Abstract
Histidine (His) is one of the standard amino acids in proteins, and plays a critical role in plant growth and development. The chemical properties of the imidazole side group allow His to participate in acid-base catalysis, and in the co-ordination of metal ions. Despite the biological importance of this molecule, His biosynthesis has been somewhat neglected in plants, in stark contrast to micro-organisms where the study of this pathway was fundamental in the discovery of operon structure and regulation by attenuation. With the recent isolation of histidinol-phosphate phosphatase, all the enzymes of His biosynthesis have now been identified in Arabidopsis, and several lines of evidence have implicated ATP-phosphoribosyl transferase (which catalyses the first committed step of the pathway) as playing an important role in the regulation of this pathway. However, little is known about the transcriptional regulation of the His biosynthetic genes, nor how demand for this amino acid is balanced with other metabolic requirements in plants. Similarly, the pathway of His catabolism has yet to be determined.
INTRODUCTION
The amino acid L-histidine (His) was discovered independently by Kossel and Hedin in 1896. Uniquely among the twenty standard amino acids, the imidazole side group of His has a pKa of approximately 6, allowing it to alternate between the protonated and unprotonated states under physiological conditions (Figure 1). This property allows His to participate in general acid-base catalysis (Fersht, 1999), and consequently it is present in the active sites of many enzymes. The unprotonated imidazole group is nucleophilic, and can thus act as a general base, for example in catalytic triads where the basic N abstracts protons from Ser, Thr or Cys residues, while the protonated form is a general acid. In addition, the unprotonated imidazole group of His plays important roles as a nucleophile in phosphoryl transfer, and in the co-ordination of metal ions in a range of metalloproteins, perhaps best known in the zinc finger motif (Fraústo da Silva and Williams, 2001; Harding, 2004). Free His also plays an important role in Ni tolerance in several Ni hyperaccumulating plant species, where it acts as a Ni-binding ligand (Krämer et al., 1996; Persans et al., 1999). Indeed, the high affinity of the Ni-His interaction has been exploited for many years in metal binding affinity chromatography where recombinant proteins containing a poly His-tag are purified using an immobilized Ni column.
Figure 1.
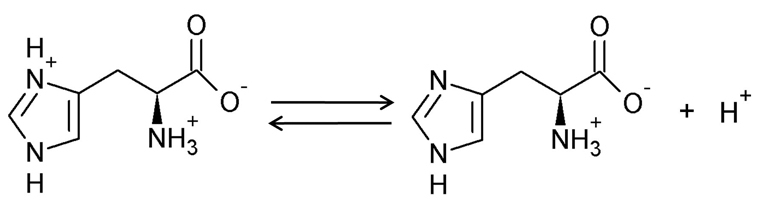
Structure of L-histidine. The imidazole side group is a weak acid with a pKa of approximately 6, allowing it to switch between the protonated and unprotonated states under cellular conditions.
While research on His biosynthesis in micro-organisms was instrumental in the discovery of operon structure, and the regulation of amino acid biosynthesis by attenuation (Ames et al., 1960; Roth and Ames, 1966), this pathway has been largely neglected in plants. Although His is an essential amino acid in humans (at least in infants), this pathway has not been the focus of attempts to increase His concentrations for nutritional purposes, as the His contents of commercially important crop plants are not generally regarded as limiting, in contrast to those of Lys, Met and Trp. Instead, a number of the enzymes involved in His biosynthesis in plants were first identified through efforts to develop selective herbicides to block this pathway (Mori et al., 1995; Ward and Ohta, 1999). Genes encoding all eight of the His biosynthetic (HISN) enzymes have now been identified from Arabidopsis (Petersen et al., 2010). In contrast to the genetic redundancy evident in the majority of amino acid biosynthetic pathways, five of the His biosynthetic enzymes are encoded by single copy genes, with only the HISN1, HISN5 and HISN6 genes duplicated in the Arabidopsis genome (Stepansky and Leustek, 2006; Petersen et al., 2010).
As would be expected, His is required throughout plant growth and development (Muralla et al., 2007). The most common phenotype in null Arabidopsis mutants defective in one of the five non-redundant HISN genes is seed abortion at the preglobular stage of embryo development, which can be rescued by supplying heterozygous plants with exogenous His (Muralla et al., 2007; Petersen et al., 2010). These data indicate that endogenous His concentrations in the surrounding heterozygous maternal tissues are insufficient to support embryonic development of null hisn mutants (Muralla et al., 2007). Homozygous null mutants obtained by His feeding of heterozygotes are able to germinate and grow, provided that His is available in the growth medium, however mature plants display a range of phenotypes including reduced apical dominance and fertility (Muralla et al., 2007). Mutants with weak hisn alleles can also display seedling morphological abnormalities including a short root phenotype in hpa1 (a mutant allele of HISN6A), and pale green cotyledons in apg10(a mutant allele of HISN3) (Noutoshi et al., 2005; Mo et al., 2006)
His biosynthesis in plants (Figure 2) occurs via the same metabolic route as in micro-organisms, beginning with the condensation of 5′-phosphoribosyl 1-pyrophosphate (PRPP) and ATP (Ohta et al., 2000; Stepansky and Leustek, 2006). His biosynthesis is tightly linked to nucleotide metabolism (Figure 3) as PRPP (derived from ribose-5-phosphate supplied by the pentose phosphate pathway) is also required for the de novo biosynthesis and salvaging of purines, pyrimidines and the pyridine nucleotide cofactors NAD and NADP (Alifano et al., 1996; Koslowsky et al., 2008). In addition, the intermediate 5′-phosphoribosyl-4-carboximide-5-aminoimidazole (AICAR) released at the branch point in the His biosynthetic pathway, enters the de novo purine biosynthetic pathway (Alifano et al., 1996; Ward and Ohta, 1999). In contrast, His biosynthesis is relatively isolated from other amino acid biosynthetic pathways, with the exception of Trp production, as PRPP is also required for the synthesis of N-(5′-phosphoribosyl) anthranilate from anthranilate (Koslowsky et al., 2008). However, the biosynthesis of His does require both Glu and Gln as N donors. The recent identification of the HISN7 enzyme in Arabidopsis, which catalyses the dephosphorylation of histidinol-P to histidinol (Petersen et al., 2010), has suggested the existence of additional potential links between His biosynthesis and other plant metabolic pathways. The recombinant HISN7 protein is able to catalyze the dephosphorylation of D-inositol-1 (or 3)-P and L-galactose-1-P in vitro (Torabinejad et al., 2009), suggesting that it may also function in myoinositol and ascorbate biosynthesis respectively, though this has yet to be demonstrated in vivo. His is the fourth most metabolically expensive amino acid to synthesize (after the aromatic amino acids Phe, Trp and Tyr), with estimates of 31–41 ATP molecules required per molecule of His produced (Alifano et al., 1996; Akashi and Gojobori, 2002; Swire, 2007). This may account for the relatively low abundance of His in proteins (where it is typically the 3rd or 4th least abundant amino acid) in line with the predictions of the cost minimization hypothesis, which seeks to explain the negative correlation between the frequency of amino acid usage and their biosynthetic cost (Seligmann, 2003).
Figure 2.
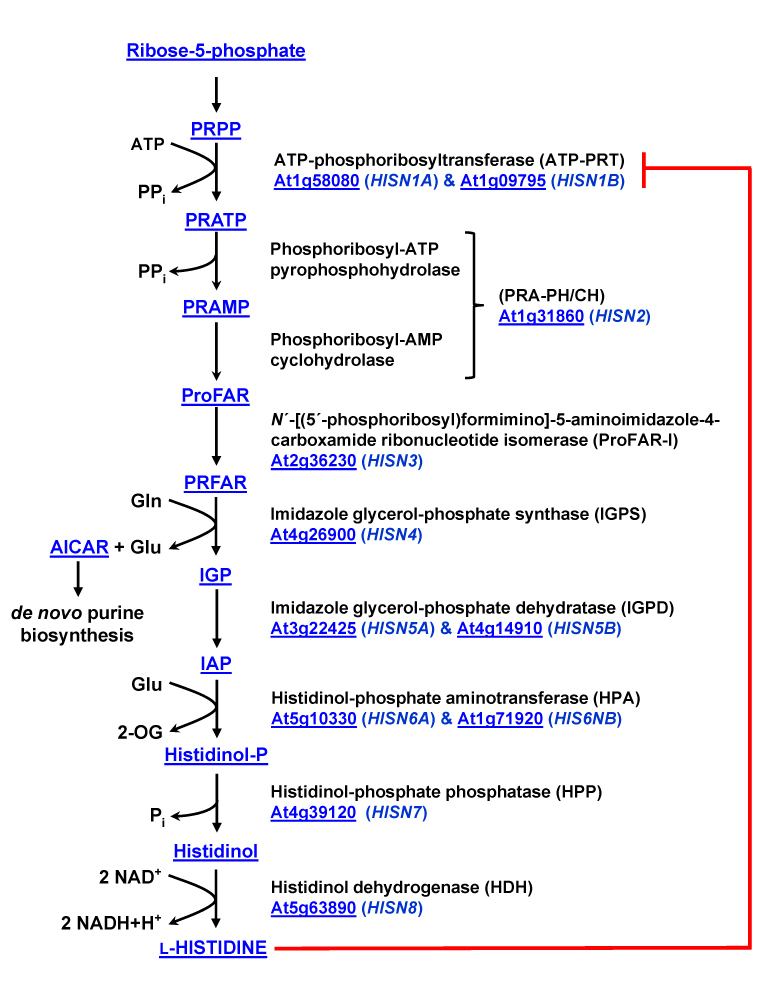
The histidine biosynthetic pathway in Arabidopsis. Abbreviations used for enzyme names are indicated in parentheses, and the corresponding Arabidopsis gene names and AGI codes are shown in blue. Allosteric inhibition of ATP-PRT activity by L-His is indicated in red. Abbreviations used for intermediates are: PRPP (5′-phosphoribosyl-1-pyrophosphate), PRATP (N′-5′-phosphoribosyl-ATP), PRAMP (N′-5′-phosphoribosyl-AMP), ProFAR (N′-[(5′-phosphoribosyl)formimino]-5-aminoimidazole-4-carboxamide) ribonucleotide, PRFAR (N′-[(5-phosphoribulosyl)formimino]-5-aminoimidazole-4-carboxamide) ribonucleotide, IGP (imidazole glycerol-phosphate), IAP (imidazole acetol-phosphate), AICAR (5′-phosphoribosyl-4-carboximide-5-aminoimidazole) and 2-OG (2-oxoglutarate). Hyperlinks to chemical structures and TAIR locus pages are provided.
Figure 3.
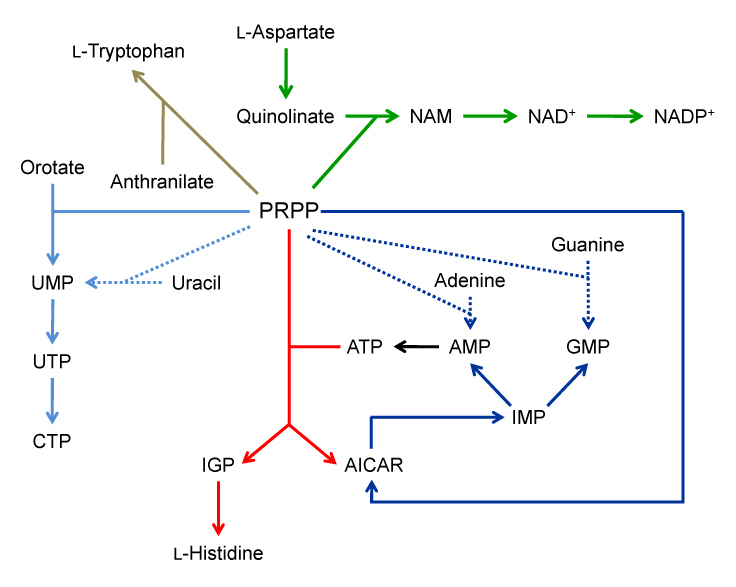
Metabolic links between His and Trp, purine, pyrimidine and pyridine nucleotide metabolism. Red arrows indicate the pathway of His biosynthesis, dark blue the de novo (solid line) and salvage (dotted line) pathways of purine biosynthesis, light blue the de novo (solid line) and salvage (dotted line) pathways of pyrimidine biosynthesis, green the pyridine nucleotide biosynthetic pathway, and brown the Trp biosynthetic pathway. For clarity only selected metabolic intermediates have been shown. Abbreviations used are: AICAR (5′-phosphoribosyl-4-carboximide-5-aminoimidazole), AMP (adenosine monophosphate), ATP (adenosine triphosphate), CTP (cytosine triphosphate), GMP (guanosine monophosphate), IGP (imidazole glycerol-phosphate), IMP (inosine monophosphate), NAD+ (nicotinamide adenine dinucleotide), NADP+ (nicotinamide adenine dinucleotide phosphate) NAM (nicotinic acid mononucleotide), PRPP (5′-phosphoribosyl-1-pyrophosphate), UMP (uridine monophosphate) and UTP (uridine triphosphate).
All the plant HISN proteins identified to date contain putative N-terminal plastid transit peptides, and several have been demonstrated to localize to the plastid (Nagai et al., 1993; Tada et al., 1995; Petersen et al., 2010), suggesting that His biosynthesis is confined to this organelle. However, His is mobile within the plant, as evidenced by its detection in both xylem and phloem exudates (Frommer et al., 1995; Krämer et al., 1996). While the identity of the protein(s) facilitating the export of His from the plastid is unknown, several proteins have been identified that can mediate the transport of His across the plasma membrane. Lysine Histidine Transporter 1 (LHT1 ; At5g40780) is a high affinity His transporter that plays important roles in His uptake from the soil across the root epidermis and in the supply of xylem-derived His to leaf mesophyll tissues (Chen and Bush, 1997; Hirner et al., 2006). Transport of His across the plasma membrane by Cationic Amino Acid Transporter 1 (CAT1; At4g21120) is sensitive to protonophores, suggesting that this protein functions as a His/proton symporter (Frommer et al., 1995).
While the enzymes involved in His biosynthesis have now been identified, we currently understand relatively little about the regulation of His biosynthesis in plants, nor how demand for this amino acid is balanced with other metabolic requirements. Similarly, the pathway of His catabolism in plants remains to be elucidated. This chapter explores our current knowledge of His biosynthesis in plants and its regulation, and identifies future avenues for research.
THE HISTIDINE BIOSYNTHETIC PATHWAY
ATP-phosphoribosyl transferase (ATP-PRT, EC 2.4.2.17) catalyses the first step in the His biosynthetic pathway, namely the condensation of ATP and PRPP to form N′-5′-phosphoribosyl-ATP (PRATP). ATP-PRT activity was first reported from extracts of several plant species by Wiater et al. (1971), but only 21 years later was the first ATP-PRT protein purified from wheat germ (Münzer et al., 1992). As with the ATP-PRT proteins from E. coli and S. typhimurium, the native wheat germ enzyme was shown to be a hexamer and required Mg2+ for activity (Voll et al., 1967; Münzer et al., 1992). There were, however, no subsequent reports of the corresponding cDNA sequence from wheat germ, and instead the first ATP-PRT encoding cDNA (THG1) was isolated from the Ni hyperaccumulator Thlaspi goesingense (Brassicaceae) through functional complementation of an E. coli hisG- mutant (Persans et al., 1999). DNA gel blot analysis suggested the presence of two genes encoding ATP-PRT in the T. goesingense genome (Persans et al., 1999). Subsequently it was demonstrated that the Arabidopsis genome also contains two ATP-PRT encoding genes, At1g58080 (HISN1A) and At1g09795 (HISN1B) (Ohta et al., 2000), as is the case in two other members of the Brassicaceae, Alyssum lesbiacum and Alyssum montanum (Ingle et al., 2005). Maximum likelihood analysis suggested that a gene duplication event occurred prior to the divergence of these three species of Brassicaceae from their last common ancestor, but after the monocot-eudicot split (Ingle et al., 2005). Analysis of purified recombinant Arabidopsis HISN1A protein revealed similar kinetic constants to those observed for the ATP-PRT enzymes from wheat germ and S. typhimurium, with Km values for ATP and PRPP of 0.6 mM and 0.13 mM respectively (Ohta et al., 2000). While ATP-PRT activity was also detected in crude extracts from E. coli expressing HISN1B, the recombinant protein could not be affinity purified due to instability (Ohta et al. 2000). Similarly, recombinant HISN1 B protein from A. lesbiacum was found to be less stable than recombinant HISN1A (R. A. Ingle & J.A.C. Smith, unpublished data), suggesting that the two ATP-PRT enzymes possess distinct biochemical properties that are evolutionary conserved. Null hisn1b mutants are viable, and display no obvious phenotype (Muralla et al., 2007), indicating that HISN1B is not essential. In contrast, null hisn1a mutants display a severe phenotype with stunted root development, and do not survive to maturity indicating that HISN1A and HISN1B are not fully redundant in Arabidopsis (Muralla et al., 2007).
The second and third reactions of the pathway are catalyzed by PRATP pyrophosphohydrolase (PRA-PH, EC 3.6.1.31) and phosphoribosyl-AMP cyclohydrolase (PRA-CH, EC 3.5.4.19) (Figure 2). There is significant variation in the domain organization of the proteins catalyzing these reactions in micro-organisms. In some bacteria these two enzymatic activities are catalyzed by a single polypeptide e.g. hisl in E coli and S. typhimurium (Chiariotti et al., 1986), while in others separate PRA-PH and PRA-CH enzymes are found e.g. hisl and hisE in the Actinobacteria (Fani et al., 2007). A third type of organization is observed in S. cerevisiae which has a trifunctional protein where the N-terminus has PRA-PH and PRA-CH activities, and the C-terminus histidinol dehydrogenase (HDH) activity (Donahue et al., 1982). Arabidopsis has a single gene, At1g31860 (HISN2), encoding a protein with both PRA-PH and PRA-CH activities, but lacking HDH activity (Fujimori and Ohta, 1998b), suggesting that the domain organization of this protein is most similar to that of the E. coli hisl protein. This was supported by the characterization of the two catalytic domains in the HISN2 protein, with the C-terminus shown to exhibit PRA-PH activity, and the N-terminus PRA-CH activity as is the case in the E. coli hisl protein (Fujimori and Ohta, 1998b). No data on the kinetic properties or 3D structure of the HISN2 protein have been reported to date. The fourth reaction is catalyzed by N′-[(5′-phosphoribosyl)formimino]-5-aminoimidazole-4-carboxamide ribonucleotide isomerase (ProFAR-I or BBMII isomerase, EC 5.3.1.16). In Arabidopsis this monofunctional enzyme is encoded by a single copy gene on chromosome II, At2g36230 (HISN3), and was identified through functional complementation of an E. coli hisA-mutant (Fujimori et al., 1998). As with HISN2, the HISN3 protein has not been characterized to date.
Imidazole glyercol-phosphate synthase (IGPS, EC 2.4.2.-) catalyses the branch point reaction of the His biosynthetic pathway resulting in the formation of IGP and 5′-phosphoribosyl-4-carboximide-5-aminoimidazole (AICAR) which enters the de novo purine biosynthetic pathway (Ward and Ohta, 1999). This is a two step process involving the transfer of an amide group from a Gln donor, followed by a cyclization reaction to generate the imidazole ring. In bacteria, the glutamine amidotransferase and cyclase activities are found on separate polypeptides, hisH and hisF, which form a heterodimer, while the HIS7 gene in S. cerevisiae encodes a single bifunctional protein (Kuenzler et al., 1993). The isolation of a single cDNA from Arabidopsis that could complement the His auxotrophy of a S. cerevisiae his7 mutant demonstrated that plants also possess a single bifunctional IGPS protein (Fujimori and Ohta, 1998a). As with the HIS7 protein from S. cerevisiae, the N-terminal domain of HISN4 (encoded by At4g26900) catalyses the amidotransferase reaction, while the C-terminal domain catalyses the cyclase reaction (Fujimori and Ohta, 1998a).
Dehydration of IGP to imidazole acetol-phosphate (IAP) is catalyzed by imidazole glycerol-phosphate dehydratase (IGPD, EC 4.2.1.19). IGPD activity was first detected by Wiater et al. (1971), and the first plant IGPD protein was purified from wheat germ by Mano et al. (1993) using a novel alkaline phosphatase-coupled assay. The wheat germ enzyme did not display histidinol-phosphate phosphatase (HPP) activity, indicating that it was a monofunctional IGPD, as is the HIS3 protein in S. cerevisiae, in contrast to the bi-functional hisB protein in E. coli which has both IGPD and HPP activities (Alifano et al., 1996). Arabidopsis has two genes encoding IGPD, At3g22425 (HIS5NA) and At4g14910 (HISN5B) (Tada et al., 1994), which display 82% identity at the amino acid level. Null hisn5a mutants are viable, indicating that HISN5A is not essential for growth and development (Muralla et al., 2007). However, no hisn5b mutants have been characterized to date, so it is unclear whether HISN5A and HISN5B are functionally redundant. All IGPD enzymes characterized to date require Mn2+ for activity (Mano et al., 1993; Tada et al., 1995), and for both the Arabidopsis and S. cerevisiae IGPD proteins it has been demonstrated that this requirement relates to the formation of catalytically active multimeric complexes. Removal of Mn2+ causes the 24 subunit holoenzyme to dissociate into inactive trimers, while re-addition leads of Mn2+ leads to reassembly of the active holoenzyme (Mano et al., 1993; Tada et al., 1995; Wilkinson et al., 1995; Glynn et al., 2005). The HISN5A protein has been crystallized and the structure determined to 3.0 Å resolution (Glynn et al., 2005). This work has demonstrated that Mn2+ is also required for formation of the active site, and that the reaction mechanism of IGPD likely proceeds through the binding of the imidazole ring of IGP to a di-Mn2+ cluster followed by conversion through diazafulvene and Δ2-enol intermediates to IAP (Glynn et al., 2005). Both the wheat germ and Arabidopsis enzymes also require a high concentration of 2-mercaptoethanol (100 mM) for maximal activity (Mano et al., 1993; Tada et al., 1995). The biochemical basis for this requirement is unclear as the mature IGPD proteins do not contain a Cys residue (Tada et al., 1994).
IAP is then transaminated (using Glu as an N donor) to histidinol-phosphate by histidinol-phosphate aminotransferase (HPA, EC 2.6.1.9). The first plant cDNA encoding HPA was identified from Nicotiana tabacum through functional complementation of an E. coli his C-mutant (El Malki et al., 1998). DNA gel blot analysis indicated the presence of two genes encoding HPA in N. tabacum, and two HPA homologues, At5g10330 (HISN6A) and At1g71920 (HISN6B) have also been identified in the Arabidopsis genome. The coding sequences of these paralogs differ by only 2 synonymous single nucleotide polymorphisms, but are highly divergent in their 5' upstream regions (Mo et al., 2006; Bikard et al., 2009). Intraspecific sequence comparisons with Arabidopsis lyrata have shown that HISN6B is the ancestral locus, with HISN6A resulting from a recent 3.3 kb duplication event centered around HISN6B (Moore and Purugganan, 2003). Whether both loci play a role in His biosynthesis in Arabidopsis is controversial however, as RT-PCR analysis indicated that, while HISN6A was expressed in all tissues, there was no evidence for transcriptional activity of HISN6B (Moore and Purugganan, 2003; Bikard et al., 2009), and null hisn6a mutants display embryonic lethality (Mo et al., 2006; Muralla et al., 2007). However, the observation that null hisn6a seeds (obtained through supplying HISN6A/hisn6a heterozygotes with exogenous His) are able to germinate and form a small rosette on media lacking His, suggests that HISN6B may be expressed at low levels in Arabidopsis (Muralla et al., 2007).
Histidinol-phosphate phosphatase (HPP, EC 3.1.3.15) was the last His biosynthetic enzyme to be identified in plants (Petersen et al., 2010), although HPP activity in plant extracts was first reported almost 40 years ago (Wiater et al., 1971). HPP enzymes isolated from micro-organisms prior to 2006 belong to one of two superfamilies, the DDDD (containing four conserved Asp residues) or PHP (polymerase and histidinol phosphatase) families. The Arabidopsis genome does not contain any genes encoding proteins with significant sequence similarity to members of either superfamily. Novel HPP proteins recently identified from several Actinobacteria including Corynebacterium glutamicum and Streptomyces coelicolor show no similarity to members of either the DDDD or PHP superfamily, and instead have high sequence identity to myoinositol monophosphatase enzymes (IMPs) (Mormann et al., 2006; Marineo et al., 2008). The Arabidopsis genome contains three putative IMPs, VTC4, myoinositol monophosphatase-like 1 (IMPL1) and IMPL2 (Torabinejad et al., 2009). Heterologous expression of IMPL2 but not IMPL1 was sufficient to rescue the His auxotrophy of an S. coelicolor hisN- mutant, and homozygous impl2 null mutants displayed embryonic lethality, which could be rescued by supplying heterozygous plants with exogenous His (Petersen et al., 2010). These data suggest that At4g39120 (IMPL2/HISN7) is likely the only HPP encoding gene in the Arabidopsis genome. The IMPL2 protein can catalyze the dephosphorylation of D-inositol-1(or 3)-P and L-galactose-1-P in vitro (Torabinejad et al., 2009), but it is not known whether it plays a role in myoinositol or ascorbate biosynthesis in vivo.
The final reactions of the His biosynthetic pathway, the two-step oxidation of histidinol to histidine via the aldehyde intermediate histidinal, are catalyzed by histidinol dehydrogenase (HDH, EC 1.1.1.23). HDH activity was first detected in extracts from several plant species, and an HDH protein from wheat germ partially purified by Wong and Mazelis (1981). HDH was subsequently purified from Brassica oleracea by affinity chromatography and shown to function as a homodimer (Nagai and Scheidegger, 1991), with the corresponding cDNA isolated shortly afterwards (Nagai et al., 1991). The cabbage HDH protein is the best characterized His biosynthetic enzyme from plants. One mol of His is produced per two mol of NAD+ reduced, and the enzyme displays a 870-fold preference for NAD+ compared to NADP+ despite its localization to the plastid (Nagai et al., 1993). As with the S. typhimurium HDH, the cabbage HDH uses a bi-uni-uni-bi-ping-pong mechanism whereby histidinol binds first, and NAD+ second, with His the last product to be released (Kheirolomoom et al., 1994). The oxidation of histidinol to histidinal is reversible, while that of histidinal to His is not, and a low dissociation constant for the histidinal-HDH complex explains the failure to detect free histidinal (Kheirolomoom et al., 1994). Site directed mutagenesis studies have established that Cys residues conserved between the cabbage and S. typhimurium HDH enzymes are not required for catalytic activity, while His261 is absolutely required as a Zn2+ binding ligand (Nagai et al., 1993; Nagai and Ohta, 1994). Subsequent NMR studies revealed that Zn2+ is present in the active site of the HDH enzyme and is required for substrate binding (Kanaori et al., 1996). A single HDH encoding gene, At5g63890 (HISN8) has been identified in the Arabidopsis genome, but the protein product has not been characterized to date.
While the route of His biosynthesis has now been established, the pathway of His catabolism in plants has not been elucidated. In micro-organisms and mammals, the major routes of His catabolism are deamination to urocanate (catalyzed by His ammonia lyase) which is then metabolized to glutamate via 4-imidazolone-5-propionate, or decarboxylation to histamine (catalyzed by His decarboxylase) (Mohammad et al., 1999). No His ammonia lyase enzyme has been identified in Arabidopsis to date, and a cDNA Initially annotated as a putative His decarboxylase was later demonstrated to encode a Ser decarboxylase, which showed no activity against His (Rontein et al., 2001).
REGULATION OF HISTIDINE BIOSYNTHESIS
While relatively little is known about the regulation of His biosynthesis in plants, several lines of evidence have indicated that ATP-PRT plays an important role in this process. The majority of enzymes catalyzing the committed step of an amino acid biosynthetic pathway are subject to allosteric inhibition by the end product of the pathway (Less and Galili, 2008) and, as is the case in micro-organisms, both ATP-PRT enzymes in Arabidopsis are subject to feedback inhibition by His (Ohta et al., 2000). However, recombinant HISN1A displayed greater sensitivity to feedback inhibition by His in crude bacterial extracts than did HISN1B, with IC50 values of 40 and 320 µM His respectively, suggesting that HISN1A might also be more susceptible to feedback inhibition in vivo. In the Ni hyperaccumulator A. lesbiacum, constitutively high mRNA levels of both HISN1A and HISN1B were found to correlate with elevated root concentrations of free His, in a comparison with closely related non-accumulating species (Ingle et al., 2005). Moreover, heterologous expression of the A. lesbiacum HISN1B cDNA under the control of the CaMV 35S promoter In Arabidopsis was sufficient to increase free His content by up to 15-fold in shoot tissue (Ingle et al., 2005). In a subsequent study, Rees et al. (2009) demonstrated that while over-expression of either the native HISN1A or HISN1B cDNA in Arabidopsis resulted in increased free His levels in shoot tissue (by up to 42-fold), over-expression of any other HISN cDNA had no effect on His content. Lastly, transgenic Arabidopsis plants expressing a PRPP synthase from Ashbya gossypii had increased levels of Trp but not His (measured as a percentage of total amino acids) (Koslowsky et al., 2008), suggesting that PRPP was not limiting for His biosynthesis. Together these data support the hypothesis that control of the pool of free His resides with ATP-PRT activity. The concentration of free His in Arabidopsis plastids has not been determined, and thus it is unclear to what extent feedback inhibition of ATP-PRT activity by His takes place in vivo, or whether the two ATP-PRT isoforms display differential sensitivity to His in vivo. However, while under normal conditions His may play a role in controlling free His content as an allosteric regulator of ATP-PRT activity, it appears that a sufficiently high degree of ATP-PRT over-expression is sufficient to overcome any such negative feedback by His and result in a substantial increase in the free His pool (Rees et al., 2009). This is in marked contrast with several other amino acid biosynthetic pathways e.g. Trp and Lys where the use of feedback insensitive forms of enzymes has been required to increase free amino acid contents in plants (Ufaz and Galili, 2008).
As is the case with the majority of amino acid biosynthetic pathways in plants, little is known about the mechanisms through which transcription of the His biosynthetic genes is regulated. The limited studies undertaken to date have indicated that the HISN genes in Arabidopsis are constitutively expressed in all tissues (being lowest in pollen), and at all developmental stages, suggesting His is produced throughout the plant (Fujimori and Ohta, 1998a; Fujimori et al., 1998; Ohta et al., 2000; Muralla et al., 2007). While no detailed study of the transcriptional regulation of the HISN genes in Arabidopsis has been undertaken, analysis of publically available microarray data using Genevestigator V3 (Hruz et al., 2008) has provided some evidence for differential expression of the HISN1 and HISN5 paralogs. Subfunctionalization has been proposed as a mechanism by which duplicated genes may be maintained in the genome. This involves the partitioning of function (or expression pattern) such that the complement of the duplicated genes represents the functional capability of the ancestral gene (Lynch and Conery, 2000; Duarte et al., 2006). In Arabidopsis, both HISN1A and HISN5A are expressed at a similar level in root and rosette tissues, while transcript levels of their paralogs HISN1B and HISN5B are >6-fold lower in root tissue than in rosette tissue. The observation that null hisn1a mutants display a severely stunted root phenotype (Muralla et al., 2007) suggests that HISN1B expression in root tissue does not provide sufficient His for root development, and is consistent with subfunctionalization of these paralogs. However, no corresponding phenotype exists in hisn5a mutants, which display normal root development despite the low level of HISN5B expression in this tissue (Muralla et al., 2007). The absence of detectable HISN6B expression in Arabidopsis (Moore and Purugganan, 2003; Bikard et al., 2009) suggests that this paralog may be(come) non-functional.
It is unclear to what extent the biosynthesis of His is co-ordinated with that of other amino acids in plants. In fungi, regulation of His biosynthesis is tightly co-ordinated with that of purine biosynthesis, through the BAS1p–BAS2p transcriptional complex which mediates the de-repression of several His biosynthetic genes in response to adenine limitation (Springer et al., 1996). In addition, His biosynthesis is regulated via the general nitrogen control mechanism by the bZIP transcription factor GCN4p. GCN4p mediates the co-ordinated de-repression of genes encoding amino acid biosynthetic enzymes (from all pathways except Cys), amino-acyl tRNA synthetases and purine biosynthetic enzymes in response to limitation for a single amino acid (Niederberger et al., 1981). Six of the HISN genes in Arabidopsis contain GCN4-like binding elements in their upstream regions (Stepansky and Leustek, 2006) which may be indicative of co-ordinate regulation of expression, but the regulatory protein(s) that bind to this motif have not been identified. The existence of a general nitrogen control mechanism in plants has been proposed, based on the observations that treatment of Arabidopsis with the IGPD inhibitor IRL1803 led to upregulation of HISN8, and of several genes involved in Lys, aromatic amino acid and purine biosynthesis, while the apg10 mutant (a weak allele of HISN3) had increased concentrations of most amino acids, including His (Guyer et al., 1995; Noutoshi et al., 2005). However, it has been suggested the response to IRL1803 may simply reflect stress imposed by His starvation rather than general control (Denby and Last, 1999). In shoot tissue of potato, a correlation between the concentrations of free His and eight other minor amino acids has been reported, but this phenomenon was not observed in wheat (Noctor et al., 2002). In contrast to results obtained following His starvation, transgenic Arabidopsis plants with elevated His content (up to 20% of the total free amino acid pool, compared to 0.6% in wild-type plants) displayed little or no differences in the concentrations of any other amino acid (Rees et al., 2009).
FUTURE PROSPECTS
The enzymes involved in His biosynthesis in Arabidopsis have now been identified (summarized in Table 1), and progress has been made in understanding how this important metabolic pathway is regulated. However, a number of key questions remain unanswered. Firstly, do the HISN1, HISN5 and HISN6 paralogs in Arabidopsis play distinct roles, consistent with the hypothesis of subfunctionalization of duplicated genes? While the lethality of the hisn1a mutants suggest that this is the case for the HISN1 paralogs, it is not clear whether the same holds for HISN5. The observation that the correlation coefficient, determined using the Arabidopsis co-expression tool (Manfield et al., 2006), for the two HISN5 paralogs is only 0.33 across a range of microarray experiments suggests that they may be differentially regulated. The isolation of a hisn5b mutant would also aid in determining whether this is the case. Confirmation of the role of HISN6B in His biosynthesis is also required, as current data are contradictory. The observation that hisn6a seedlings are viable (at least over the short-term) suggests that HISN6B is a functional HPA, yet RT-PCR analyses have failed to detect HISN6B mRNA in Arabidopsis. Detailed expression studies (tissue, developmental stage and environmental stimuli) of all three pairs of paralogs would aid in determining whether subfunctionalization has occurred. Secondly, His biosynthesis must be balanced with other metabolic demands in plants, but how is this achieved? For example, is there co-ordinate regulation of purine and His biosynthesis in plants, given their close metabolic links? Lastly, what is the route of His catabolism in plants? Since it has been suggested that the transcriptional regulation of catabolic genes plays an important role in the modulation of amino acid metabolism in response to environmental stimuli (Less and Galili, 2008), identification of the gene(s) involved in His catabolism is a priority for future research.
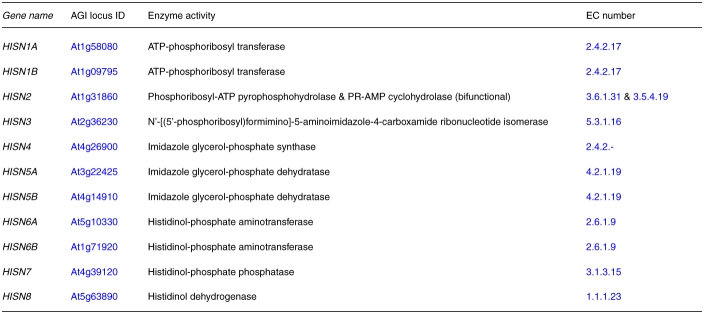
Table 1.
Arabidopsis loci and enzyme activities mentioned in this review article.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Research Foundation (NRF), South Africa and the University of Cape Town.
Footnotes
First published on February 2, 2011
Citation: Robert A. Ingle. (2011) Histidine Biosynthesis. The Arabidopsis Book 9:e0141. doi:10.1199/tab.0141
REFERENCES
- Akashi H., Gojobori T. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 2002;99:3695–3700. doi: 10.1073/pnas.062526999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alifano P., Fani R., Liò P., Lazcano A., Bazzicalupo M., Carlomagno M.S., Bruni C.B. Histidine biosynthetic pathway and genes: structure, regulation, and evolution. Microbiol. Rev. 1996;60:44–69. doi: 10.1128/mr.60.1.44-69.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames B.N., Garry B., Herzenberg L.A. The genetic control of the enzymes of histidine biosynthesis in Salmonella typhimurium. J. Mol. Biol. 1960;33:533–546. doi: 10.1099/00221287-22-2-369. [DOI] [PubMed] [Google Scholar]
- Bikard D., Patel D., Le Mette C., Giorgi V., Camilleri C., Bennett M.J., Loudet O. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science. 2009;323:623–626. doi: 10.1126/science.1165917. [DOI] [PubMed] [Google Scholar]
- Chen L., Bush D.R. LHT1, a lysine- and histidine-specific amino acid transporter in Arabidopsis. Plant Physiol. 1997;115:1127–1134. doi: 10.1104/pp.115.3.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiariotti L., Alifano P., Carlomagno M.S., Bruni C.B. Nucleotide sequence of the Escherichia coli hisD gene and of the Escherichia coli and Salmonella typhimurium hislE region. Mol. Gen. Genet. 1986;203:382–388. doi: 10.1007/BF00422061. [DOI] [PubMed] [Google Scholar]
- Denby K.J., Last R.L. Diverse regulatory mechanisms of amino acid biosynthesis in plants. Genet. Eng. (NY) 1999;21:173–189. doi: 10.1007/978-1-4615-4707-5_9. [DOI] [PubMed] [Google Scholar]
- Donahue T.F., Farabaugh P.J., Fink G.R. The nucleotide sequence of the HIS4 region of yeast. Gene. 1982;18:47–59. doi: 10.1016/0378-1119(82)90055-5. [DOI] [PubMed] [Google Scholar]
- Duarte J.M., Cui L., Wall P.K., Zhang Q., Zhang X., Leebens-Mack J., Ma H., Altman N., dePamphilis C.W. Expression pattern shifts following duplication indicative of subfunctionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol. Biol. Evol. 2006;23:469–478. doi: 10.1093/molbev/msj051. [DOI] [PubMed] [Google Scholar]
- El Malki F., Frankard V., Jacobs M. Molecular cloning and expression of a cDNA sequence encoding histidinol phosphate aminotransferase from Nicotiana tabacum. Plant Mol. Biol. 1998;37:1013–1022. doi: 10.1023/a:1006007125448. [DOI] [PubMed] [Google Scholar]
- Fani R., Brilli M., Fondi M., Lió P. The role of gene fusions in the evolution of metabolic pathways: the histidine biosynthesis case. BMC Evol. Biol. 2007;7:S4. doi: 10.1186/1471-2148-7-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. Structure and Mechanism in Protein Science. W.H. Freeman and Company; New York: 1999. [Google Scholar]
- Fraústo da Silva J.J.R., Williams R.J.P. The Biological Chemistry of the Elements: The Inorganic Chemistry of Life. Oxford University Press; Oxford: 2001. [Google Scholar]
- Frommer W.B., Hummel S., Unseld M., Ninnemann O. Seed and vascular expression of a high affinity transporter for cationic amino acids in Arabidopsis. Proc. Natl. Acad. Sci. USA. 1995;92:12036–12040. doi: 10.1073/pnas.92.26.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori K., Ohta D. An Arabidopsis cDNA encoding a bifunctional glutamine amidotransferase/cyclase suppresses the histidine auxotrophy of a Saccharomyces cerevisiae his7 mutant. FEBS Lett. 1998a;428:229–234. doi: 10.1016/s0014-5793(98)00535-3. [DOI] [PubMed] [Google Scholar]
- Fujimori K., Ohta D. Isolation and characterization of a histidine biosynthetic gene in Arabidopsis encoding a polypeptide with two separate domains for phosphoribosyl-ATP pyrophosphohydrolase and phosphoribosyl-AMP cyclohydrolase. Plant Physiol. 1998b;118:275–283. doi: 10.1104/pp.118.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori K., Tada S., Kanai S., Ohta D. Molecular cloning and characterization of the gene encoding N'-[(5'-phosphoribosyl)formimino]-5-aminoimidazole-4-carboxamide ribonucleotide (BBM II) isomerase from Arabidopsis thaliana. Mol. Gen. Genet. 1998;259:216–223. doi: 10.1007/s004380050807. [DOI] [PubMed] [Google Scholar]
- Glynn S.E., Baker P.J., Sedelnikova S.E., Davies C.L., Eadsforth T.C., Levy C.W., Rodgers H.F., Blackburn G.M., Hawkes T.R., Viner R., Rice D.W. Structure and mechanism of imidazoleglycerol-phosphate dehydratase. Structure. 2005;13:1809–1817. doi: 10.1016/j.str.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Guyer D., Patton D., Ward E. Evidence for cross-pathway regulation of metabolic gene expression in plants. Proc. Natl. Acad. Sci. USA. 1995;92:4997–5000. doi: 10.1073/pnas.92.11.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding M.M. The architecture of metal coordination groups in proteins. Acta Cryst. 2004;D60:849–859. doi: 10.1107/S0907444904004081. [DOI] [PubMed] [Google Scholar]
- Hirner A., Ladwig L., Stransky H., Okumoto S., Keinath M., Harms A., Frommer W.B., Koch W. Arabidopsis LHT1 is a highaffinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. Plant Cell. 2006;18:1931–1946. doi: 10.1105/tpc.106.041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., Widmayer P., Gruissem W., Zimmermann P. Genevestigator V3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics. 2008. p. 420747. [DOI] [PMC free article] [PubMed]
- Ingle R.A., Mugford S.T., Rees J.D., Campbell M.M., Smith J.A.C. Constitutively high expression of the histidine biosynthetic pathway contributes to nickel tolerance in hyperaccumulator plants. Plant Cell. 2005;17:2089–2106. doi: 10.1105/tpc.104.030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaori K., Uodome N., Nagai A., Ohta D., Ogawa A., Iwasaki G., Nosaka A.Y. Cd-113 nuclear magnetic resonance studies of cabbage histidinol dehydrogenase. Biochemistry. 1996;35:5949–5954. doi: 10.1021/bi951659y. [DOI] [PubMed] [Google Scholar]
- Kheirolomoom A., Mano J., Nagai A., Ogawa A., Iwasaki G., Ohta D. Steady-state kinetcis of cabbage histidinol dehydrogenase. Arch. Biochem. Biophys. 1994;312:493–500. doi: 10.1006/abbi.1994.1337. [DOI] [PubMed] [Google Scholar]
- Koslowsky S., Riegler H., Bergmüller E., Zrenner R. Higher biomass accumulation by increasing phosphoribosylpyrophosphate synthetase activity in Arabidopsis thaliana and Nicotiana tabacum. Plant Biotech. J. 2008;6:281–294. doi: 10.1111/j.1467-7652.2007.00314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer U., Cotter-Howells J.D., Charnock J.M., Baker A.J.M., Smith J.A.C. Free histidine as a metal chelator in plants that accumulate nickel. Nature. 1996;379:635–638. [Google Scholar]
- Kuenzler M., Balmelli C., Egli C.M., Paravinci G., Braus G.H. Cloning, primary structure, and regulation of the HIS7 gene encoding a bifunctional glutamine amidotransferase:cylcase from Saccharomyces cerevisiae. J. Bacteriol. 1993;175:5548–5558. doi: 10.1128/jb.175.17.5548-5558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Less H., Galili G. Principal transcriptional programs regulating plant amino acid metabolism in response to abiotic stresses. Plant Physiol. 2008;147:316–330. doi: 10.1104/pp.108.115733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J.S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Manfield I.J., Jen C.H., Pinney J.W., Michalopoulos I., Bradford J.R., Gilmartin P.M., Westhead D.R. Arabidopsis Co-expression Tool (ACT): web server tools for microarray-based gene expression analysis. Nucleic Acids Res. 2006;34:W504–W509. doi: 10.1093/nar/gkl204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano J., Hatano M., Koizumi S., Tada S., Hashimoto M., Scheidegger A. Purification and properties of a monofunctional imidazoleglycerol-phosphate dehydratase from wheat. Plant Physiol. 1993;103:733–739. doi: 10.1104/pp.103.3.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marineo S., Cusimano M.G., Limauro D., Coticchio G., Puglia A.M. The histidinol phosphate phosphatase involved in histidine biosynthetic pathway is encoded by SCO5208 (hisN) in Streptomyces coelicolor A3(2) Current Microbiol. 2008;56:6–13. doi: 10.1007/s00284-007-9014-7. [DOI] [PubMed] [Google Scholar]
- Mo X., Zhu Q., Li X., Li J., Zeng Q., Rong H., Zhang H., Wu P. The hpa1 mutant of Arabidopsis reveals a crucial role of histidine homeostasis in root meristem maintenance. Plant Physiol. 2006;141:1425–1435. doi: 10.1104/pp.106.084178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad T., Morrison H., HogenEsch H. Urocanic acid photochemistry and photobiology. Photochem. Photobiol. 1999;69:115–135. [PubMed] [Google Scholar]
- Moore R.C., Purugganan M.D. The early stages of genome duplication. Proc. Natl. Acad. Sci. USA. 2003;100:15682–15687. doi: 10.1073/pnas.2535513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori I., Fonnepfister R., Matsunaga S., Tada S., Kimura Y., Iwasaki G., Mano J., Hatano M., Nakano T., Koizumi S., Scheidegger A., Hayakawa K., Ohta D. A novel class of herbicides - specific inhibitors of imidazoleglycerol phosphate dehydratase. Plant Physiol. 1995;107:719–723. doi: 10.1104/pp.107.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mormann S., Lömker A., Rückert C., Gaigalat L., Tauch A., Pühler A., Kalinowski J. Random mutagenesis in Corynebacterium glutamicum ATCC 13032 using an IS6100-based transposon vector identified the last unknown gene in the histidine biosynthesis pathway. BMC Genomics. 2006;7:205. doi: 10.1186/1471-2164-7-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münzer S., Hashimoto-Kumpaisal R., Scheidegger A., Ohta D. Purification and properties of ATP-phosphoribosyl transferase from wheat germ. In: Murata N, editor. Research in Photosynthesis. Kluwer Academic Publishers; Dordrecht, The Netherlands: 1992. pp. 91–94. [Google Scholar]
- Muralla R., Sweeney C., Stepansky A., Leustek T., Meinke D. Genetic dissection of histidine biosynthesis in Arabidopsis. Plant Physiol. 2007;144:890–903. doi: 10.1104/pp.107.096511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai A., Ohta D. Histidinol dehydrogenase loses its catalytic function through the mutation of His261 Asn due to its inability to ligate the essential Zn. J. Biochem. 1994;115:22–25. doi: 10.1093/oxfordjournals.jbchem.a124298. [DOI] [PubMed] [Google Scholar]
- Nagai A., Scheidegger A. Purification and characterization of histidinol dehydrogenase from cabbage. Arch. Biochem. Biophys. 1991;284:127–132. doi: 10.1016/0003-9861(91)90274-m. [DOI] [PubMed] [Google Scholar]
- Nagai A., Suzuki K., Ward E., Moyer M., Mano J., Beck J., Tada S., Hashimoto M., Chang J., Ryals J., Scheidegger A., Ohta D. Histidinol dehydrogenase in higher plants. Purification, cloning and expression. In: Murata, N, editor. Research in photosynthesis. Kluwer Academic Publishers; Dordrecht, The Netherlands: 1993. pp. 95–98. [Google Scholar]
- Nagai A., Ward E., Beck J., Tada S., Chang J. Y., Scheidegger A., Ryals J. Structural and functional conservation of histidinol dehydrogenase between plants and microbes. Proc. Natl. Acad. Sci. USA. 1991;88:4133–4137. doi: 10.1073/pnas.88.10.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederberger P., Miozzari G., Hutter R. Biological role of the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1981;1:584–593. doi: 10.1128/mcb.1.7.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G., Novitskaya L., Lea P.J., Foyer C.H. Co-ordination of leaf minor amino acid contents in crop species: significance and interpretation. J. Exp. Bot. 2002;53:939–945. doi: 10.1093/jexbot/53.370.939. [DOI] [PubMed] [Google Scholar]
- Noutoshi Y., Ito T., Shinozaki K. ALBINO AND PALE GREEN 10 encodes BBMII isomerase involved in histidine biosynthesis in Arabidopsis thaliana. Plant Cell Physiol. 2005;46:1165–1172. doi: 10.1093/pcp/pci119. [DOI] [PubMed] [Google Scholar]
- Ohta D., Fujimori K., Mizutani M., Nakayama Y., Kunpaisal-Hashimoto R., Munzer S., Kozaki A. Molecular cloning and characterization of ATP-phosphoribosyl transferase from Arabidopsis, a key enzyme in the histidine biosynthetic pathway. Plant Physiol. 2000;122:907–914. doi: 10.1104/pp.122.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persans M.W., Yan X., Patnoe J.-M.M.L., Krämer U., Salt D.E. Molecular dissection of the role of histidine in nickel hyperaccumulation in Thlaspi goesingense (Halacsy). Plant Physiol. 1999;121:1117–1126. doi: 10.1104/pp.121.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen L.N., Marineo S., Mandala S., Davids F., Sewell B.T., Ingle R.A. The missing link in plant histidine biosynthesis: Arabidopsis myoinositol monophosphatase-like2 encodes a functional histidinol-phosphate phosphatase. Plant Physiol. 2010;152:1186–1196. doi: 10.1104/pp.109.150805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees J.D., Ingle R.A., Smith J.A.C. Relative contributions of nine genes in the pathway of histidine biosynthesis to control of free histidine concentrations in Arabidopsis thaliana. Plant Biotech. J. 2009;7:499–511. doi: 10.1111/j.1467-7652.2009.00419.x. [DOI] [PubMed] [Google Scholar]
- Rontein D., Nishida I., Tashiro G., Yoshioka K., Wu W.-I., Voelker D.R., Basset G., Hanson A.D. Plants synthesize ethanolamine by direct decarboxylation of serine using a pyridoxal phosphate enzyme. J. Biol. Chem. 2001;276:35523–35529. doi: 10.1074/jbc.M106038200. [DOI] [PubMed] [Google Scholar]
- Roth J.R., Ames B.N. Histidine regulatory mutants having altered histidyl-tRNA synthetase. J. Mol. Biol. 1966;22:325–334. doi: 10.1016/0022-2836(66)90135-5. [DOI] [PubMed] [Google Scholar]
- Seligmann H. Cost-minimization of amino acid usage. J. Mol. Evol. 2003;56:151–161. doi: 10.1007/s00239-002-2388-z. [DOI] [PubMed] [Google Scholar]
- Springer C., Künzler M., Balmelli T., Braus G.H. Amino acid and adenine cross-pathway regulation act through the same 5′-TGACTC-3′motif in the yeast HIS7 promoter. J. Biol. Chem. 1996;271:29637–29643. doi: 10.1074/jbc.271.47.29637. [DOI] [PubMed] [Google Scholar]
- Stepansky A., Leustek T. Histidine biosynthesis in plants. Amino Acids. 2006;30:127–142. doi: 10.1007/s00726-005-0247-0. [DOI] [PubMed] [Google Scholar]
- Swire J. Selection on synthesis cost affects interprotein amino acid usage in all three domains of life. J. Mol. Evol. 2007;64:558–571. doi: 10.1007/s00239-006-0206-8. [DOI] [PubMed] [Google Scholar]
- Tada S., Hatano M., Nakayama Y., Volrath S., Guyer D., Ward E., Ohta D. Insect cell expression of recombinant imidazoleglycerolphosphate dehydratase of Arabidopsis and wheat and inhibition by triazole herbicides. Plant Physiol. 1995;109:153–159. doi: 10.1104/pp.109.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada S., Volrath S., Guyer D., Scheidegger A., Ryals J., Ohta D., Ward E. Isolation and characterization of cDNAs encoding imidazoleglycerolphosphate dehydratase from Arabidopsis thaliana. Plant Physiol. 1994;105:579–583. doi: 10.1104/pp.105.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabinejad J., Donahue J.L., Gunesekera B.N., Allen-Daniels M.J., Gillaspy G.E. VTC4 Is a bifunctional enzyme that affects myoinositol and ascorbate biosynthesis in plants. Plant Physiol. 2009;150:951–961. doi: 10.1104/pp.108.135129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ufaz S., Galili G. Improving the content of essential amino acids in crop plants: goals and opportunities. Plant Physiol. 2008;147:954–961. doi: 10.1104/pp.108.118091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voll M.J., Appella E., Martin R. Purification and composition studies of phosphoribosyladenosine triphosphate: pyrophosphate phosphoribosyltransferase, the first enzyme of histidine biosynthesis. J. Biol. Chem. 1967;242:1760–1767. [PubMed] [Google Scholar]
- Ward E., Ohta D. Histidine biosynthesis. In: Singh BK, editor. Plant amino acids: Biochemistry and biotechnology. Marcel Dekker; New York: 1999. pp. 293–303. [Google Scholar]
- Wiater A., Krajewska-Grynkiewicz K., Kloptowski T. Histidine biosynthesis and its regulation in plants. Acta Biochim. Pol. 1971;18:299–307. [PubMed] [Google Scholar]
- Wilkinson K.W., Baker P.J., Rice D.W., Rodgers H. F., Stillman T.J., Hawkes T., Thomas P., Edwards L. Crystallization and analysis of the subunit assembly and quaternary structure of imidazoleglycerol phosphate dehydratase from Saccharomyces cerevisiae. Acta Cryst. 1995;D51:845–847. doi: 10.1107/S0907444995001569. [DOI] [PubMed] [Google Scholar]
- Wong Y.S., Mazelis M. Detection and properties of L-histidinol dehydrogenase in wheat germ. Phytochemistry. 1981;20:1831–1834. [Google Scholar]