Abstract
Phosphonates are compounds that contain the chemically stable carbon–phosphorus (C–P) bond. They are widely distributed amongst more primitive life forms including many marine invertebrates and constitute a significant component of the dissolved organic phosphorus reservoir in the oceans. Virtually all biogenic C–P compounds are synthesized by a pathway in which the key step is the intramolecular rearrangement of phosphoenolpyruvate to phosphonopyruvate. However C–P bond cleavage by degradative microorganisms is catalyzed by a number of enzymes – C–P lyases, C–P hydrolases, and others of as-yet-uncharacterized mechanism. Expression of some of the pathways of phosphonate catabolism is controlled by ambient levels of inorganic P (Pi) but for others it is Pi-independent. In this report we review the enzymology of C–P bond metabolism in bacteria, and also present the results of an in silico investigation of the distribution of the genes that encode the pathways responsible, in both bacterial genomes and in marine metagenomic libraries, and their likely modes of regulation. Interrogation of currently available whole-genome bacterial sequences indicates that some 10% contain genes encoding putative pathways of phosphonate biosynthesis while ∼40% encode one or more pathways of phosphonate catabolism. Analysis of metagenomic data from the global ocean survey suggests that some 10 and 30%, respectively, of bacterial genomes across the sites sampled encode these pathways. Catabolic routes involving phosphonoacetate hydrolase, C–P lyase(s), and an uncharacterized 2-aminoethylphosphonate degradative sequence were predominant, and it is likely that both substrate-inducible and Pi-repressible mechanisms are involved in their regulation. The data we present indicate the likely importance of phosphonate-P in global biogeochemical P cycling, and by extension its role in marine productivity and in carbon and nitrogen dynamics in the oceans.
Keywords: phosphonate, C–P bond, marine bacteria, phosphorus cycling
Phosphorus in the Oceans
Phosphorus (P) is usually found in the biosphere in its highest oxidation state (+5) as orthophosphate (Pi), which is thought to be its most bioavailable form. P availability has long been identified as a key determinant of marine productivity (Dyhrman and Haley, 2006; Paytan and McLaughlin, 2007; Dyhrman et al., 2009) and its cycling is closely linked to carbon (C) and nitrogen (N) dynamics in the oceans. In oligotrophic regions, and seasonally in the surface waters of a broad range of marine environments, Pi concentrations can be extremely low, e.g., most of the central oceanic gyres have levels close to the limit of chemical detection (<1 nM). In these instances recent climatically influenced extension of the N-fixation season appears to have led to a shift from an N- to a P-limited ecosystem (Karl et al., 2001). Correlated with this has been an increase in levels of Dissolved Organic Phosphorus (DOP), relative to those of Pi. Several recent studies suggest that DOP is a significant additional P source that is available to the marine microbiota in a variety of trophic regimes (Björkman and Karl, 2003; Luo et al., 2009, 2011; Martinez et al., 2010). For example a study in the oligotrophic North Pacific subtropical gyre showed that DOP accounted for up to 80% of total soluble P and was rapidly utilized by the microbial fraction (Björkman and Karl, 2003). It may thus play an important role in determining the composition and community dynamics of marine microbial populations.
Phosphonates in the Marine Environment
An important component of marine DOP is phosphonate-P, which occurs widely in nature in a range of biogenic molecules that contain the highly stable and apparently ancient direct carbon–phosphorus (C–P) bond, rather than the more common C–O–P bond found in phosphate esters (Ternan et al., 1998; Quinn et al., 2007; White and Metcalf, 2007). Phosphonates are known to be produced by many primitive life forms, including a wide range of marine invertebrates in which phosphonate-P may comprise as much as 50% of the total P content (Quin, 1965, 2000). The presence of phosphonates at levels of some 10% of total P in cells of the bloom-forming marine cyanobacterium Trichodesmium erythraeum IMS 101 has also recently been demonstrated (Dyhrman et al., 2009). Natural-product phosphonates such as 2-aminoethylphosphonic acid (2-AEP; ciliatine) or 2-amino-3-phosphonopropionic acid (phosphonoalanine) are normally found as side groups on exopolysaccharides or glycoproteins, or in the polar head groups of membrane phosphonolipids (Hilderbrand, 1983); their presence may increase structural rigidity or protect against enzymatic degradation, since the C–P bond is resistant to the action of phosphatases. However a growing range of small-molecule phosphonates are also known to be produced (reviewed by Metcalf and van der Donk, 2009); many are likely to serve as antimetabolites (e.g., fosfomycin, fosmidomycin, alafosfalin, and phosphinothricin) due to their structural analogies to phosphate esters or carboxylic acids.
The qualitative composition and quantitative importance of phosphonate-P in the oceans have been difficult to establish definitively because of analytical limitations. Nonetheless studies using 31P-NMR indicate that C–P compounds comprise more than 25% of ultrafiltered DOP, and 23% of total particulate P (Clark et al., 1998; Kolowith et al., 2001). In situ evidence for high rates of phosphonate remineralization has also been presented, for example by Benitez-Nelson et al. (2004). Moreover a number of recent field, laboratory, and genome-enabled studies (Dyhrman et al., 2006; Quinn et al., 2007; Karl et al., 2008; Gilbert et al., 2009; Ilikchyan et al., 2009; Martinez et al., 2010; Thomas et al., 2010; Luo et al., 2011) also suggest that phosphonate-P may constitute a significant P source in the oceans. Additional – if circumstantial – evidence for this view is provided by the fact that the “streamlined” genome of SAR 11, the most abundant bacterial clade on earth, contains (and expresses in situ) phosphonate transport genes (Sowell et al., 2009). Despite this growing body of information, however, our understanding of the factors that influence the production and consumption of phosphonates in the oceans remains limited (Dyhrman et al., 2007, 2009).
Biosynthesis of Phosphonates
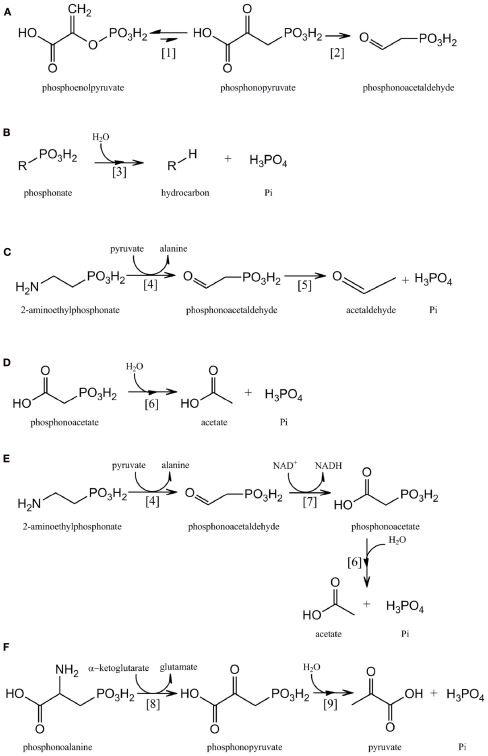
Virtually all known pathways of phosphonate biosynthesis share a central C–P bond-forming reaction in which an intramolecular rearrangement of the intermediary metabolite phosphoenolpyruvate (PEP) is catalyzed by the enzyme PEP phosphomutase (Ppm; EC 5.4.2.9), with phosphoryl group transfer leading to formation of phosphonopyruvate (Bowman et al., 1988; Seidel et al., 1988; Metcalf and van der Donk, 2009; Figure 1A). Since the equilibrium of the reaction carried out by PEP phosphomutase strongly favors formation of the C–O–P bond of PEP, however, most C–P bond-synthesizing microorganisms contain a coupled enzyme, phosphonopyruvate decarboxylase (Ppd; EC 4.1.1.82), which subsequently converts phosphonopyruvate to phosphonoacetaldehyde in an irreversible reaction (Zhang et al., 2003; Figure 1A). Phosphonoacetaldehyde then forms the central metabolite in a range of subsequent biosynthetic sequences (Metcalf and van der Donk, 2009); for example in the biosynthesis of 2-AEP it is transaminated by the enzyme 2-AEP–pyruvate transaminase (EC 2.6.1.37; Barry et al., 1988).
Figure 1.
Pathways for the microbial metabolism of phosphonates: (A) phosphonate biosynthesis by phosphoenolpyruvate phosphomutase [1] and phosphonopyruvate decarboxylase [2]. (B) C–P lyase [3] pathway. (C) 2-AEP catabolism by 2-AEP transaminase [4] and phosphonoacetaldehyde hydrolase [5]. (D) Phosphonoacetate catabolism by phosphonoacetate hydrolase [6]. (E) 2-AEP catabolism by 2-AEP transaminase [4], phosphonoacetaldehyde dehydrogenase [7], and phosphonoacetate hydrolase [6]. (F) Phosphonoalanine catabolism by a phosphonoalanine transaminase [8] and phosphonopyruvate hydrolase [9].
Enzymes of Phosphonate Catabolism in Microorganisms
The importance of phosphonates as a P reservoir in the global ocean is by now well-established but information on the molecular strategies employed by marine microorganisms for their utilization remains incomplete. Current understanding of the catabolic activities involved is summarized below.
C–P lyase
The predominant route for microbial utilization of phosphonates has been thought to be the “C–P lyase” pathway (Ternan et al., 1998) and its presence and activity in marine microorganisms has recently been demonstrated (e.g., Dyhrman et al., 2006). However the broad-specificity C–P lyase multienzyme complex has not yet been fully characterized (Hove-Jensen et al., 2011) although a major technical obstacle to its elucidation has very recently been overcome with the first report (Kamat et al., 2011) of the in vitro reconstitution of the pathway in Escherichia coli and identification of the reaction intermediates. In E. coli the C–P lyase complex is comprised of 14 gene products which together catalyze the difficult dephosphonation reaction in a range of structurally diverse phosphonates which may contain either activated or unactivated C–P bonds (Wackett et al., 1987; Yakovleva et al., 1998). In each case the product is the corresponding hydrocarbon and Pi (Figure 1B). Of these 14 genes, phnCDE are thought to encode an ATP-binding cassette transporter, and phnF a repressor protein, while the products of phnNOP are thought to have regulatory or other accessory roles (Hove-Jensen et al., 2010, 2011). Seven gene products (PhnGHIJKLM) constitute the “core” components of the C–P lyase reaction pathway, with PhnJ now known to catalyze its central reaction, the S-adenosyl-l-methionine-dependent radical cleavage of 5-phosphoribosyl-1-phosphonate to produce 5-phosphoribosyl-1,2-cyclic phosphate and the corresponding alkane (Hove-Jensen et al., 2011; Jochimsen et al., 2011; Kamat et al., 2011). Variant forms of the C–P lyase enzyme complex with different substrate ranges are also known to exist (e.g., Kertesz et al., 1991; Sviridov et al., 2012), and other genes of as-yet unknown function have frequently been found associated with the C–P lyase operon (see Occurrence of the phnJ Component of the C-P Lyase Operon in Sequenced Bacterial Genomes). In addition the role of PhnK remains unknown (Kamat et al., 2011).
In all bacteria studied to date the genes that encode the C–P lyase pathway(s) have been found to be arranged in tandem and transcribed from a single promoter in an operon that is a member of the Pho regulon – a suite of genes whose products include proteins required for Pi-scavenging, and for the catabolism of alternative P sources. Expression of the genes of the Pho regulon is induced under conditions of Pi-starvation but is Pi-repressible (Quinn et al., 2007). Thus in T. erythraeum the genes of the C–P lyase pathway were expressed in situ at Pi concentrations of ∼6 nmol l−1, but not under Pi-replete conditions (Dyhrman et al., 2006). Similarly the aerobic production of methane from methylphosphonate by mixed marine microbial communities (likely to be mediated by C–P lyase) was demonstrated under Pi-stressed conditions (Karl et al., 2008).
All C–P bond cleavage enzymes had originally been thought to be similarly subject to Pho regulon control; however, it seems likely that in this event phosphonates could be utilized by microorganisms only as a source of P, since any Pi released during their catabolism in excess of that required for balanced cellular growth would repress any further degradation (McGrath et al., 1997; Quinn et al., 2007). Indeed, further phosphonate uptake by cells would also be repressed because the relevant high-affinity transporters are also Pho-regulated. However field and laboratory observations (Benitez-Nelson et al., 2004; Beversdorf et al., 2010) suggest that phosphonate degradation in the oceans continues under conditions of Pi sufficiency. And indeed three inducible enzymes capable of C–P bond cleavage have been identified and characterized (Quinn et al., 2007). Unlike C–P lyase, which is capable of the cleavage of both activated and unactivated C–P bonds, each of these phosphonohydrolases is uniquely specific to a natural-product phosphonate that contains an electron-withdrawing carbonyl group in the beta position to the P atom. Expression of all three enzymes has been shown to be Pi-insensitive and hence independent of Pho regulation – thus they are likely to play a role in the marine environment that is complementary to that of C–P lyase.
Phosphonoacetaldehyde hydrolase
The phnX gene product phosphonoacetaldehyde hydrolase (EC 3.11.1.1; phosphonatase) cleaves the C–P bond of its unique substrate to yield acetaldehyde and Pi (LaNauze et al., 1977; Dumora et al., 1989); it is encoded within a degradative operon for the mineralization of 2-aminoethylphosphonic acid (2-AEP, ciliatine). 2-AEP replaces its structural analog ethanolamine phosphate as a component of the phosphonoglycolipids common in the cell membrane of many invertebrates and is considered – along with its N-methylated derivatives – to be the most abundant and ubiquitous marine C–P compound (Hilderbrand, 1983; Hori et al., 1984; Quin and Quin, 2001). The mechanism of C–P bond cleavage by phosphonatase, a member of the halo acid dehalogenase superfamily, involves the formation of a Schiff base intermediate between a lysine residue at the active site of the enzyme and the phosphonoacetaldehyde carbonyl group; this activates the phosphonate group for attack by an active site nucleophile (Baker et al., 1998; Morais et al., 2004). The phosphonatase pathway of 2-AEP degradation (Figure 1C) also includes a 2-AEP:pyruvate aminotransferase (EC 2.6.1.37), encoded by phnW, whose products are phosphonoacetaldehyde and alanine (Kim et al., 2002). The Pi-starvation-inducible expression of this pathway as a part of the Pho regulon has been described in Enterobacter aerogenes and Salmonella typhimurium, neither of which is able to utilize 2-AEP as sole carbon and energy source (Jiang et al., 1995). However in other bacterial strains, e.g., Pseudomonas putida NG2 and Pseudomonas aeruginosa PAO1 that are able to mineralize 2-AEP as sole C, N, and P source, expression of the operon is substrate-inducible, mediated by the product of an adjacent gene that encodes a LysR-like transcriptional activator (LTTR; Ternan and Quinn, 1998a; Quinn et al., 2007).
Phosphonoacetate hydrolase
The phnA gene product, phosphonoacetate hydrolase, was identified in an activated sludge isolate of Pseudomonas fluorescens that was capable of utilizing phosphonoacetate as sole C and P source (McMullan et al., 1992). The enzyme is a member of the alkaline phosphatase superfamily and forms acetate and Pi from its substrate following metal-assisted hydrolytic cleavage of the C–P bond (McGrath et al., 1995; Agarwal et al., 2011; Kim et al., 2011; Figure 1D). In its original host phosphonoacetate hydrolase, together with an associated phosphonate transporter, was found to be encoded within an operon whose expression was regulated by a phosphonoacetate-responsive LTTR (Kulakova et al., 2001, 2009). More recently, however, the phnA gene has been found in other bacterial strains within an operon that also encodes 2-AEP transaminase (PhnW) and a novel NAD+-dependent phosphonoacetaldehyde dehydrogenase (PhnY) whose product is phosphonoacetate (Borisova et al., 2011; Kim et al., 2011; Figure 1E). The enzyme sequence encoded by this operon thus represents an alternative route of 2-AEP mineralization to that of the phosphonatase pathway (see Phosphonoacetaldehyde Hydrolase), and is likely to be a major route of biogenic phosphonoacetate production (Cooley et al., 2011).
Phosphonopyruvate hydrolase
This enzyme was identified by Ternan and Quinn (1998b) in cells of a Burkholderia strain that utilized phosphonoalanine (2-amino-3-phosphonopropionic acid) as sole C, N, and P source. Phosphonoalanine, the phosphonate analog of aspartic acid, is formed by the transamination of phosphonopyruvate (Horigane et al., 1989). It is a constituent of membrane phosphonolipids and is widely distributed amongst marine invertebrates (Kittredge and Hughes, 1964); its degradative pathway involves an initial transamination to phosphonopyruvate, followed by cleavage of the C–P bond by a metal-dependent phosphonohydrolase to yield pyruvate and Pi (Figure 1F). Phosphonopyruvate hydrolase (EC 3.11.1.3) is a member of the phosphoenolpyruvate phosphomutase/isocitrate lyase superfamily and was shown in Variovorax sp. Pal2 to be encoded within an operon that also contains phosphonoalanine transaminase and phosphonate transporter genes; its expression is regulated by an adjacent phosphonoalanine- and phosphonopyruvate-responsive LTTR (Kulakova et al., 2003, 2009). Phosphonopyruvate hydrolase shares 41% sequence identity with the C–P bond-forming enzyme, phosphoenolpyruvate phosphomutase (see Biosynthesis of Phosphonates above); a single amino acid substitution at the active site is likely to be responsible for the functional divergence of the two enzymes (Chen et al., 2006).
Other enzymes of phosphonate catabolism
There is also evidence for the existence of a number of less well-characterized C–P bond cleavage activities that are likely to be of significance in marine environments:
-
(a)
Martinez et al. (2010) have demonstrated by functional gene complementation that a fragment of marine genomic DNA that encodes a putative 2-oxoglutarate dioxygenase (PhnY*) and a phosphohydrolase (PhnZ) conferred on E. coli the ability to utilize 2-AEP as a sole P source; however the biochemical reactions catalyzed by these genes remain uncharacterized. [It will be important to avoid future confusion between the activity encoded by phnY* and the unrelated phosphonoacetaldehyde dehydrogenase (see Phosphonoacetate Hydrolase above) recently described in Sinorhizobium meliloti by Borisova et al. (2011) which has also been designated as PhnY, both here and in a number of annotated genomes].
-
(b)
McGrath et al. (1998) have reported the utilization of the phosphonate antibiotic fosfomycin (2-epoxypropylphosphonic acid) as the sole C and P source by a Sinorhizobium huakuii isolate. The catabolic reaction sequence involved has been further elucidated (McGrath et al., 2009, 2011), and although the enzyme responsible for the C–P bond cleavage step has not yet been fully described its substrate is likely to be (R)-1-hydroxy-2-oxopropylphosphonic acid. Given that S. huakuii cells release essentially quantitative levels of Pi when using fosfomycin as a sole C source, expression of the C–P cleavage enzyme is clearly Pho-independent.
-
(c)
Mendz et al. (2005) and Ford et al. (2010) have presented evidence for the existence of novel, as-yet-undescribed pathways of Pi-insensitive phosphonate catabolism in Campylobacter and Helicobacter spp. The existence of further uncharacterized, but likely unrelated, C–P cleavage activities has also been reported in cells of Synechococcus spp. (Gomez-Garcia et al., 2011).
Distribution of the Genes of Phosphonate Metabolism Amongst Sequenced Bacteria
BLAST analysis of the occurrence of the genes of phosphonate metabolism amongst the bacterial genome sequences deposited in the NCBI GenBank database as at the beginning of 2011 was carried out. The query sequences from deduced peptides of confirmed function used to interrogate the database are listed in Table 1. A minimum expectation value of 1 × 10−10 and a minimum of 10% sequence identity over the entire sequence length were employed as criteria for homolog selection. In addition each gene identity was confirmed by inspection of its genomic context.
Table 1.
Polypeptides used as query sequences for bioinformatic analyses.
| Pathway | Enzyme | Accession number | Microbial origin | Reference |
|---|---|---|---|---|
| Phosphonate biosynthesis | Ppd | YP_099861.1 | Bacteroides fragilis YCH46 | Kuwahara et al. (2004) |
| C–P lyase | Complex subunit PhnJ | NP_418522.1 | Escherichia coli str. K-12 substr. MG1655 | Bury-Mone et al. (2009) |
| Phosphonoacetaldehyde hydrolase | PhnX | NP_250002.1 | Pseudomonas aeruginosa PAO1 | Cirz et al. (2006) |
| Phosphonoacetate hydrolase | PhnA | AAC15507.1 | Pseudomonas fluorescens 23F | Kulakova et al. (2001) |
| Phosphonopyruvate hydrolase | PalA | AAO24736.1 | Variovorax sp. Pal2 | Kulakova et al. (2009) |
| Phosphonate transport | Transporter subunit PhnD | NP_418529.1 | Escherichia coli str. K-12 substr. MG1655 | Bury-Mone et al. (2009) |
| Potential new degradation pathway (Martinez et al. (2010)) | PhnY* | ACU83549 | Uncultured bacterium HF130_AEPn_1 | Martinez et al. (2010) |
| PhnZ | ACU83550 | Uncultured bacterium HF130_AEPn_1 | Martinez et al. (2010) | |
| Protein of unknown function | Complex subunit DUF6 | YP_0010234431 | Methylibium petroleiphilum PM1 | |
| related to C–P lyase | Complex subunit DUF1045 | ABQ32108.1 | Acidiphilium cryptum JF-5 |
Out of the 1384 sequenced genomes in the database, 664 (48%) contained homologs that indicated possession of one or more of the known pathways of phosphonate metabolism (Table 2); these included 121 of the 190 marine bacterial genomes in the database. Fourfold more strains contained known genes associated with phosphonate catabolism than with C–P bond formation. Of the putative phosphonate degradative pathways detected, those involving C–P lyase and phosphonoacetaldehyde hydrolase comprised some 83% of the total. The abundance of phnX homologs encoding the substrate-specific phosphonoacetaldehyde hydrolase, and of homologs that encode the PhnD transporter which shows highest affinity for 2-AEP (Rizk et al., 2006) suggests that 2-AEP is likely to have been an important source of P through a long period of evolutionary history. An analysis of the distribution of each of the pathways and of the variation in their associated operons is presented below.
Table 2.
Homologs retrieved from the NCBI bacterial genomes database that indicate possession of pathways of phosphonate metabolism.
| Pathway | Query polypeptide | “Hits” in sequenced strains | “Hits” in marine strains |
|---|---|---|---|
| Phosphonate biosynthesis | Ppd | 129 (9.3%)a | 7 (3.6%)b |
| C–P lyase | Complex subunit PhnJ | 223 (16.1%) | 44 (23.2%) |
| Phosphonoacetaldehyde hydrolase | PhnX | 235 (17.0%) | 50 (26.3%) |
| Phosphonoacetate hydrolase | PhnA | 89 (6.4%) | 6 (3.2%) |
| Phosphonopyruvate hydrolase | PalA | 3 (0.2%) | 0 |
| Phosphonate transport | Transporter subunit PhnD | 294 (21.2%) | 67 (35.3%) |
| Potential new 2-AEP degradation pathway (Martinez et al., 2010) | PhnY* | 5 (0.4%) | 4 (2.1%) |
| PhnZ | 5 (0.4%) | 4 (2.1%) | |
| Unknown function related to C–P lyase | Complex subunit DUF6 | 9 (0.7%) | 4 (2.1%) |
| Complex subunit DUF1045 | 78 (5.6%) | 22 (11.6%) |
aExpressed as a percentage of the 1384 sequenced bacterial genomes contained in the database.
bExpressed as a percentage of the 190 sequenced marine bacterial genomes contained in the database.
Occurrence in sequenced bacterial genomes of the phosphoenolpyruvate phosphomutase/phosphonopyruvate decarboxylase operon for phosphonate biosynthesis
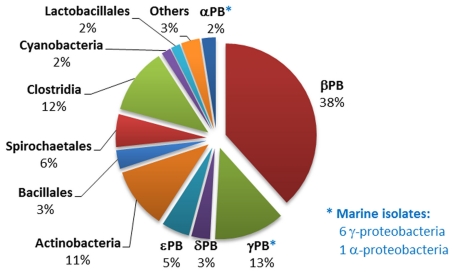
129 sequenced strains (almost 10% of the genomes contained in the NCBI GenBank database) contained the ppd gene indicative of the PEP phosphomutase/phosphonopyruvate decarboxylase pathway of phosphonate biosynthesis; seven of these were of marine origin. Of the 129 strains, 48 also possessed a pathway for phosphonate degradation, and 10 the associated phnCDE operon encoding a phosphonate transporter linked to phosphonate catabolism (Huang et al., 2005). The phosphonate biosynthetic pathway was distributed amongst distant bacterial groups, but predominantly within the members of the proteobacteria, actinobacteria, and clostridia (Figure 2). Analysis of their ppm/ppd operon structures failed to reveal any clearly conserved gene composition, which may reflect the wide range of phosphonate metabolites that are produced by microorganisms (Circello et al., 2010; Johannes et al., 2010; Lee et al., 2010). Nevertheless, two enzymes frequently found to be encoded within the operon (in addition to PEP phosphomutase and phosphonopyruvate decarboxylase) were 2-AEP aminotransferase (PhnW) – observed in 60 strains – which plays a key role in the production of 2-AEP, and an acetaldehyde dehydrogenase (found in 16 strains), which shares strong homology (44% identity, 60% similarity) with the phosphonoacetaldehyde dehydrogenase (PhnY) known to be involved in the production of phosphonoacetate from phosphonoacetaldehyde and found in the phosphonoacetate hydrolase pathway of 2-AEP degradation (Borisova et al., 2011; see Phosphonoacetate Hydrolase). In addition nine Gram-negative bacterial strains possessed a copy of the phosphonate biosynthetic operon that was flanked by genes encoding enzymes involved in exopolysaccharide biosynthesis; it is known that the capsular polysaccharide B complex associated with virulence in Bacteroides fragilis is characterized by its high content of 2-AEP.
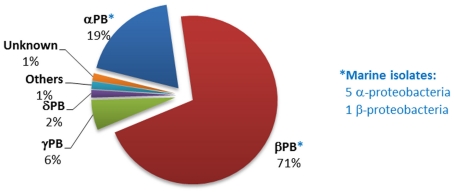
Figure 2.
Distribution of phosphonopyruvate decarboxylase (ppd) gene homologs within sequenced bacterial genomes. The numbers and affiliations of marine bacterial strains that contain the ppd gene, out of the 190 sequenced marine bacterial genomes publically available, are indicated to the right of the Figure.
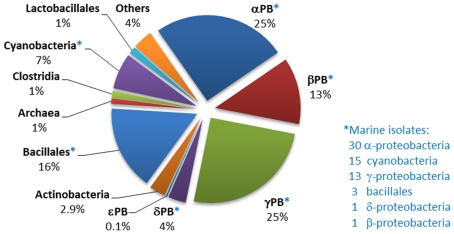
Occurrence of the phnJ component of the C–P lyase operon in sequenced bacterial genomes
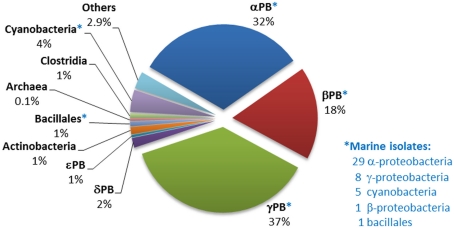
Analysis of the distribution of phnJ, which encodes a catalytic component of the C–P lyase multienzyme system, indicates that C–P lyase is (after “phosphonatase,” see Occurrence in Sequenced Bacterial Genomes of the phnX Gene Encoding Phosphonoacetaldehyde Hydrolase) the second most abundant pathway for phosphonate degradation amongst sequenced bacteria, and that it is widely distributed amongst diverse bacterial groups (Figure 3). Its 223 homologs, including 44 from marine strains, occur mainly in the alpha-, beta-, and gamma-proteobacteria. (Interestingly, however, a copy of the entire operon, and a second copy of its phnCDE genes, were found in the genome of the archaeon Haloquadratum walsbyi; further analysis suggests that these copies have separate proteobacterial origins).
Figure 3.
Distribution of phnJ homologs within sequenced bacterial genomes. The numbers and affiliations of marine bacterial strains that contain the phnJ gene, out of the 190 sequenced marine bacterial genomes publically available, are indicated to the right of the Figure.
Great diversity was found in the composition and architecture of the C–P lyase operon (including insertions, substitutions, and deletions of components); this confirms a similar observation by Huang et al. (2005) and suggests that the operon has been subject to multiple horizontal gene transfers. All these genetic variations could represent potential variations in the specificity or functionality of the reaction sequence encoded, and thus provide possible evolutionary advantages to their hosts. For example domain of unknown function (DUF) 1045, which is not conventionally designated as one of the 14 genes of the C–P lyase operon, was in fact present in 78 of the 223 operons identified (almost all from the alpha- and beta-proteobacteria, and including 26 of the 44 marine strains in which the operon was found). In 86% of instances DUF1045 flanked or lay close to phnN which is thought to encode a “feeder” activity for the C–P bond cleavage pathway (Hove-Jensen et al., 2011). Expression of both DUF1045 and phnN has been found to be highly induced in the presence of methylphosphonate or the absence of Pi in Acidithiobacillus ferrooxidans (Vera et al., 2008).
Genomic data thus appear to provide support for existing biochemical evidence (see C–P Lyase above) that the C–P lyases may be considered as a family of closely related multienzyme systems that nevertheless vary in, for example, their substrate specificities. The high incidence of “optional” genes suggests that these may provide additional functional features. In terms of their regulation, ∼90% of C–P lyase operons were apparently controlled by flanking GntR regulators, confirming numerous findings that this pathway is inducible only under conditions of Pi-starvation (Wackett et al., 1987). Surprisingly, however, genes predicted to encode alternative regulators (including those from the LysR, IclR, DeoR, LuxR, MarR, ArsR, TetR, and TonB families) were found adjacent to C–P lyase operons in some 6% of cases, while in a small number of others no putative regulatory genes could be identified.
Occurrence in sequenced bacterial genomes of the phnX gene encoding phosphonoacetaldehyde hydrolase
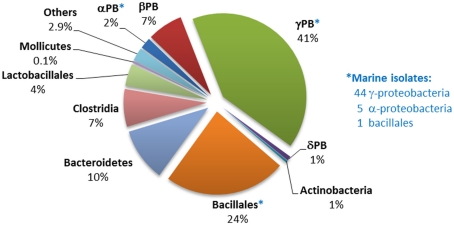
Genes encoding the phosphonatase pathway of 2-AEP degradation, which comprises 2-AEP-pyruvate transaminase (PhnW) and phosphonoacetaldehyde hydrolase (PhnX; see Phosphonoacetaldehyde Hydrolase), were found in 235 bacterial genomes of which 50 were of marine origin, mostly from within the alpha- and gamma-proteobacteria. It is thus the most abundant phosphonate degradation pathway amongst the 1384 bacteria whose sequences are currently available; this may reflect the fact that 2-AEP is thought to be the most widely distributed biogenic phosphonate in the environment (White and Metcalf, 2007). The distribution of genes encoding the phosphonatase operon is shown in Figure 4; some 44% belonged to the gamma-proteobacteria, of whom the majority were potential pathogens (Salmonella, Vibrio, etc.) that have been implicated in gastrointestinal diseases. Predicted phnX-containing strains belonging to the bacillales, bacteroidetes, and clostridia are also associated with similar pathologies. To date, there is no direct evidence to link phosphonoacetaldehyde hydrolase activity to pathogenesis; however it has been reported that low Pi conditions can enhance the virulence of pathogenic species such as P. aeruginosa and Salmonella typhimurimum (Beier and Gross, 2006; Lamarche et al., 2008). Additionally, mutations in Pho regulon genes have been thought to be responsible for the decrease of colonization rates by intestinal strains of E. coli and Vibrio cholerae (von Krüger et al., 1999, 2006). Since phnX in most gamma-proteobacteria was associated with GntR regulators and hence likely to be expressed under Pi limiting conditions (Gebhard and Cook, 2008), it is conceivable that phosphonoacetaldehyde hydrolase may play a role in the infection process.
Figure 4.
Distribution of phnX homologs within sequenced bacterial genomes. The numbers and affiliations of marine bacterial strains that contain the phnX gene, out of the 190 sequenced marine bacterial genomes publically available, are indicated to the right of the Figure.
The phosphonatase operons in Bacillaceae and clostridial strains were observed to be flanked by deoR, and in bacteroidetes by araC regulatory sequences. Only in alpha-proteobacteria (mostly marine isolates), and in beta-, and gamma-proteobacteria (all pseudomonads) were the phosphonatase pathways associated with LysR type regulators. No such strains have been implicated in gastrointestinal infections.
Several features of the phosphonatase operon structures provide strong evidence that PhnX is encoded as part of a genomic island which can be easily transferred amongst bacteria. “Pick-and-mix” horizontal gene transfer is suggested not only by the broad range of regulatory systems apparently involved in the control of the operon expression (including GntR, LysR, DeoR, AraC, and TetR), but also by the insertion of genes such as that encoding 4-aminobutyrate transferase, or DUF6 homologs (the latter is found in some marine strains; DUF6 can also occur within some C–P lyase operons). Furthermore the various rearrangements of the operon components (phnW,X and the phosphonate transporter genes phnC,D,E) that were observed is a feature that is almost entirely absent in pathways of apparently shorter evolutionary history, e.g., that involving phnA (see Occurrence in Sequenced Bacterial Genomes of the phnA Gene Encoding Phosphonoacetate Hydrolase).
Occurrence in sequenced bacterial genomes of the phnA gene encoding phosphonoacetate hydrolase
The phnA gene was much less widely distributed in sequenced bacterial genomes than those encoding the C–P lyase or phosphonoacetaldehyde hydrolase pathways. Figure 5 shows the affiliation of the 69 bacterial strains that encoded the 89 phnA homologs detected during the bioinformatic analysis (these were predominantly Burkholderia spp. within the beta-proteobacteria). Only six strains were identified as marine isolates; however five of these are members of the Roseobacter clade, which is known to be the predominant bacterial group in the near-surface waters of the oceans.
Figure 5.
Distribution of phnA homologs within sequenced bacterial genomes. The numbers and affiliations of marine bacterial strains that contain the phnA gene, out of the 190 sequenced marine bacterial genomes publically available, are indicated to the right of the Figure.
The limited distribution of phnA within other phyla suggests that phosphonate degradation via phosphonoacetate hydrolase is a pathway of recent development or that it lacks the transferability observed in the cases of the C–P lyase and phosphonoacetaldehyde hydrolase pathways. Nevertheless, the phnA gene was frequently duplicated within the genomes of the species that possessed it (e.g., in 20 different strains of beta-proteobacteria). Only three sequenced strains possessed duplications of phnX, and five strains a duplication of the C–P lyase operon. Mechanisms of duplication have been suggested to consolidate the existence of pathways in the host genome, as has been reported for phthalate metabolism in Rhodococcus sp. strain DK17 (Choi et al., 2007).
In contrast with the C–P lyase and phosphonatase pathways with their apparently longer evolutionary histories, analysis of those operons that contained phnA homologs indicated that these were almost exclusively controlled by only two kinds of regulators: GntR or LysR. Of these, GntR regulators were exclusively encoded in gamma-proteobacteria, with LysR present in all alpha- and some beta-proteobacterial strains (including all six marine strains identified). Rearrangements in the structure of the operons that contained phnA were very limited; most (71 out of 89) contained the phnY gene that encodes phosphonoacetaldehyde dehydrogenase and phnW that encodes 2-AEP aminotransferase, suggesting that phosphonoacetate hydrolase (phnA) is more commonly a component of a degradative sequence for the catabolism of 2-AEP (see Phosphonoacetate Hydrolase), than directly inducible by phosphonoacetate as is the case in the operon originally characterized in P. fluorescens 23F by Kulakova et al. (2001), in which neither phnW or phnY is present.
Distribution in sequenced bacterial strains of the pph gene encoding phosphonopyruvate hydrolase
In contrast to the wide distribution exhibited by the phosphonate degradative sequences analyzed above, pph was found to be present in only three sequenced bacterial strains, all Burkholderia spp. In each instance the associated operon was controlled by a LysR regulator and contained a copy of the phnW gene coding for 2-AEP-aminotransferase. This suggests that the operon associated with phosphonopyruvate hydrolase activity may be of relatively recent origin and that it has not been subject to extensive gene transfer.
Distribution in sequenced bacterial strains of additional genes involved in phosphonate metabolism
In addition to the pathways responsible for the breakdown of phosphonates, the distribution of genes that encode other activities associated with their metabolism was analyzed.
The gene that encodes the PhnD periplasmic binding protein of the phosphonate PhnCDE transporter was very widely distributed across bacteria (Figure 6), even more so than those encoding the C–P lyase catalytic protein, PhnJ, or phosphonoacetaldehyde hydrolase. Numbers of predicted homologs were particularly high amongst marine bacteria; of the 294 strains in the NCBI database that were confirmed to possess a phnD homolog, 67 were of marine origin. In 177 of the strains identified, phnD was juxtaposed with a phosphonate degradative gene (thus 60% of strains capable of transporting phosphonates into the cell have an apparent ability to degrade them). On the other hand, 117 other strains that contained phnD did not encode any known pathway of phosphonate degradation, suggesting that not all phosphonate degradation pathways have yet been discovered (as has been proposed by Palenik et al., 2003). It is also possible that phosphonates are not always used as a P source, but after transport into the cell may be incorporated into phosphonolipids, phosphonoproteins, or other macromolecules by anabolic routes. And conversely, 289 strains that possessed genes associated with a known phosphonate degradation pathway showed no evidence of possessing the PhnCDE route of phosphonate acquisition – suggesting the existence of as-yet unknown uptake routes.
Figure 6.
Distribution of phnD homologs within sequenced bacterial genomes. The numbers and affiliations of marine bacterial strains that contain the phnD gene, out of the 190 sequenced marine bacterial genomes publically available, are indicated to the right of the Figure.
An apparent example of a recent pathway of phosphonate metabolism is encoded by the operon designated as PhnY*/PhnZ and discovered by Martinez et al. (2010) in a clone from a fosmid library from the Sargasso Sea. The enzymes of the pathway have not yet been characterized, but it was experimentally proven that they conferred the capacity to consume phosphonates as a P source on an E. coli phn mutant strain (Martinez et al., 2010). PhnY*is thought to be a 2-oxoglutarate dioxygenase, and PhnZ a protein of the HD phosphohydrolase family (Martinez et al., 2010). Bioinformatic analysis indicated that only five sequenced bacterial strains possessed a copy of this operon. Interestingly, four of these were isolated from the marine environment, suggesting that this pathway may have evolved in the oceans and may now play a significant role in P acquisition in marine microorganisms. Three of the strains (Plesiocystis pacifica SIR-1, and Prochlorococcus marinus str. MIT9301 and MIT9303) possess a copy of the phosphonate transporter operon phnCDE but had been considered to lack any recognized phosphonate degradation pathway. As is the case with the phosphonopyruvate hydrolase pathway of C–P bond cleavage, the PhnY*/PhnZ pathway appears to have undergone no extensive evolutionary rearrangement. In this paper the phnY gene described by Martinez et al. (2010) is designated as phnY*.
Abundance and Distribution of the Genes of Phosphonate Metabolism in Marine Metagenomic Databases
The analysis presented in Section “Distribution of the Genes of Phosphonate Metabolism Amongst Sequenced Bacteria” is based on the information currently available on sequenced bacterial genomes which remains heavily biased in favor of species of clinical, or other applied significance. Hence little can be extrapolated to the ecological significance of the various pathways of phosphonate metabolism identified. In an attempt to address this shortcoming, query sequences indicative of the various phosphonate metabolic pathways were used to interrogate the specialist genomic database for marine bacteria, CAMERA (Nealson and Venter, 2007), using the protocol of Lim et al. (2007). An indication was thus obtained of the abundance of the genes encoding those pathways in genomic DNA recovered at the 83 sites from the surface of the world’s oceans sampled by the global ocean survey (GOS) Expedition (Table 3). The results from the CAMERA BLAST analysis (including those for some additional activities associated with bacterial P metabolism) were also normalized against the number of “hits” obtained for RecA; recA is frequently used as a single-copy-per-genome control in distribution and prevalence analyses (Moran et al., 2004; Karl et al., 2008; Table 4).
Table 3.
Presence of genes encoding pathways of phosphonate metabolism at the 83 oceanic sites sampled by the GOS Expedition.
| Pathway | Query polypeptide | % of sampled GOS sites |
|---|---|---|
| Phosphonate biosynthesis | Phosphonopyruvate decarboxylase | 80.7 |
| C–P lyase | Complex subunit PhnJ | 30.1 |
| Phosphonoacetaldehyde hydrolase | PhnX | 28.9 |
| Phosphonoacetate hydrolase | PhnA | 78.3 |
| Phosphonopyruvate hydrolase | PalA | 1.2 |
| Phosphonate transporter | Transporter subunit PhnD | 72.3 |
| Potential new degradation pathway (Martinez et al., 2010) | PhnY* | 51.8 |
| PhnZ | 77.1 | |
| Repair and maintenance of DNA (Control) | RecA | 100 |
Table 4.
Abundance of phosphonate (and related) metabolic pathways in bacterial genomes sampled by the Global Ocean Survey.
| Pathway | Query polypeptide | Abundance of pathway (%) |
|---|---|---|
| Phosphonate biosynthesis | Phosphonopyruvate decarboxylase | 10.2 |
| C–P lyase | Complex subunit PhnJ | 5.1 |
| Phosphonoacetaldehyde hydrolase | PhnX | 1.8 |
| Phosphonoacetate hydrolase | PhnA | 11.2 |
| Phosphonopyruvate hydrolase | PalA | 0.1 |
| Phosphonate transporter | Transporter subunit PhnD | 5.5 |
| Potential new degradation pathway (Martinez et al., 2010) | PhnY* | 1.6 |
| PhnZ | 9.4 | |
| Alkaline phosphatase A | PhoA | 5.9 |
| Alkaline phosphatase D | PhoD | 4.6 |
| Phosphatase X | PhoX | 3.5 |
| Repair and maintenance of DNA (Control) | RecA | 100 |
The ppd gene that encodes the phosphonate biosynthetic enzyme phosphonopyruvate decarboxylase was found at 67 sites, almost 81% of those sampled by the GOS Expedition, and in the equivalent of 10% of sampled genomes. This suggests that natural-product phosphonates may be widely produced in the oceanic environment, and correlates with the potential biological advantages that these molecules may provide as structural components or secondary metabolites. To date, no experimental data has been presented on the ability of marine bacterial strains to synthesize phosphonates, although a recent study using solid-state NMR spectroscopy has demonstrated that C–P compounds comprised almost 10% of the cellular particulate phosphorus pool in cultured strains of T. erythraeum, suggesting that this cyanobacterium might be capable of C–P bond formation (Dyhrman et al., 2009).
In respect of phosphonate catabolism, bioinformatic analysis indicated a wide distribution of the phosphonate transporter subunit, PhnD. The confirmed presence of the phnD gene at 72% of the sites, and in some 5% of the genome-equivalents predicted, suggests that phosphonates are not only being synthesized by bacteria in the oceans but also that exogenous C–P compounds can be taken up for catabolism (or other metabolic activities) at significant levels. This is consistent with recent in situ expression studies on phnD in marine picocyanobacteria by Ilikchyan et al. (2009).
In total, some 28% of bacterial genome-equivalents sampled during the GOS were found to contain genes encoding one or more pathways of C–P bond cleavage (Table 4); however the apparent relative abundances of phosphonate degradation pathways contrasted with those found amongst sequenced bacteria in the GenBank database. Thus genes encoding C–P lyase and phosphonoacetaldehyde hydrolase had a more limited presence in the oceanic environment, being present at only 30 and 29% respectively of the sites sampled by the GOS (Table 3), and in 5 and 2% of sampled genomes (Table 4). However the phnA gene that encodes phosphonoacetate hydrolase was more widely distributed in the oceans, being present at almost over 78% of the sites sampled by the GOS Expedition (Table 3), and in an estimated 11% of genome-equivalents (Table 4). Similarly, the genes that encode the PhnY*/PhnZ phosphonate catabolic pathway (Martinez et al., 2010) were of limited occurrence in sequenced bacterial genomes but were amongst the most widely distributed of those analyzed in the GOS database; as noted above, four of the five sequenced isolates that possess phnY*/phnZ are marine strains (see Distribution in Sequenced Bacterial Strains of Additional Genes Involved in Phosphonate Metabolism).
This discrepancy between the apparent abundances of the various pathways of phosphonate catabolism and their prevalence in sequenced bacterial genomes may be in part explained by the fact that the majority of sequenced strains that possess a copy of the C–P lyase and phosphonoacetaldehyde hydrolase operons belong to the gamma-proteobacteria and other classes of prokaryotes associated with gastrointestinal pathogenesis, whereas those strains found to possess the phosphonoacetate hydrolase and PhnY*/PhnZ pathways were in many cases alpha-proteobacteria. Most of these had been isolated from the marine environment; they comprise the predominant class of prokaryotes in the upper layer of the sea surface (Wagner-Döbler and Biebl, 2006; Moran et al., 2007). Furthermore, since all those sequenced marine strains that possessed a phnA (or a phnX) – encoded pathway of 2-AEP degradation had an associated lysR gene encoding a transcriptional activator it is possible that this form of inducible regulation might be of competitive advantage, allowing cells to consume phosphonates as an additional source of C, (N), and P independently of ambient Pi concentrations. Given that expression of the genes that encode the C–P lyase operon in all cases studied appears to Pi-starvation-inducible, possession of a Pi-insensitive mechanism of phosphonate catabolism could be of particular relevance in niches of localized organic enrichment within the bulk oligotrophic oceans, such as “marine snow.”
The limited available data from other studies are broadly consistent with our findings. Thus a metagenomic study of a coastal marine mesocosm by Gilbert et al. (2009) found that phnA was contained in >3% of genomes sampled while analysis of a metagenomic database derived from a 4000-m depth profile sampled in the North Pacific Subtropical Gyre (Luo et al., 2011) found that phnA was present in approximately 32% of genomes sampled throughout the water column, but that C–P lyase genes were confined to P-limited surface waters. Martinez et al. (2010) in metagenomic analyses of depth profile samples from the Sargasso Sea and Hawaii Ocean Time Series found that phnA was particularly abundant in deeper waters (present in an average of 10% of genomes across all depths at the two sites). The phnY* and phnZ genes were present on average in 3 and 10% respectively of genomes at these sites (Martinez et al., 2010). Finally, the likely relative importances of phosphonate-P and ester-P within marine DOP can perhaps be gauged by comparison of the broadly similar abundances in GOS metagenomic samples of genes that encode C–P cleavage enzymes and those that encode the three major marine alkaline phosphatases (Table 4).
Summary/Conclusion
There is growing awareness of the importance of phosphonates in the P economy of the oceans, although analytical constraints continue to limit our knowledge of the identities, abundances, and biogenic origins of marine C–P compounds, and of the dynamics of their biotransformations. Genomic, metagenomic (and increasingly, metatranscriptomic) evidence all point to the widespread utilization of phosphonate-P by the marine microflora; the indications these provide that regulation of C–P bond cleavage activity may be niche-dependent require further exploration. In addition there is need for further experimental proof of strong in silico suggestions that marine bacteria may be of importance as C–P producers, in addition to their better-known role as phosphonate consumers.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank CONACYT, Mexico, for a studentship to Juan F. Villarreal-Chiu.
References
- Agarwal V., Borisova S. A., Metcalf W. W., van der Donk W. A., Nair S. K. (2011). Structural and mechanistic insights into C-P bond hydrolysis by phosphonoacetate hydrolase. Chem. Biol. 18, 1230–1240 10.1016/j.chembiol.2011.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A. S., Ciocci M. J., Metcalf W. W., Kim J., Babbitt P. C., Wanner B. L., Martin B. M., Dunaway-Mariano D. (1998). Insights into the mechanism of catalysis by the P-C bond-cleaving enzyme phosphonoacetaldehyde hydrolase derived from gene sequence analysis and mutagenesis. Biochemistry 37, 9305–9315 10.1021/bi981223t [DOI] [PubMed] [Google Scholar]
- Barry R. J., Bowman E., McQueney M., Dunaway-Mariano D. (1988). Elucidation of the 2-aminoethylphosphonate biosynthetic pathway in Tetrahymena pyriformis. Biochem. Biophys. Res. Commun. 153, 177–182 10.1016/S0006-291X(88)81205-1 [DOI] [PubMed] [Google Scholar]
- Beier D., Gross R. (2006). Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 9, 143–152 10.1016/j.mib.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Benitez-Nelson C. R., O’Neill L., Kolowith L. C., Pellechia P., Thunell R. (2004). Phosphonates and particulate organic phosphorus cycling in an anoxic marine basin. Limnol. Oceanogr. 49, 1593–1604 10.4319/lo.2004.49.5.1593 [DOI] [Google Scholar]
- Beversdorf L. J., White A. E., Björkman K. M., Letelier R. M., Karl D. M. (2010). Phosphonate metabolism of Trichodesmium IMS101 and the production of greenhouse gases. Limnol. Oceanogr. 55, 1768–1778 10.4319/lo.2010.55.4.1768 [DOI] [Google Scholar]
- Björkman K. M., Karl D. M. (2003). Bioavailability of dissolved organic phosphorus in the euphotic zone at Station ALOHA, North Pacific Subtropical Gyre. Limnol. Oceanogr. 48, 1049–1057 10.4319/lo.2003.48.3.1049 [DOI] [Google Scholar]
- Borisova S. A., Christman H. D., Metcalf M. E. M., Zulkepli N. A., Zhang J. K., van der Donk W. A., Metcalf W. W. (2011). Genetic and biochemical characterization of a pathway for the degradation of 2-aminoethylphosphonate in Sinorhizobium meliloti 1021. J. Biol. Chem. 286, 22283–22290 10.1074/jbc.M111.237735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman E., McQueney M., Barry R. J., Dunaway-Mariano D. (1988). Catalysis and thermodynamics of the phosphoenolpyruvate/phosphopyruvate rearrangement. Entry into the phosphonate class of naturally occurring organophosphorus compounds. J. Am. Chem. Soc. 110, 5575–5576 10.1021/ja00224a054 [DOI] [Google Scholar]
- Bury-Mone S., Nomane Y., Reymond N., Barbet R., Jacquet E., Imbeaud S., Jacq A., Bouloc P. (2009). Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet. 5, e1000651. 10.1371/journal.pgen.1000651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. C., Han Y., Niu W., Kulakova A. N., Howard A., Quinn J. P., Dunaway-Mariano D., Herzberg O. (2006). Structure and kinetics of phosphonopyruvate hydrolase from Variovorax sp. Pal2: new insight into the divergence of catalysis within the PEP mutase/isocitrate lyase superfamily. Biochemistry 45, 11491–11504 10.1021/bi0521792 [DOI] [PubMed] [Google Scholar]
- Choi K. Y., Kim D., Chae J. C., Zylstra G. J., Kim E. (2007). Requirement of duplicated operons for maximal metabolism of phthalate by Rhodococcus sp. strain DK17. Biochem. Biophys. Res. Commun. 357, 766–771 10.1016/j.bbrc.2007.04.009 [DOI] [PubMed] [Google Scholar]
- Circello B. T., Eliot A. C., Lee J. H., van der Donk W. A., Metcalf W. W. (2010). Molecular cloning and heterologous expression of the dehydrophos biosynthetic gene cluster. Chem. Biol. 17, 402–411 10.1016/j.chembiol.2010.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirz R. T., O’Neill B. M., Hammond J. A., Head S. R., Romesberg F. E. (2006). Defining the Pseudomonas aeruginosa SOS response and its role in the global response to the antibiotic ciprofloxacin. J. Bacteriol. 188, 7101–7110 10.1128/JB.00807-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark L. L., Ingall E. D., Benner R. (1998). Marine phosphorus is selectively remineralized. Nature 393, 426–426 10.1038/30881 [DOI] [Google Scholar]
- Cooley N. A., Kulakova N. A., Villarreal-Chiu J. F., Gilbert J. A., McGrath J. W., Quinn J. P. (2011). Phosphonoacetate biosynthesis: in vitro detection of a novel NADP+-dependent phosphonoacetaldehyde-oxidizing activity in cell-extracts of a marine Roseobacter. Microbiology 80, 335–340 10.1134/S0026261711030076 [DOI] [PubMed] [Google Scholar]
- Dumora C., Lacoste A. M., Cassaigne A. (1989). Phosphonoacetaldehyde hydrolase from Pseudomonas aeruginosa – purification, properties and comparison with Bacillus cereus enzyme. Biochim. Biophys. Acta 997, 193–198 10.1016/0167-4838(89)90186-6 [DOI] [PubMed] [Google Scholar]
- Dyhrman S., Ammerman J. W., Van Mooy B. A. (2007). Microbes and the marine phosphorus cycle. Oceanography 20, 110–116 10.5670/oceanog.2007.54 [DOI] [Google Scholar]
- Dyhrman S., Chappell P. D., Haley S. T., Moffett J. W., Orchard E. D., Waterbury J. B., Webb E. A. (2006). Phosphonate utilization by the globally important marine diazotroph Trichodesmium. Nature 439, 68–71 10.1038/nature04203 [DOI] [PubMed] [Google Scholar]
- Dyhrman S., Haley S. (2006). Phosphorus scavenging in the unicellular marine diazotroph Crocosphaera watsonii. Appl. Environ. Microbiol. 72, 1452–1458 10.1128/AEM.72.1.252-260.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyhrman S. T., Benitez-Nelson C. R., Orchard E. D., Haley S. T., Pellechia P. J. (2009). A microbial source of phosphonates in oligotrophic marine systems. Nat. Geosci. 2, 696–699 10.1038/ngeo639 [DOI] [Google Scholar]
- Ford J. L., Kaakoush N. O., Mendz G. L. (2010). Phosphonate metabolism in Helicobacter pylori. Antonie Van Leeuwenhoek 97, 51–60 10.1007/s10482-009-9387-7 [DOI] [PubMed] [Google Scholar]
- Gebhard S., Cook G. M. (2008). Differential regulation of high-affinity phosphate transport systems of Mycobacterium smegmatis: identification of PhnF, a repressor of the phnDCE operon. J. Bacteriol. 190, 1335–1343 10.1128/JB.01764-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J. A., Thomas S., Cooley N. A., Kulakova A., Field D., Booth T., McGrath J. W., Quinn J. P., Joint I. (2009). Potential for phosphonoacetate utilization by marine bacteria in temperate coastal waters. Environ. Microbiol. 11, 111–125 10.1111/j.1462-2920.2008.01745.x [DOI] [PubMed] [Google Scholar]
- Gomez-Garcia M. R., Davison M., Blain-Hartnung M., Grossman A. R., Bhaya D. (2011). Alternative pathways for phosphonate metabolism in thermophilic cyanobacteria from microbial mats. ISME J. 5, 141–149 10.1038/ismej.2010.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilderbrand R. L. (1983). The Role of Phosphonates in Living Systems. Boca Raton, FL: CRC Press Inc [Google Scholar]
- Hori T., Horiguchi M., Hayashi A. (1984). Biochemistry of Natural C-P Compounds. Kyoto: Maruzen Ltd [Google Scholar]
- Horigane A., Horiguchi M., Matsumoto T. (1989). Metabolism of 2-amino-3-phosphono [3-14C] propionic acid in cell-free preparations of Tetrahymena. Biochim. Biophys. Acta 618, 383–388 [PubMed] [Google Scholar]
- Hove-Jensen B., McSorley F. R., Zechel D. L. (2011). Physiological role of PhnP-specified phosphoribosyl cyclic phosphodiesterase in catabolism of organophosphonic acids by the carbon-phosphorus lyase pathway. J. Am. Chem. Soc. 133, 3617–3624 10.1021/ja1102713 [DOI] [PubMed] [Google Scholar]
- Hove-Jensen B., Rosenkrantz T. J., Zechel D. L., Willemoës M. (2010). Accumulation of intermediates of the carbon-phosphorus lyase pathway for phosphonate degradation in phn mutants of Escherichia coli. J. Bacteriol. 192, 370–374 10.1128/JB.01131-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Su Z., Xu Y. (2005). The evolution of microbial phosphonate degradative pathways. J. Mol. Evol. 61, 682–690 10.1007/s00239-004-0349-4 [DOI] [PubMed] [Google Scholar]
- Ilikchyan I. N., McKay R. M., Zehr J. P., Dyhrman S. T., Bullerjahn G. S. (2009). Detection and expression of the phosphonate transporter gene phnD in marine and freshwater picocyanobacteria. Environ. Microbiol. 11, 1314–1324 10.1111/j.1462-2920.2009.01869.x [DOI] [PubMed] [Google Scholar]
- Jiang W. H., Metcalf W. W., Lee K. S., Wanner B. L. (1995). Molecular cloning, mapping, and regulation of Pho regulon genes for phosphonate breakdown by the phosphonatase pathway of Salmonella typhimurium LT2. J. Bacteriol. 177, 6411–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochimsen B., Lolle S., McSorley F. R., Nabi M., Stougaard J., Zechel D. L., Hove-Jensen B. (2011). Five phosphonate operon gene products as components of a multi-subunit complex of the carbon-phosphorus lyase pathway. Proc. Natl. Acad. Sci. U.S.A. 108, 11393–11398 10.1073/pnas.1104922108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes T. W., DeSieno M. A., Griffin B. M., Thomas P. M., Kelleher N. L., Metcalf W. W., Zhao H. (2010). Deciphering the late biosynthetic steps of antimalarial compound FR-900098. Chem. Biol. 17, 57–64 10.1016/j.chembiol.2009.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat S. S., Williams H. J., Raushel F. M. (2011). Intermediates in the transformation of phosphonates to phosphate by bacteria. Nature 480, 570–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl D. M., Beversdorf L. J., Björkman K. M., Church M. J., Martinez A., DeLong E. F. (2008). Aerobic production of methane in the sea. Nat. Geosci. 1, 473–478 10.1038/ngeo234 [DOI] [Google Scholar]
- Karl D. M., Björkman K. M., Dore J. E., Fujieki L., Hebel D. V., Houlihan T., Letelier R. M., Tupas L. M. (2001). Ecological nitrogen-to-phosphorus stoichiometry at station ALOHA. Deep Sea Res. II 48, 1449–1470 10.1016/S0967-0645(00)00149-1 [DOI] [Google Scholar]
- Kertesz M., Elgorriaga A., Amrhein N. (1991). Evidence for two distinct phosphonate-degrading enzymes (C-P lyases) in Arthrobacter sp. GLP-1. Biodegradation 2, 53–59 10.1007/BF00122425 [DOI] [PubMed] [Google Scholar]
- Kim A., Benning M. M., OkLee S., Quinn J. P., Martin B. M., Holden H. M., Dunaway-Mariano D. (2011). Divergence of chemical function in the alkaline phosphatase superfamily: structure and mechanism of the C-P bond cleaving enzyme phosphonoacetate hydrolase. Biochemistry 50, 3481–3494 10.1021/bi101659u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A. D., Baker A. S., Dunaway-Mariano D., Metcalf W. W., Wanner B. L., Martin B. M. (2002). The 2-aminoethylphosphonate-specific transaminase of the 2-aminoethylphosphonate degradation pathway. J. Bacteriol. 184, 4134–4140 10.1128/JB.184.15.4134-4140.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittredge J. S., Hughes R. R. (1964). The occurrence of alpha-amino-beta-phosphonopropionic acid in the Zoanthid, Zoanthus sociatus, and the ciliate, Tetrahymena pyriformis. Biochemistry 3, 991–996 10.1021/bi00895a026 [DOI] [PubMed] [Google Scholar]
- Kolowith L. C., Ingall E. D., Benner R. (2001). Composition and cycling of marine organic phosphorus. Limnol. Oceanogr. 46, 309–320 10.4319/lo.2001.46.2.0309 [DOI] [Google Scholar]
- Kulakova A. N., Kulakov L. A., Akulenko N. V., Ksenzenko V. N., Hamilton J. T. G., Quinn J. P. (2001). Structural and functional analysis of the phosphonoacetate hydrolase (phnA) gene region in Pseudomonas fluorescens 23F. J. Bacteriol. 183, 3268–3275 10.1128/JB.183.11.3268-3275.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakova A. N., Kulakov L. A., McGrath J. W., Quinn J. P. (2009). The construction of a whole-cell biosensor for phosphonoacetate, based on the LysR-like transcriptional regulator PhnR from Pseudomonas fluorescens 23F. Microb. Biotechnol. 2, 234–240 10.1111/j.1751-7915.2008.00082.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakova A. N., Wisdom G. B., Kulakov L. A., Quinn J. P. (2003). The purification and characterization of phosphonopyruvate hydrolase, a novel carbon-phosphorus bond cleavage enzyme from Variovorax sp. Pal2. J. Biol. Chem. 278, 23426–23431 10.1074/jbc.M301871200 [DOI] [PubMed] [Google Scholar]
- Kuwahara T., Yamashita A., Hirakawa H., Nakayama H., Toh H., Okada N., Kuhara S., Hattori M., Hayashi T., Ohnishi Y. (2004). Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. U.S.A. 101, 14919–14924 10.1073/pnas.0307609100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarche M. G., Wanner B. L., Crepin S., Harel J. (2008). The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol. Rev. 32, 461–473 10.1111/j.1574-6976.2008.00101.x [DOI] [PubMed] [Google Scholar]
- LaNauze J. M., Coggins J. R., Dixon H. B. F. (1977). Aldolase-like imine formation in mechanism of action of phosphonoacetaldehyde hydrolase. Biochem. J. 165, 409–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H., Bae B., Kuemin M., Circello B. T., Metcalf W. W., Nair S. K., van der Donk W. A. (2010). Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 17557–17562 10.1073/pnas.0911528107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B. L., Yeung P., Cheng C., Hill J. E. (2007). Distribution and diversity of phytate-mineralizing bacteria. ISME J. 1, 321–330 [DOI] [PubMed] [Google Scholar]
- Luo H., Benner R., Long R. A., Hu H. (2009). Subcellular localization of marine bacterial alkaline phosphatases. Proc. Natl. Acad. Sci. U.S.A. 106, 21219–21233 10.1073/pnas.0810731105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Zhang H., Long R. A., Benner R. (2011). Depth distributions of alkaline phosphatase and phosphonate utilization genes in the North Pacific Subtropical Gyre. Aquat. Microb. Ecol. 62, 61–69 10.3354/ame01458 [DOI] [Google Scholar]
- Martinez A., Tyson G. W., DeLong E. F. (2010). Widespread known and novel phosphonate utilization pathways in marine bacteria revealed by functional screening and metagenomic analyses. Environ. Microbiol. 12, 222–238 10.1111/j.1462-2920.2009.02062.x [DOI] [PubMed] [Google Scholar]
- McGrath J. W., Hammerschmidt F., Kählig H., Wuggenig F., Lamprecht G., Quinn J. P. (2011). Studies on the biodegradation of fosfomycin: synthesis of (13) C-labeled intermediates, feeding experiments with Rhizobium huakuii PMY1, and isolation of labeled amino acids from cell mass by HPLC. Chemistry 17, 13341–13348 10.1002/chem.201100725 [DOI] [PubMed] [Google Scholar]
- McGrath J. W., Hammerschmidt F., Preusser W., Quinn J. P., Schweifer A. (2009). Studies on the biodegradation of fosfomycin: growth of Rhizobium huakuii PMY1 on possible intermediates synthesised chemically. Org. Biomol. Chem. 7, 1944–1953 10.1039/b821829c [DOI] [PubMed] [Google Scholar]
- McGrath J. W., Hammerschmidt F., Quinn J. P. (1998). Biodegradation of phosphonomycin by Rhizobium huakuii PMY1. Appl. Environ. Microbiol. 64, 356–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J. W., Ternan N. G., Quinn J. P. (1997). Utilization of organophosphonates by environmental microorganisms. Lett. Appl. Microbiol. 24, 69–73 10.1046/j.1472-765X.1997.00350.x [DOI] [Google Scholar]
- McGrath J. W., Wisdom G. B., McMullan G., Larkin M. J., Quinn J. P. (1995). The purification and properties of phosphonoacetate hydrolase, a novel carbon-phosphorus bond cleavage enzyme from Pseudomonas fluorescens. Eur. J. Biochem. 234, 225–230 10.1111/j.1432-1033.1995.225_c.x [DOI] [PubMed] [Google Scholar]
- McMullan G., Harrington F., Quinn J. P. (1992). Metabolism of phosphonoacetate as the sole carbon and phosphorus source by an environmental bacterial isolate. Appl. Environ. Microbiol. 58, 1364–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendz G. L., Mégraud F., Korolik V. (2005). Phosphonate catabolism by Campylobacter spp. Arch. Microbiol. 183, 113–120 10.1007/s00203-004-0752-7 [DOI] [PubMed] [Google Scholar]
- Metcalf W. W., van der Donk W. A. (2009). Biosynthesis of phosphonic and phosphinic acid natural products. Annu. Rev. Biochem. 78, 65–94 10.1146/annurev.biochem.78.091707.100215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais M. C., Zhang G. F., Zhang W. H., Olsen D. B., Dunaway-Mariano D., Allen K. N. (2004). X-ray crystallographic and site-directed mutagenesis analysis of the mechanism of Schiff-base formation in phosphonoacetaldehyde hydrolase catalysis. J. Biol. Chem. 279, 9353–9361 10.1074/jbc.M312345200 [DOI] [PubMed] [Google Scholar]
- Moran M. A., Belas R., Schell M. A., González J. M., Sun F., Sun S., Binder B. J., Edmonds J., Ye W., Orcutt B., Howard E. C., Meile C., Palefsky W., Goesmann A., Ren Q., Paulsen I., Ulrich L. E., Thompson L. S., Saunders E., Buchan A. (2007). Ecological genomics of marine Roseobacters. Appl. Environ. Microbiol. 73, 4559–4569 10.1128/AEM.02580-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran M. A., Buchan A., Gonzalez J. M., Heidelberg J. F., Whitman W. B., Kiene R. P., Henriksen J. R., King G. M., Belas R., Fuqua C., Brinkac L., Lewis M., Johri S., Weaver B., Pai G., Eisen J. A., Rahe E., Sheldon W. M., Ye W., Miller T. R., Carlton J., Rasko D. A., Paulsen I. T., Ren Q., Daugherty S. C., Deboy R. T., Dodson R. J., Durkin A. S., Madupu R., Nelson W. C., Sullivan S. A., Rosovitz M. J., Haft D. H., Selengut J., Ward N. (2004). Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432, 910–913 10.1038/nature03170 [DOI] [PubMed] [Google Scholar]
- Nealson K. H., Venter J. C. (2007). Metagenomics and the global ocean survey: what’s in it for us, and why should we care? ISME J. 1, 185–190 10.1038/ismej.2007.43 [DOI] [PubMed] [Google Scholar]
- Palenik B., Brahamsha B., Larimer F. W., Land M., Hauser L., Chain P., Lamerdin J., Regala W., Allen E. E., McCarren J., Paulsen I., Dufresne A., Partensky F., Webb E. A., Waterbury J. (2003). The genome of a motile marine Synechococcus. Nature 424, 1037–1042 10.1038/nature01943 [DOI] [PubMed] [Google Scholar]
- Paytan A., McLaughlin A. (2007). The oceanic phosphorus cycle. Chem. Rev. 107, 563–576 10.1021/cr0503613 [DOI] [PubMed] [Google Scholar]
- Quin L. D. (1965). The presences of compounds with a carbon-phosphorus bond in some marine invertebrates. Biochemistry 4, 324–330 10.1021/bi00878a022 [DOI] [Google Scholar]
- Quin L. D. (2000). A Guide to Organophosphorus Chemistry. New York: John Wiley & Sons [Google Scholar]
- Quin L. D., Quin G. S. (2001). Screening for carbon-bound phosphorus in marine animals by high-resolution P-31 NMR spectroscopy: coastal and hydrothermal vent invertebrates. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 128, 173–185 10.1016/S1096-4959(00)00310-9 [DOI] [PubMed] [Google Scholar]
- Quinn J. P., Kulakova A. N., Cooley N. A., McGrath J. W. (2007). New ways to break an old bond: the bacterial carbon–phosphorus hydrolases and their role in biogeochemical phosphorus cycling. Environ. Microbiol. 9, 2392–2400 10.1111/j.1462-2920.2007.01397.x [DOI] [PubMed] [Google Scholar]
- Rizk S. S., Cuneo M. J., Hellinga H. W. (2006). Identification of cognate ligands for the Escherichia coli phnD protein product and engineering of a reagentless fluorescent biosensor for phosphonates. Protein Sci. 15, 1745–1751 10.1110/ps.062135206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. M., Freeman S., Seto H., Knowles J. R. (1988). Phosphonate biosynthesis: isolation of the enzyme responsible for the formation of a carbon-phosphorus bond. Nature 335, 457–458 10.1038/335457a0 [DOI] [PubMed] [Google Scholar]
- Sowell S. M., Wilhelm L. J., Norbeck A. D., Lipton M. S., Nicora C. D., Barofsky D. F., Carlson C. A., Smith R. D., Giovanonni S. J. (2009). Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 3, 93–105 10.1038/ismej.2008.83 [DOI] [PubMed] [Google Scholar]
- Sviridov A. V., Shushkova T. V., Zelenkova N. F., Vinokurova N. G., Morgunov I. G., Ermakova I. T., Leontievsky A. A. (2012). Distribution of glyphosate and methylphosphonate catabolism systems in soil bacteria Ochrobactrum anthropi and Achromobacter sp. Appl. Microbiol. Biotechnol. 93, 787–796 10.1007/s00253-011-3485-y [DOI] [PubMed] [Google Scholar]
- Ternan N. G., McGrath J. W., McMullan G., Quinn J. P. (1998). Organophosphonates: occurrence, synthesis and biodegradation by microorganisms. World J. Microbiol. Biotechnol. 14, 635–647 10.1023/A:1008848401799 [DOI] [Google Scholar]
- Ternan N. G., Quinn J. P. (1998a). Phosphate starvation-independent 2-aminoethylphosphonic acid biodegradation in a newly isolated strain of Pseudomonas putida, NG2. Syst. Appl. Microbiol. 21, 346–352 10.1016/S0723-2020(98)80043-X [DOI] [PubMed] [Google Scholar]
- Ternan N. G., Quinn J. P. (1998b). In vitro cleavage of the carbon-phosphorus bond of phosphonopyruvate by cell extracts of an environmental Burkholderia cepacia isolate. Biochem. Biophys. Res. Commun. 248, 378–381 10.1006/bbrc.1998.8962 [DOI] [PubMed] [Google Scholar]
- Thomas S., Burdett H., Temperton B., Wick R., Snelling D., McGrath J. W., Quinn J. P., Munn C., Gilbert J. A. (2010). Evidence for phosphonate usage in the coral holobiont. ISME J. 4, 459–461 10.1038/ismej.2009.129 [DOI] [PubMed] [Google Scholar]
- Vera M., Pagliai F., Guiliani N., Jerez C. A. (2008). The chemolithoautotroph Acidithiobacillus ferrooxidans can survive under phosphate-limiting conditions by expressing a C-P lyase operon that allows it to grow on phosphonates. Appl. Environ. Microbiol. 74, 1829–1835 10.1128/AEM.02101-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Krüger W. M., Humphreys S., Ketley J. M. (1999). A role for the PhoBR regulatory system homologue in the Vibrio cholerae phosphate-limitation response and intestinal colonization. Microbiology 145, 2463–2475 [DOI] [PubMed] [Google Scholar]
- von Krüger W. M., Lery L. M. S., Soares M. R., de Neves-Manta F. S., Batista e Silva C. M., Neves-Ferreira A. G., Perales J., Bisch P. M. (2006). The phosphate-starvation response in Vibrio cholerae O1 and a phoB mutant under proteomic analysis: disclosing functions involved in adaptation, survival and virulence. Proteomics 6, 1495–1511 10.1002/pmic.200500238 [DOI] [PubMed] [Google Scholar]
- Wackett L. P., Shames S. L., Venditti C. P., Walsh C. T. (1987). Bacterial carbon-phosphorus lyase-products, rates, and regulation of phosphonic and phosphinic acid metabolism. J. Bacteriol. 169, 710–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner-Döbler I., Biebl H. (2006). Environmental biology of the marine Roseobacter lineage. Annu. Rev. Microbiol. 60, 255–280 10.1146/annurev.micro.60.080805.142115 [DOI] [PubMed] [Google Scholar]
- White A. K., Metcalf W. W. (2007). Microbial metabolism of reduced phosphorus compounds. Annu. Rev. Microbiol. 61, 379–400 10.1146/annurev.micro.61.080706.093357 [DOI] [PubMed] [Google Scholar]
- Yakovleva G. M., Kim S. K., Wanner B. L. (1998). Phosphate-independent expression of the carbon-phosphorus lyase activity of Escherichia coli. Appl. Microbiol. Biotechnol. 49, 573–578 10.1007/s002530051215 [DOI] [PubMed] [Google Scholar]
- Zhang G., Dai J., Lu Z., Dunaway-Mariano D. (2003). The phosphonopyruvate decarboxylase from Bacteroides fragilis. J. Biol. Chem. 278, 41302–41308 10.1074/jbc.M305976200 [DOI] [PubMed] [Google Scholar]