PA3 and 4 of Wnt1Cre;R26R;RosaNotch embryos as compared to two streams of migrating cardiac NCCs in the Wnt1Cre;R26R control (A, B, D, E, arrows). (G-I) Reduced cardiac NCC contribution is apparent in the Wnt1Cre;RosaNotch and Wnt1Cre;RBP-Jf/f OFT, as compared to Cre-negative control littermates. (J-O) Reduction of NCC contribution in the enteric nervous system of the gut is apparent in Wnt1Cre;RosaNotch and Wnt1Cre;RBP-Jf/f embryos (arrows). OFT, outflow tract. White brackets show migration distance. Dashed lines outline the OFT (G-I) and enteric nervous system of the midgut (J-L). PAs are labeled numerically in panels A-C and G-I. (n=6 for each genotype at each age analyzed)
NCCs arising at the level of somites 1-7 contribute to the enteric nervous system (ENS) consisting of neurons and glia that innervate the gastrointestinal tract (Farlie et al., 2004). As compared to control embryos, the midgut ENS is almost completely lacking NCCs in Wnt1Cre;R26R;RosaNotch embryos (Fig. 4J-O, Fig. S3B). Likewise, NCC migration into the midgut ENS also is significantly reduced in Wnt1Cre;R26R;RBP-Jf/f embryos, as measured by β-Gal positive NCCs (Fig. 4L,O, Fig. S3B). Therefore, gain or loss of Notch signaling in the NC results in NCC-deficiency in the ENS, similar to that observed in the cardiac OFT.
Notch signaling in NCCs regulates pharyngeal arch artery development and smooth muscle cell differentiation
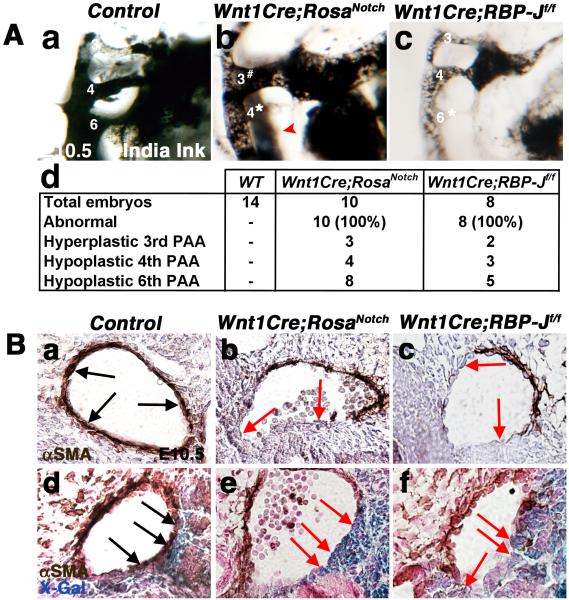
Pharyngeal arch artery (PAA) malformations are characteristic of NC deficiency in mouse models and human syndromes (Lindsay et al., 2001; Porras and Brown, 2008; Wurdak et al., 2005). Cardiac NCCs invest PAAs 3, 4, and 6, providing growth support via differentiation into PAA tunica media smooth muscle cells (Kirby and Waldo, 1995). To visualize and evaluate the growth and remodeling of PAAs, intracardial ink injections were performed in E10.5 embryos (McCright et al., 2002). PAA malformations observed in embryos with increased or decreased Notch signaling in the NC include hyperplastic 3rd, hypoplastic and non-patent 4th, as well as hypoplastic 6th PAA (Fig. 5Aa-c). PAA defects were noted in 100% of the embryos with either gain or loss of Notch signaling in NCC (Fig. 5Ad). Most, but not all, embryos had more than one PAA anomaly, with hypoplasia or aplasia of the 6th PAA being the most frequent, but the combinations of malformations observed were variable. Taken together, these studies demonstrate that canonical Notch signaling in NCCs is required for normal PAA development. In addition, increased Notch signaling affects the development of these NC-derived structures, resulting in PAA anomalies similar to those observed with decreased Notch signaling.
Fig. 5.
Pharyngeal arch artery malformations and deficient smooth muscle cell differentiation are evident in E10.5 Wnt1Cre;RosaNotch and Wnt1Cre;RBP-Jf/f embryos. (A) Ink injected intracardially into E10.5 embryos illustrates pharyngeal arch artery (PAA) defects including hyperplastic 3rd PAA, hypoplastic 4th PAA, and hypoplasia of the 6th PAA (red arrowhead) in Wnt1Cre;RosaNotch embryos (b). Hypoplasia of the 6th PAA is apparent in Wnt1Cre;RBP-Jf/f hearts (c), as compared to Cre-negative control littermates (a). The frequency and spectrum of PAA defects in Wnt1Cre;RosaNotch and Wnt1Cre;RBP-Jf/f embryos with number of embryos analyzed is shown in (d). All of the mutant embryos were affected with one or more PAA abnormality and the incidence of specific PAA malformations is indicated. (B) Decreased PAA4 anti-αSMA reactivity is apparent in Wnt1Cre;RosaNotch and Wnt1Cre;RBP-Jf/f embryos, as compared to controls (a-c, arrows). Double labeling of X-Gal labeled NCCs and anti-αSMA illustrates a lack of NC-derived smooth muscle cell differentiation in PAA3, as compared to controls (d-f, arrows). # increased PAA size; * reduced PAA size; hypo, hypoplastic. (n=6 for each genotype)
Cardiac NCCs differentiate into smooth muscle cells that surround the developing PAAs to provide stability and support for proper blood flow in the vessels (Bergwerff et al., 1999; High et al., 2007; Jiang et al., 2000). Therefore, we examined PAA smooth muscle cell differentiation by immunostaining for α smooth muscle actin (SMA) in embryos with gain or loss of Notch signaling in NCCs. In E10.5 control embryos, NCC derivatives differentiate into smooth muscle cells surrounding the PAAs apparent in a multi-cell layer of concentric and organized SMA expression (Fig. 5Ba, arrows). However, in both Wnt1Cre;RosaNotch and Wnt1Cre;RBP-Jf/f embryos, SMA staining is observed only partially around PA4 (Fig. 5Bb-c, arrows). Deficient smooth muscle cell differentiation is also apparent in Wnt1Cre;RosaNotch and Wnt1Cre;RBP-Jf/f PAAs in E12.5 sections, as compared to controls (data not shown). Co-staining for β-Gal and SMA in Wnt1Cre;R26R;RosaNotch and Wnt1Cre;R26R;RBP-Jf/f embryos demonstrates that β-Gal-positive NCCs are present in the PAA, but these cells do not express SMA (Fig. 5Bd-f). In addition, not all of the smooth muscle cells of the PAA are NC-derived, and the cells that do not express β-Gal in Wnt1Cre;R26R;RosaNotch and Wnt1Cre;R26R;RBP-Jf/f embryos do express SMA. Therefore, NCCs with increased or decreased Notch signaling, that have properly migrated to the PAAs, do not differentiate into smooth muscle cells. These results demonstrate a cell-autonomous role for Notch signaling in controlling NCC-specific smooth muscle differentiation in the PAA. Importantly, too much or too little Notch signaling in NCCs inhibits smooth muscle cell differentiation in vivo.
Notch signaling promotes neural crest cell proliferation
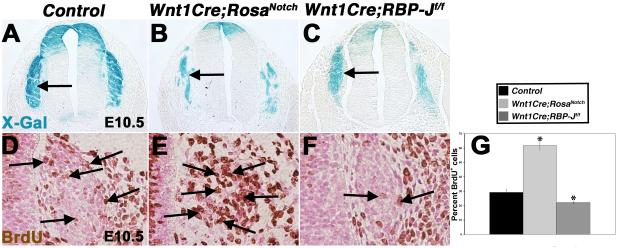
NCCs migrate to discrete locations in the embryo where they proliferate prior to NCC differentiation (Murphy et al., 1994; Nagy and Goldstein, 2006). To investigate if altered Notch pathway activation affects NCC proliferation, immunohistochemistry for BrdU incorporation was performed on Wnt1Cre;R26R;RosaNotch and Wnt1Cre;R26R;RBP-Jf/f embryos at E10.5. At this stage, the dorsal root ganglia (DRG), derived from trunk NC, are apparent lateral to the neural tube (Fig. 6A-C). Wnt1Cre;RosaNotch mice have a 2-fold increase in proliferation of cells in the DRG, while Wnt1Cre;RBP-Jf/f DRG have a significant decrease in proliferation, as determined by BrdU staining compared to control (Fig. 6A-G, arrows). Cell death also was investigated as a potential contributing factor to the NCC deficiency observed in Wnt1Cre;RosaNotch and Wnt1Cre;RBP-Jf/f embryos. Apoptosis, as determined by anti-cleaved caspase 3 antibody reactivity, revealed no change in cell death with either gain or loss of Notch signaling in the NC, as compared to controls (Fig. S4). Therefore, Notch signaling promotes cell proliferation, and loss of Notch signaling results in decreased proliferation of NCCs in the DRG.
Fig. 6.
Notch signaling promotes cell proliferation of neural crest derivatives. (A-C) The DRG (arrows) are NCC-derived structures, as evident by X-Gal staining of Wnt1Cre;R26R, Wnt1Cre;R26R;RosaNotch, and Wnt1Cre;R26R;RBP-Jf/f E10.5 sections. (n=8 for each genotype) (D-G) Notch activation results in increased NCC proliferation, while Notch pathway inactivation results in significant reduction of NCC proliferation in the DRG, as determined by BrdU incorporation (arrows), compared to Wnt1Cre-negative control littermates. (n=6 for each genotype) * p ≤ 0.01
Notch signaling is necessary and sufficient to promote the glial cell fate, while repressing the neuronal cell fate, in cranial and dorsal root ganglia
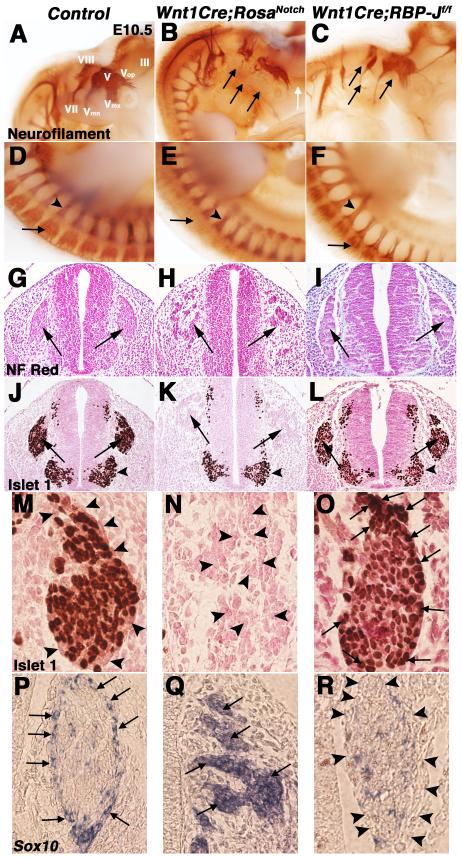
NCCs of the peripheral nervous system contribute to glial cells and proximal neurons of the cranial and dorsal root ganglia (D’Amico-Martel and Noden, 1983; Le Douarin and Smith, 1988). The distribution and differentiation of NC-derived neurons in cranial ganglia and DRG was examined at E10.5 in embryos with increased or decreased Notch pathway activity. Whole-mount immunohistochemistry with anti-neurofilament 2H3 antibody was used to detect differentiated neurons (Swiatek and Gridley, 1993). Wnt1Cre;RosaNotch embryos exhibit defective cranial gangliogenesis, including severely reduced trigeminal (V), geniculate (VII), and vestibulo/cochlear (VIII) ganglia, with decreased neural projections, as compared to controls (Fig. 7A-B, arrows). The mandibular (Vmn) and maxillary (Vmx) segments of the trigeminal ganglia in Wnt1Cre;RosaNotch embryos are absent, as is the oculomotor nerve (III), as compared to controls (Fig. 7A-B, arrows). Likewise, Wnt1Cre;RosaNotch embryos lack anti-neurofilament antibody staining in the DRG, indicative of deficient neuronal differentiation (Fig. 7D-E, arrows). In Wnt1Cre;RBP-Jf/f embryos, the cranial ganglia appear to form normally, but the size of distal neural projections is reduced (Fig. 7C, arrows), consistent with a previous report (Hu et al., 2011). Together, these data demonstrate that gain of Notch signaling in the NC results in hypoplasia and mispatterning of cranial ganglia, as well as decreased neuronal differentiation in the DRG. In contrast, patterning and neuronal differentiation of cranial ganglia and DRG in embryos with loss of Notch signaling in the NC is relatively normal, but specific neuronal elements are reduced in size.
Fig. 7.
Notch signaling regulates neuronal and glial cell fates in cranial and dorsal root gangliogenesis. (A-F) Whole-mount anti-neurofilament 2H3 staining illustrates defective and reduced V, VII, and VIII cranial ganglia branching (black arrows) and loss of III formation (white arrow) in a Wnt1Cre;RosaNotch embryo (B) compared to a Wnt1Cre-negative RosaNotch control littermate (A). Wnt1Cre;RosaNotch embryos also lack dorsal root ganglia (DRG) neurofilament staining (arrow in E) at E10.5. Neurofilament patterning appears normal in Wnt1Cre;RBP-Jf/f embryos but specific neuronal projections appear to be reduced in size (C, F, arrows), as compared to wildtype littermate controls (A, D). (n=5 embryos were analyzed for each genotype) Arrowheads in D-F note spinal nerve neurofilament marker expression. (G-I) Nuclear fast red stained sections of E10.5 Wnt1Cre;RosaNotch embryos demonstrate defective DRG morphogenesis (H, arrows), while Wnt1Cre;RBP-Jf/f DRG appear normal (I, arrows). (J-L) Lack of neurogenic marker Islet1 protein expression is apparent in Wnt1Cre;RosaNotch DRG (K, arrows). Arrowheads in J-L denote Islet1 expression in non-NC derived motor neurons. (M-R) 60x objective magnification of control DRG show Islet1-positive cells in the center of the DRG with Sox10-expressing glial cells surrounding (M, P, arrowheads). Wnt1Cre;RosaNotch DRG lack Islet1 (N, arrowheads), but have increased Sox10 expression in the DRG (Q, arrows). Wnt1Cre;RBP-Jf/f DRG have reduced Sox10 (R, arrowheads) and increased Islet1 expression (O, arrows) in the DRG outer cell layer. (n=6 embryos were analyzed for each genotype)
The NC-derived DRG includes progenitors of neurons and glial cells that differentiate from a common NCC-progenitor pool (Stemple and Anderson, 1992). Condensed DRG are apparent in control and Wnt1Cre;RBP-Jf/f embryos at E10.5, but the DRG of Wnt1Cre;RosaNotch embryos are disorganized with fragmented cellular condensations (Fig. 7G-I, arrows). Neuronal progenitors in the DRG express Islet1 in DRG condensations in control and Wnt1Cre;RBP-Jf/f embryos (Fig. 7J, L, arrows). In contrast, there is essentially no Islet1 antibody reactivity in the fragmented Wnt1Cre;RosaNotch DRG (Fig. 7K, arrows). Therefore, increased Notch signaling results in a lack of neurons and abnormal DRG morphogenesis.
Sox10 expression is essential for the generation of glial cells in the DRG, and glial progenitors expressing Sox10 normally surround the Islet1-positive neuronal DRG cells (Fig. 7M, P) (Britsch et al., 2001). In Wnt1Cre;RosaNotch embryos, the DRG exhibit a dramatic increase in glial cells apparent in increased Sox10 expression, accompanying the essential absence of neurons, evident in loss of Islet1 expression (Fig. 7M-N, P-Q). In contrast, the Wnt1Cre;RBP-Jf/f DRG contain fewer Sox10 expressing cells surrounding the Islet1 positive DRG cells, indicative of fewer supporting glial cells as compared to controls (Fig. 7M, P, O, R, arrows and arrowheads). However, there is no reduction in the total number of DRG cells in Wnt1Cre;RBP-Jf/f embryos relative to controls (data not shown). Analysis of neuronal progenitors demonstrates that there are additional Islet1-positive cells on the periphery of the Wnt1Cre;RBP-Jf/f DRG, where the Sox10-positive glial cells are normally present (Fig. 7O, arrows). Therefore, there is an increase of neuronal cells and an accompanied decrease of glial cells in the Wnt1Cre;RBP-Jf/f DRG, as compared to controls (Fig. 7M, O, P, R), consistent with a previous report (Hu et al., 2011). Likewise, increased Sox10-expressing glial cells are apparent in Wnt1Cre;RosaNotch cranial ganglia, while Sox10 expression is reduced in Wnt1Cre;RBP-Jf/f mice cranial ganglia, as compared to controls (data not shown). Therefore, activation of Notch signaling in cranial ganglia and DRG results in decreased neuronal and increased glial cells, while loss of Notch signaling results in a reduction of glial cells with an increase of neuronal cells.
Discussion
Here we demonstrate that the Notch signaling pathway has multiple critical roles in NCC migration, proliferation, differentiation, and cell fate determination in vivo. Cre-mediated gain or loss of Notch signaling in NCCs and their derivatives leads to craniofacial, cardiovascular, and neurogenic abnormalities that are reminiscent of NC ablation models and pathological presentation of NC-related diseases including DiGeorge syndrome (High and Epstein, 2008; Kirby et al., 1983; Lindsay et al., 2001; Porras and Brown, 2008). While gain or loss of Notch signaling in NCCs results in cranial, cardiac, and trunk abnormalities, the mechanisms of the malformations differ. Gain of Notch signaling in NCCs results in abnormal and deficient cell migration, increased cell proliferation, decreased PAA smooth muscle cell differentiation, and promotion of a glial cell fate in cranial ganglia and DRG. Loss of Notch signaling in NCCs results in decreased cell proliferation, deficient migration, decreased PAA smooth muscle differentiation, and promotion of a neuronal cell fate, with accompanying loss of glial cells, in the DRG. Together, these studies demonstrate multiple functions for proper levels of Notch signaling in NCCs during cranial, cardiac, and trunk development.
Manipulation of Notch signaling in NCCs leads to craniofacial malformations; however, there is remarkably little evidence for the necessity for endogenous Notch signaling in cranial NCC migration or initial contributions to developing head structures. Gain of Notch signaling in NCCs results in severe craniofacial malformations including cleft face and palate, exencephaly, and micrognathia as a result of deficient cranial NCC migration and lack of differentiation. In striking contrast, the craniofacial malformations in Wnt1Cre;RBP-Jf/f mice are much less severe with apparently normal development up to E14.5. In addition, no craniofacial abnormalities were noted in Wnt1Cre;Notch1fl/fl embryos. Therefore, canonical Notch signaling is not required for cranial NCC migration or initial contributions to the skull and face, consistent with reported lack of defective craniofacial development in Xenopus or zebrafish embryos with altered Notch signaling in NCCs (Cornell and Eisen, 2005). The increased severity of craniofacial defects in the Wnt1Cre;RosaNotch relative to Wnt1Cre;RBP-Jf/f embryos could be due to signaling through a RBP-J-independent noncanonical pathway, but this signaling mechanism has not yet been implicated in embryonic neural crest development (D’Souza, et al., 2008). At later stages of development, loss of Notch signaling affects cranial frontal suture formation as well as frontal bone formation. These defects are likely due to known requirements for Notch signaling in proliferation and differentiation of osteochondro progenitors (Dong et al., 2010; Mead and Yutzey, 2009; Zanotti and Canalis, 2010). Together, these studies demonstrate that endogenous canonical Notch signaling has a relatively minor role in cranial NCC migration and differentiation in vivo, in contrast to more severe malformations observed in cardiac and trunk NC derivatives.
Cardiac NCCs contribute to the septation and proper alignment of the cardiac OFT. Increased Notch pathway activation in NCCs results in a lack of OFT septation evident in PTA and embryonic lethality at approximately E14.5. Embryos with loss of RBP-J or specific loss of Notch1 in the NC have less severe cardiac malformations and are viable throughout gestation. However, these mice do not survive after birth likely due to insufficient cardiovascular function associated with conotruncal anomalies and ventricular septal defects prevalent in these animals. Fetuses with loss of RBP-J in NCCs exhibit DORV and OA, whereas loss of Notch1 leads to only OA. The reduced severity of Wnt1Cre;Notch1fl/fl relative to Wnt1Cre;RBP-Jf/f OFT malformations suggests that other Notch receptors, potentially Notch2, or noncanonical ligands could be involved in cardiac NCC regulation (Varadkar et al., 2008; D’Souza et al., 2008). Therefore, precise levels of Notch signaling are required in NCC for conotruncal septation and alignment of the developing OFT.
Gain or loss of Notch signaling results in deficient NCC migration and contribution to the cardiac OFT and ENS. A previous study reported that loss of Notch signaling in the NCCs, utilizing a dominant-negative Notch pathway co-activator (Wnt1Cre;DNMAML), results in cardiovascular defects, predominantly pulmonary stenosis (High et al., 2007). However, no defects in NCC migration were observed in these mice, and they were viable after birth. In the current study, decreased NCC migration in Wnt1Cre;RBP-Jf/f embryos is associated with a range of OFT alignment malformations, including DORV, and these mice do not survive after birth. Therefore, the loss of RBP-J results in a more severe NC deficiency than the expression of DNMAML in these analyses. In addition, the trajectory of NCC migration is affected by induction of Notch signaling. Normally, the NCCs migrate into the PAs in two predominant streams, however, in the PAs of Wnt1Cre;RosaNotch animals, migrating NCCs do not organize into distinct streams and are present in greater numbers. Likewise, NCC migration into the cardiac OFT and ENS is deficient in embryos with increased Notch signaling in NCCs. These data support a previous study suggesting that Notch signaling is required for proper migration of enteric NCCs (Okamura and Saga, 2008). Therefore, it would be of interest to examine the role of Notch signaling in Hirschsprung disease, characterized by a lack of NCC-derived ganglia in the gut (Southard-Smith et al., 1998). Together, these data indicate that proper levels of Notch signaling are required for normal NCC migration and cellular contributions to the heart and ENS.
In the DRG, Notch activation in NCCs results in increased proliferation, while loss of Notch signaling results in decreased NCC proliferation. These results point to a cell autonomous role for Notch signaling in promoting cell proliferation of NC derivative cell types. Similarly, Notch pathway activation through forced expression of NICD promotes cell proliferation, and Notch inhibition reduces cell proliferation in cultured quail NCCs (Wakamatsu et al., 2000). In many cases, increased Notch signaling maintains multi-potential progenitor cells in a proliferative, undifferentiated state, as is apparent in Wnt1Cre;RosaNotch NCCs (Hansson et al., 2004; Hurlbut et al., 2007). In some cell types, increased Notch signaling leads to cell death, but we observed no change in apoptosis as a result of manipulation of Notch signaling in NCCs. In Wnt1Cre;RBP-Jf/f mice, it is likely that decreased NCC proliferation results in an overall decreased number of NCCs, leading to characteristic cardiac and pharyngeal malformations associated with NCC deficiency. In contrast, increased Notch signaling results in increased proliferation and more cells adjacent to the neural tube and PAAs. However, these cells have deficient migration and contribution to the cardiac OFT and ENS that is the likely cause of malformations characteristic of NC deficiency in Wnt1Cre;RosaNotch animals.
Terminal NCC differentiation occurs as cells reach their target destination. Smooth muscle differentiation of NCCs is necessary to stabilize the PAAs and, in the absence of smooth muscle, the PAAs are rapidly destabilized and become hypo- or aplastic (Waldo et al., 1996). Here we show that gain or loss of Notch signaling in cardiac NCCs results in decreased smooth muscle cell differentiation of NC derivative cells in multiple PAAs. This is the first demonstration that high levels of activated Notch signaling in NC derivatives inhibit smooth muscle differentiation in the PAAs by a cell autonomous mechanism in vivo. This inhibition could occur through Notch induction of Hairy/Enhancer-of-split related with YRPW motif (Hey) transcription factors that have been shown to inhibit myocardin in smooth muscle cell cultures (Gridley, 2007; Proweller et al., 2005; Tang et al., 2008). Loss of RBP-J also leads to decreased NC-derived smooth muscle differentiation in the PAAs, consistent with cell culture studies demonstrating that Notch induction of RBP-J transcriptional activity promotes expression of smooth muscle differentiation genes (Tang et al., 2010). The observed decrease in smooth muscle differentiation with NCC loss of RBP-J is consistent with a previous study in which inhibition of Notch signaling via DNMAML in Pax3Cre mice results in decreased smooth muscle gene expression in a subset of the PAAs (High et al., 2007). Taken together, these studies demonstrate that high levels of Notch signaling inhibit smooth muscle differentiation, while loss of Notch signaling also leads to reduced smooth muscle cell differentiation in vivo. Therefore, precise levels of Notch signaling in cardiac NCCs are required for PAA smooth muscle cell differentiation in post-migratory NCCs.
NCC derivatives contribute to glial and neuronal cells of the peripheral nervous system, including those in the cranial ganglia and DRG, via a binary cell fate decision (Taylor et al., 2007). Here, we report that Notch signaling is necessary and sufficient for the generation of glial cells in cranial ganglia and DRG. In addition, we observe that Notch signaling inhibits neurogenesis, as indicated by loss of neuronal differentiation in mice with increased Notch activity in NCC-derivatives. Likewise, there is an apparent conversion of glial progenitors to a neuronal cell fate with loss of Notch function. The reduction in glial cells in Wnt1Cre;RBP-Jf/f mice suggests that endogenous canonical Notch signaling is required to promote gliogenesis in the DRG, consistent previous reports (Taylor et al., 2007; Hu et al., 2011). In cultured avian NCCs, Notch activation suppresses neurogenic differentiation of NCCs and accelerates glial differentiation via promotion of Sox10 expression, but this has not been demonstrated previously in vivo (Lassiter et al., 2010; Morrison et al., 2000; Wakamatsu et al., 2000). Our data demonstrate that increased Notch signaling in NCCs promotes gliogenesis through the promotion of Sox10, while inhibiting neurogenesis in cranial ganglia and DRG, in vivo in mice. This is the first in vivo demonstration in a mammalian system of an apparent cell fate conversion of DRG progenitors regulated by increased Notch pathway activity. Together, these studies support a mechanism whereby Notch signaling controls the cell fate of murine NCC-derived DRG cells through the promotion of gliogenesis, while inhibiting neurogenesis, in vivo.
The comparison of Notch pathway over-expression with loss of endogenous Notch signaling elucidates the multiple roles of Notch signaling in NCC development. We report that Notch signaling regulates NCC migration, proliferation, differentiation, and cell fate decisions in mammalian NC, and that perturbations in these processes result in malformations characteristic of neurocristopathies. Congenital defects associated with DiGeorge Syndrome, including craniofacial, cardiac OFT, and thymic malformations (data not shown), are all observed in mice with gain or loss of Notch signaling in the NC (Lindsay et al., 2001). Related smooth muscle cell deficiencies also occur in human CADASIL and Alagille Syndromes that result from Notch pathway gene mutations (Gridley, 2003). Therefore, Notch signaling may have an underappreciated role in human neurocristopathy syndromes, and future studies of the specific requirements of Notch signaling in the NC could have potential therapeutic applications.
Experimental Procedures
Mouse strains
Wnt1Cre (Danielian et al., 1998), RosaNotch (Murtaugh et al., 2003), RBP-Jf/f (Han et al., 2002), Notch1fl/fl (Cheng et al., 2007), and Rosa26R (Soriano, 1999) mouse lines and genotyping have been described. RBP-Jf/f mice were obtained with permission from the Riken BioResource Center, Kyoto. Wnt1Cre transgenic mice were interbred with RosaNotch, RBP-Jf/f, or Notch1fl/fl mice to obtain Wnt1Cre;RosaNotch, Wnt1Cre;RBP-Jf/f, and Wnt1Cre;Notch1fl/fl mice. Rosa26R mice were utilized to monitor NCC expression. Control mice used in these studies are wild type, RosaNotch, or RBP-Jf/+ littermates without the Wnt1Cre transgene. E0.5 was designated by the morning presence of a vaginal plug. Animals were maintained in accordance with the National Institutes of Health Guide for the Care and Use Laboratory Animals, and were used according to Institutional Animal Care and Use Committee approved protocols.
Skeletal preparation and histology
Whole mount skeletal preparations were stained with Alcian blue 8GX (Sigma-Aldrich) and Alizarin red S (Sigma-Aldrich) as previously described (Kuczuk and Scott, 1984; Mead and Yutzey, 2009).
In situ hybridization
The generation of Sox9, Twist1, Sox10, and Crabp1 RNA probes has been described (Dixon et al., 2006; Ma et al., 2005; Mead and Yutzey, 2009; Ruberte et al., 1991). In situ hybridizations of whole embryos or histological sections were performed as previously described (Lincoln et al., 2004; Shelton and Yutzey, 2007).
β-Galactosidase detection and calculation of neural crest lineage migration distance
Whole-mount X-Gal staining for β-Galactosidase (β-Gal) activity was performed on Wnt1Cre;R26R embryos as described with modifications (Lincoln et al., 2004; Sanes et al., 1986). Whole embryos were developed in staining solution in the dark for 2 hours at 37°C, fixed in 4% PFA in PBS, and washed in methanol, isopropanol, and tetrahydronaphthalene (Fisher Scientific) to clear the embryo. After whole-mount photography, embryos were paraffin embedded, sectioned at 10μm, dehydrated, and mounted in Cytoseal 60 (Richard-Allen Scientific). Neural crest migration distance in the cardiac OFT was calculated as the linear extent of X-Gal staining in the OFT/total length of the OFT for each genotype using ImageJ analysis of photomicrographs of whole embryos. There were no statistically significant differences in the total length of the OFT among genotypes. Neural crest migration distance in the gut was similarly calculated as the length of X-gal staining in the gut/total length of the gut using Image J analysis.
Intracardial ink injections
India Ink was injected to visualize the cardiac OFT and PAAs, as described with modifications (McCright et al., 2002). Briefly, 1:1 PBS / India Ink (American MasterTech) was injected into the left ventricle of E10.5 embryos using a Pico-Injector (Harvard Apparatus) with a microinjection needle. The embryos were fixed in 4% PFA /PBS overnight, dehydrated in ethanol, and cleared in 1:1 benzyl benzoate (Acros Organics) / methyl salicylate (Acros Organics). Pharyngeal arch artery malformations were assessed photographically (Macatee et al., 2003).
Immunohistochemistry
Immunohistochemical staining on sections was performed as described previously (Mead and Yutzey, 2009). Sections were processed with an ultrasensistive ABC IgG Staining Kit (Pierce) and visualized with diaminobenzidine (DAB) staining. Antisera used include, anti-cleaved caspase-3 (1:100; Cell Signaling), anti-α smooth muscle actin (Sigma), and anti-Islet1 (1:50; 39.4D5 Developmental Studies Hybridoma Bank). Monoclonal anti-neurofilament antibody 2H3 (1:1000; Developmental Studies Hybridoma Bank) was utilized for whole-mount immunohistochemistry as described (Swiatek and Gridley, 1993). Alcian blue staining of histological sections was performed as previously described (Mead and Yutzey, 2009).
BrdU labeling and calculation of proliferative indices
Pregnant mice were injected intraperitoneally with 5-bromo-2-deoxyuridine (BrdU) labeling reagent (Invitrogen) at 0.1ml /10g body weight and sacrificed 2 hours later. BrdU staining was performed following the manufacturer’s protocol (Invitrogen BrdU staining kit). The proliferation indices of experimental and control sections were determined from photomicrographs as percent BrdU positive nuclei / total number of nuclei.
Statistical Analysis
Statistical significance was determined by Student’s t-test. Data are reported as mean with Standard Error of the Mean (SEM). p ≤ 0.01
Supplementary Material
Acknowledgments
We thank Doug Melton (RosaNotch), Riken BioResource Center, Kyoto (RBP-Jf/f), and Raphael Kopan (Notch1fl/fl) for mice used in this study; Kristin Melton (Crabp1 and Sox10) and Jim Martin (Twist1) for RNA probes; Kristin Melton, Ashley Cast and Pauline Parisot for technical assistance; and Kenneth Campbell, Nadean Brown, Ron Waclaw, Yutaka Yamanaka, Samantha Brugmann, and Robert Hinton for helpful discussions. This work was supported by NIH R01 HL094319 (KEY) and AHA-Great Rivers Affiliate Predoctoral Fellowship 09PRE2060551 (TJM).
References
- Bergwerff M, DeRuiter MC, Hall S, Poelmann RE, Gittenberger-de Groot AC. Unique vascular morphology of the fourth aortic arches: possible implications for pathogenesis of type-B aortic arch interruption and anomalous right subclavian artery. Cardiovasc. Res. 1999;44:185–196. doi: 10.1016/s0008-6363(99)00186-8. [DOI] [PubMed] [Google Scholar]
- Bolos V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr. Rev. 2007;28:339–363. doi: 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001;15:66–78. doi: 10.1101/gad.186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Cheng HT, Kim M, Valerius MT, Surendran K, Schuster-Gossler K, Gossler A, McMahon AP, Kopan R. Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron. Development. 2007;134:801–811. doi: 10.1242/dev.02773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell RA, Eisen JS. Notch in the pathway: the roles of Notch signaling in neural crest development. Semin. Cell Dev. Biol. 2005;16:663–672. doi: 10.1016/j.semcdb.2005.06.009. [DOI] [PubMed] [Google Scholar]
- D’Amico-Martel A, Noden DM. Contributions of placodal and neural crest cells to avian cranial peripheral ganglia. Am. J. Anat. 1983;166:445–468. doi: 10.1002/aja.1001660406. [DOI] [PubMed] [Google Scholar]
- D’Souza B, Miyamoto A, Weinmaster G. The many facets of Notch ligands. Oncogene. 2008;27:5148–5167. doi: 10.1038/onc.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Dixon J, Jones NC, Sandell LL, Jayasinghe SM, Crane J, Rey JP, Dixon MJ, Trainor PA. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc. Natl. Acad. Sci. U S A. 2006;103:13403–13408. doi: 10.1073/pnas.0603730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Jesse AM, Kohn A, Gunnell LM, Honjo T, Zuscik MJ, O’Keefe RJ, Hilton MJ. RBPj{kappa}-dependent Notch signaling regulates mesenchymal progenitor cell proliferation and differentiation during skeletal development. Development. 2010;137:1461–1471. doi: 10.1242/dev.042911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farlie PG, McKeown SJ, Newgreen DF. The neural crest: basic biology and clinical relationships in the craniofacial and enteric nervous systems. Birth Defects Res. C Embryo Today. 2004;72:173–189. doi: 10.1002/bdrc.20013. [DOI] [PubMed] [Google Scholar]
- Gridley T. Notch signaling and inherited disease syndromes. Hum. Mol. Genet. 2003;12(Spec No 1):R9–13. doi: 10.1093/hmg/ddg052. [DOI] [PubMed] [Google Scholar]
- Gridley T. Notch signaling in vascular development and physiology. Development. 2007;134:2709–2718. doi: 10.1242/dev.004184. [DOI] [PubMed] [Google Scholar]
- Han H, Tanigaki K, Yamamoto N, Kuroda K, Yoshimoto M, Nakahata T, Ikuta K, Honjo T. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 2002;14:637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- Hansson EM, Lendahl U, Chapman G. Notch signaling in development and disease. Semin. Cancer Biol. 2004;14:320–328. doi: 10.1016/j.semcancer.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Heeg-Truesdell E, LaBonne C. A slug, a fox, a pair of sox: transcriptional responses to neural crest inducing signals. Birth Defects Res. C Embryo Today. 2004;72:124–139. doi: 10.1002/bdrc.20011. [DOI] [PubMed] [Google Scholar]
- High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nat. Rev. 2008;9:49–61. doi: 10.1038/nrg2279. [DOI] [PubMed] [Google Scholar]
- High FA, Zhang M, Proweller A, Tu L, Parmacek MS, Pear WS, Epstein JA. An essential role for Notch in neural crest during cardiovascular development and smooth muscle differentiation. J. Clin. Invest. 2007;117:353–363. doi: 10.1172/JCI30070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu ZL, Shi M, Huang Y, Zheng MH, Pei Z, Chen JY, Han H, Ding YQ. The role of the transcription factor Rbpj in the development of dorsal root ganglia. Neural Dev. 2011;6:14. doi: 10.1186/1749-8104-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlbut GD, Kankel MW, Lake RJ, Artavanis-Tsakonas S. Crossing paths with Notch in the hyper-network. Curr. Opin. Cell Biol. 2007;19:166–175. doi: 10.1016/j.ceb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Hutson MR, Kirby ML. Model systems for the study of heart development and disease. Cardiac neural crest and conotruncal malformations. Semin. Cell Dev. Biol. 2007;18:101–110. doi: 10.1016/j.semcdb.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Kato H, Sakai T, Tamura K, Minoguchi S, Shirayoshi Y, Hamada Y, Tsujimoto Y, Honjo T. Functional conservation of mouse Notch receptor family members. FEBS. Lett. 1996;395:221–224. doi: 10.1016/0014-5793(96)01046-0. [DOI] [PubMed] [Google Scholar]
- Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- Kirby ML, Gale TF, Stewart DE. Neural crest cells contribute to normal aorticopulmonary septation. Science. 1983;220:1059–1061. doi: 10.1126/science.6844926. [DOI] [PubMed] [Google Scholar]
- Kirby ML, Waldo KL. Neural crest and cardiovascular patterning. Circ. Res. 1995;77:211–215. doi: 10.1161/01.res.77.2.211. [DOI] [PubMed] [Google Scholar]
- Knecht AK, Bronner-Fraser M. Induction of the neural crest: a multigene process. Nat. Rev. 2002;3:453–461. doi: 10.1038/nrg819. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall RA. More complicated than it looks: assembly of Notch pathway transcription complexes. Oncogene. 2008;27:5099–5109. doi: 10.1038/onc.2008.223. [DOI] [PubMed] [Google Scholar]
- Kuczuk MH, Scott WJ., Jr. Potentiation of acetazolamide induced ectrodactyly in SWV and C57BL/6J mice by cadmium sulfate. Teratology. 1984;29:427–435. doi: 10.1002/tera.1420290314. [DOI] [PubMed] [Google Scholar]
- Lassiter RN, Ball MK, Adams JS, Wright BT, Stark MR. Sensory neuron differentiation is regulated by notch signaling in the trigeminal placode. Dev. Bio. 2010;344:836–848. doi: 10.1016/j.ydbio.2010.05.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin NM, Smith J. Development of the peripheral nervous system from the neural crest. Annu. Rev. Cell Dev. Biol. 1988;4:375–404. doi: 10.1146/annurev.cb.04.110188.002111. [DOI] [PubMed] [Google Scholar]
- Le Lievre CS, Le Douarin NM. Mesenchymal derivatives of the neural crest: analysis of chimaeric quail and chick embryos. J. Embryol. Exp. Morphol. 1975;34:125–154. [PubMed] [Google Scholar]
- Lincoln J, Alfieri CM, Yutzey KE. Development of heart valve leaflets and supporting apparatus in chicken and mouse embryos. Dev. Dyn. 2004;230:239–250. doi: 10.1002/dvdy.20051. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, Bradley A, Baldini A. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- Loomes KM, Stevens SA, O’Brien ML, Gonzalez DM, Ryan MJ, Segalov M, Dormans NJ, Mimoto MS, Gibson JD, Sewell W, Schaffer AA, Nah HD, Rappaport EF, Pratt SC, Dunwoodie SL, Kusumi K. Dll3 and Notch1 genetic interactions model axial segmental and craniofacial malformations of human birth defects. Dev. Dyn. 2007;236:2943–2951. doi: 10.1002/dvdy.21296. [DOI] [PubMed] [Google Scholar]
- Ma L, Lu MF, Schwartz RJ, Martin JF. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;132:5601–5611. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- Macatee TL, Hammond BP, Arenkiel BR, Francis L, Frank DU, Moon AM. Ablation of specific expression domains reveals discrete functions of ectoderm- and endoderm-derived FGF8 during cardiovascular and pharyngeal development. Development. 2003;130:6361–6374. doi: 10.1242/dev.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmigere F, Ernfors P. Specification and connectivity of neuronal subtypes in the sensory lineage. Nat. Rev. Neurosci. 2007;8:114–127. doi: 10.1038/nrn2057. [DOI] [PubMed] [Google Scholar]
- McCright B, Lozier J, Gridley T. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development. 2002;129:1075–1082. doi: 10.1242/dev.129.4.1075. [DOI] [PubMed] [Google Scholar]
- Mead TJ, Yutzey KE. Notch pathway regulation of chondrocyte differentiation and proliferation during appendicular and axial skeleton development. Proc. Natl. Acad. Sci. U S A. 2009;106:14420–14425. doi: 10.1073/pnas.0902306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Perez SE, Qiao Z, Verdi JM, Hicks C, Weinmaster G, Anderson DJ. Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell. 2000;101:499–510. doi: 10.1016/s0092-8674(00)80860-0. [DOI] [PubMed] [Google Scholar]
- Murphy M, Reid K, Ford M, Furness JB, Bartlett PF. FGF2 regulates proliferation of neural crest cells, with subsequent neuronal differentiation regulated by LIF or related factors. Development. 1994;120:3519–3528. doi: 10.1242/dev.120.12.3519. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. U S A. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy N, Goldstein AM. Endothelin-3 regulates neural crest cell proliferation and differentiation in the hindgut enteric nervous system. Dev. Biol. 2006;293:203–217. doi: 10.1016/j.ydbio.2006.01.032. [DOI] [PubMed] [Google Scholar]
- Okamura Y, Saga Y. Notch signaling is required for the maintenance of enteric neural crest progenitors. Development. 2008;135:3555–3565. doi: 10.1242/dev.022319. [DOI] [PubMed] [Google Scholar]
- Porras D, Brown CB. Temporal-spatial ablation of neural crest in the mouse results in cardiovascular defects. Dev. Dyn. 2008;237:153–162. doi: 10.1002/dvdy.21382. [DOI] [PubMed] [Google Scholar]
- Proweller A, Pear WS, Parmacek MS. Notch signaling represses myocardin-induced smooth muscle cell differentiation. J. Biol. Chem. 2005;280:8994–9004. doi: 10.1074/jbc.M413316200. [DOI] [PubMed] [Google Scholar]
- Ruberte E, Dolle P, Chambon P, Morriss-Kay G. Retinoic acid receptors and cellular retinoid binding proteins. II. Their differential pattern of transcription during early morphogenesis in mouse embryos. Development. 1991;111:45–60. doi: 10.1242/dev.111.1.45. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Rubenstein JL, Nicolas JF. Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 1986;5:3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Shelton EL, Yutzey KE. Tbx20 regulation of endocardial cushion cell proliferation and extracellular matrix gene expression. Dev. Biol. 2007;302:376–388. doi: 10.1016/j.ydbio.2006.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Southard-Smith EM, Kos L, Pavan WJ. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat. Genet. 1998;18:60–64. doi: 10.1038/ng0198-60. [DOI] [PubMed] [Google Scholar]
- Stemple DL, Anderson DJ. Isolation of a stem cell for neurons and glia from the mammalian neural crest. Cell. 1992;71:973–985. doi: 10.1016/0092-8674(92)90393-q. [DOI] [PubMed] [Google Scholar]
- Swiatek PJ, Gridley T. Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc finger gene Krox20. Genes Dev. 1993;7:2071–2084. doi: 10.1101/gad.7.11.2071. [DOI] [PubMed] [Google Scholar]
- Tang Y, Urs S, Boucher J, Bernaiche T, Venkatesh D, Spicer DB, Vary CP, Liaw L. Notch and transforming growth factor-beta (TGFbeta) signaling pathways cooperatively regulate vascular smooth muscle cell differentiation. J. Biol. Chem. 2010;285:17556–17563. doi: 10.1074/jbc.M109.076414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Urs S, Liaw L. Hairy-related transcription factors inhibit Notch-induced smooth muscle alpha-actin expression by interfering with Notch intracellular domain/CBF-1 complex interaction with the CBF-1-binding site. Circ. Res. 2008;102:661–668. doi: 10.1161/CIRCRESAHA.107.165134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MK, Yeager K, Morrison SJ. Physiological Notch signaling promotes gliogenesis in the developing peripheral and central nervous systems. Development. 2007;134:2435–2447. doi: 10.1242/dev.005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor PA. Specification of neural crest cell formation and migration in mouse embryos. Semin. Cell Dev. Biol. 2005;16:683–693. doi: 10.1016/j.semcdb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Tsarovina K, Schellenberger J, Schneider C, Rohrer H. Progenitor cell maintenance and neurogenesis in sympathetic ganglia involves Notch signaling. Mol. Cell Neurosci. 2008;37:20–31. doi: 10.1016/j.mcn.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Varadkar P, Kraman M, Despres D, Ma G, Lozier J, McCright B. Notch2 is required for the proliferation of cardiac neural crest-derived smooth muscle cells. Dev. Dyn. 2008;237:1144–1152. doi: 10.1002/dvdy.21502. [DOI] [PubMed] [Google Scholar]
- Wakamatsu Y, Maynard TM, Weston JA. Fate determination of neural crest cells by NOTCH-mediated lateral inhibition and asymmetrical cell division during gangliogenesis. Development. 2000;127:2811–2821. doi: 10.1242/dev.127.13.2811. [DOI] [PubMed] [Google Scholar]
- Waldo K, Miyagawa-Tomita S, Kumiski D, Kirby ML. Cardiac neural crest cells provide new insight into septation of the cardiac outflow tract: aortic sac to ventricular septal closure. Dev. Biol. 1998;196:129–144. doi: 10.1006/dbio.1998.8860. [DOI] [PubMed] [Google Scholar]
- Waldo KL, Kumiski D, Kirby ML. Cardiac neural crest is essential for the persistence rather than the formation of an arch artery. Dev. Dyn. 1996;205:281–292. doi: 10.1002/(SICI)1097-0177(199603)205:3<281::AID-AJA8>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Williams R, Lendahl U, Lardelli M. Complementary and combinatorial patterns of Notch gene family expression during early mouse development. Mech. Dev. 1995;53:357–368. doi: 10.1016/0925-4773(95)00451-3. [DOI] [PubMed] [Google Scholar]
- Wurdak H, Ittner LM, Lang KS, Leveen P, Suter U, Fischer JA, Karlsson S, Born W, Sommer L. Inactivation of TGFbeta signaling in neural crest stem cells leads to multiple defects reminiscent of DiGeorge syndrome. Genes Dev. 2005;19:530–535. doi: 10.1101/gad.317405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotti S, Canalis E. Notch and the skeleton. Mol. Cell Biol. 2010;30:886–896. doi: 10.1128/MCB.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.