Abstract
Hemolytic uremic syndrome (HUS) is a potentially life-threatening condition. It often occurs after gastrointestinal infection with E. coli O157:H7, which produces Shiga toxins (Stx) that cause hemolytic anemia, thrombocytopenia, and renal injury. Stx-mediated changes in endothelial phenotype have been linked to the pathogenesis of HUS. Here we report our studies investigating Stx-induced changes in gene expression and their contribution to the pathogenesis of HUS. Stx function by inactivating host ribosomes but can also alter gene expression at concentrations that minimally affect global protein synthesis. Gene expression profiling of human microvascular endothelium treated with Stx implicated a role for activation of CXCR4 and CXCR7 by their shared cognate chemokine ligand (stromal cell–derived factor-1 [SDF-1]) in Stx-mediated pathophysiology. The changes in gene expression required a catalytically active Stx A subunit and were mediated by enhanced transcription and mRNA stability. Stx also enhanced the association of CXCR4, CXCR7, and SDF1 mRNAs with ribosomes. In a mouse model of Stx-mediated pathology, we noted changes in plasma and tissue content of CXCR4, CXCR7, and SDF-1 after Stx exposure. Furthermore, inhibition of the CXCR4/SDF-1 interaction decreased endothelial activation and organ injury and improved animal survival. Finally, in children infected with E. coli O157:H7, plasma SDF-1 levels were elevated in individuals who progressed to HUS. Collectively, these data implicate the CXCR4/CXCR7/SDF-1 pathway in Stx-mediated pathogenesis and suggest novel therapeutic strategies for prevention and/or treatment of complications associated with E. coli O157:H7 infection.
Introduction
Hemolytic uremic syndrome (HUS) is a dramatic illness characterized by nonimmune hemolytic anemia, thrombocytopenia, and acute renal failure (1). A major breakthrough in our understanding of this disease occurred 3 decades ago, with the epidemiological association of HUS, hemorrhagic colitis, and stool isolates of E. coli, specifically serotype O157:H7. Since then, whenever there are reports of food- or water-source epidemics of virulent E. coli, the public and governments immediately become engaged, even more so since children are especially at risk of diarrhea-associated HUS, a leading cause of acquired pediatric acute renal failure. E. coli O157:H7 produce Shiga toxins (Stx), also known as Shiga-like toxins or verotoxins. These protein exotoxins are strongly implicated in the pathogenesis of this microangiopathy.
Beginning in May 2011, a large outbreak of diarrhea-associated HUS was documented in deaths in Northern Germany (2, 3). Rather than the typical enterohemorrhagic (EHEC) E. coli O157:H7, the serotype for this epidemic was an enteroaggregative (EAEC) E. coli O104:H4 that had acquired the stx2 gene, presumably via horizontal gene transfer (4, 5). This especially virulent strain was associated with a high incidence of HUS that resulted in over 40 deaths (2, 3). Sadly, this new epidemic underscores the seminal role that Stx play in the pathophysiology of diarrhea-associated HUS.
The family of Stx comprises various forms, including Stx1, Stx1c, Stx2, Stx2c, Stx2d, Stx2e, and Stx2f (6). Stx2 is the variant associated with more severe human disease (7, 8). It has long been appreciated that Stx are enzymatic ribosome-inactivating proteins that enter cells via binding to globotriaosylceramide (Gb3) (9) and catalyze the N-glycosidase–mediated removal of a single adenine residue within the 28S ribosomal RNA of the large 60S ribosome subunit (10). However, it is becoming increasingly apparent that Stx can have profound effects on endothelial gene expression and phenotype at concentrations that have only minimal effects on new protein synthesis (11). The extent to which Stx-induced phenotypic changes are caused by inhibition of global protein synthesis instead of activation of specific cellular pathways is currently unclear. We have previously shown that low concentrations of Stx selectively increase specific mRNA levels in endothelial cells (11). In these studies, Stx was found to potently increase preproendothelin-1 mRNA, the precursor of the endothelial vasoconstrictor endothelin-1 (ET-1). Moreover, the observed upregulation of ET-1 was attributed to Stx-induced stabilization of the transcript. Thus, low levels of Stx may act to modulate gene expression and endothelial phenotype in HUS.
To gain a greater understanding of Stx biology, we developed an in vitro model of extremely sensitive human microvascular endothelial cells (HMVECs) and performed gene expression profiling after Stx treatment. This agnosic approach identified perturbation of the CXCR4/CXCR7/ stromal cell–derived factor-1 (CXCR4/CXCR7/SDF-1) pathway, which consists of 2 G protein–coupled receptors (CXCR4, CXCR7) and their shared cognate chemokine ligand (SDF-1/CXCL12), as a key target of Stx-induced changes in mRNA expression. An emerging theme indicates that decreased and increased activity of the CXCR4/SDF-1 axis is relevant to diseases of the vasculature. For instance, CXCR4 and/or SDF-1 deficiency have been shown to cause perinatal lethality in mice, due, in part, to a defective vasculature (12–14). Cxcr4–/– and Sdf1–/– mice exhibit defective vascular remodeling in the gastrointestinal tract (12, 13) and disorganized glomerular blood vessels in the kidneys (14). In the latter study, podocyte-derived SDF-1 was shown to be required for proper patterning of CXCR4+ endothelial cells. This axis has also been shown to be fundamental in postnatal angiogenesis and vascular repair (15–18). In contrast, an overactive CXCR4/SDF-1 pathway has been associated with various forms of kidney pathology, including glomerulopathies (19–21). For example, podocyte-specific expression of CXCR4 was shown to disrupt glomerular function (19). Taken together, these findings suggest that tight regulation of this axis is essential for homeostasis.
Given its central roles in kidney and endothelial biology, the CXCR4/CXCR7/SDF-1 pathway was of particular interest in our studies on Stx-mediated pathophysiology. In the studies reported here, we have defined a mechanism by which Stx affects gene expression, especially engagement of select target RNAs on ribosomes in Stx-treated cells. In addition, we have developed and characterized a murine model of pathogenesis and discovered a key role for this important biological pathway in mediating experimental Stx-induced disease. Finally, the clinical relevance of the work was addressed. We found elevated levels of SDF-1 in plasma samples from children who were destined to develop HUS as a complication of E. coli O157:H7 infection. Taken together, our data implicate an important role for the CXCR4/CXCR7/SDF-1 pathway in the pathogenesis of HUS.
Results
Low concentrations of Stx affect global metabolism in the microvascular endothelium.
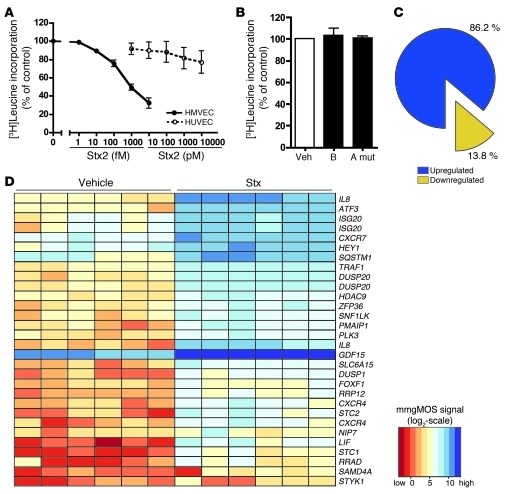
To study Stx-mediated pathogenesis of HUS, we established an in vitro model using primary HMVECs. The Stx sensitivity of HMVECs isolated from multiple donors was assessed after 24 hours of exposure to Stx1 or Stx2 (Figure 1A and Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI57313DS1). HMVECs showed a concentration-dependent inhibition of leucine incorporation into nascent proteins, with decreased levels of protein synthesis observed at concentrations as low as 10 fM Stx2. The IC50 for leucine incorporation in HMVECs was 170.9 ± 50.3 fM Stx2, more than 3 × 105 times less than the IC50 observed for aortic endothelial cells used in previous studies (11). Similar results were obtained using [3H]methionine incorporation as an alternate measure of nascent protein synthesis (Supplemental Figure 1B). Stx2 also inhibited DNA and RNA synthesis, although this inhibition required higher concentrations of toxin (Supplemental Figure 1, C and D). Protein synthesis was also measured in HUVECs, a classic in vitro endothelial model. Consistent with previous reports (22, 23), protein synthesis inhibition in this macrovascular endothelial cell type was shown to be approximately 105 times less sensitive to Stx (Figure 1A), highlighting important phenotypic differences, especially sensitivity to Stx, among endothelial cells cultured from different vascular beds. Global protein synthesis was unaffected in cells treated with the receptor-binding B subunit alone or a holotoxin containing 2 amino acid substitutions in the enzymatic A subunit (Figure 1B). Lactate dehydrogenase (LDH) release by cells treated with Stx2 demonstrated minimal or no cytotoxicity with 1,000 fM or 10 fM, respectively (Supplemental Figure 1E). Annexin V staining also indicated only minor apoptosis (1,000 fM, 24 hours), which could be blocked by the pan-caspase inhibitor, ZVAD (Supplemental Figure 1F).
Figure 1. Stx have potent effects on endothelial cell metabolism and phenotype.
HMVECs or HUVECs were treated with vehicle (Veh) or the indicated concentrations of (A) Stx2, (B) Stx1 B subunit alone (B) (1,000 fM), or holotoxin with a double mutation in the A subunit (A mut) (1,000 fM) for 24 hours. One hour before harvest, 1 μCi [3H]leucine was added to assess de novo protein synthesis. Radioactivity incorporated into TCA-precipitable material was quantitated, and data were normalized to vehicle-treated cells. The mean ± SEM of 14 experiments (HMVECs with Stx2), 5 experiments (HUVECs), or 3 experiments (mutants), triplicate determinations, is shown. (C) HMVECs (n = 6 replicates) were treated with Stx2 (10 fM, 24 hours) or vehicle and subjected to Affymetrix GeneChip gene expression profiling. By combining probe sets identified as differentially expressed by all preprocessing algorithms and removing redundant probe sets, i.e., probe sets mapping to the same gene, it was determined that 86.2% of differentially expressed genes (318 out of 369 unique genes) were upregulated by treatment with Stx. (D) The top 30 differentially expressed probe sets (based on magnitude of differential expression) were summarized in a heat map with hierarchical clustering. Processed signal intensities (computed using the mmgMOS preprocessing algorithm) are depicted on a log2 scale, with rows representing differentially expressed probe sets and columns representing Stx- or vehicle-treated samples. Note that several probe sets correspond to the same genes.
Stx potently affect microvascular endothelial gene expression.
To better understand the role of Stx in pathogenesis, we conducted gene expression profiling studies on HMVECs treated with Stx2 (10 fM, 24 hours; n = 6). The approach in this study was to focus on a concentration of Stx2 that had relatively minor effects on overall protein synthesis. The reasoning behind this approach is twofold. Because free Stx has never been detected in human blood, it has been speculated that endothelial cells are exposed to very small amounts of Stx at any given time. More importantly, at low concentrations of Stx, the majority of the cellular translation machinery is still functional, so changes in mRNA levels are more likely to correspond to changes in protein levels.
The endothelial gene expression pattern observed was highly gene specific, as Stx altered expression of only 369 (2.5%) of the 14,500 genes represented on the microarray (Supplemental Table 1). Of the 369 unique, differentially expressed genes, 318 (86.2%) were upregulated (Figure 1C), compared with only 51 (13.8%) that were downregulated. A variety of preprocessing steps yielded comparable findings. Quantitative RT-PCR (qRT-PCR) was used to independently verify the microarray results for 11 genes, including those that were upregulated and downregulated. In addition to validating genes that displayed some of the highest absolute fold changes, we also confirmed increased expression for genes that were at the user-defined 2-fold threshold used in the statistical analysis of the arrays (Supplemental Figure 2). For example, ICAM1, predicted to be induced approximately 2 fold by our microarray analysis, was also confirmed by qRT-PCR.
The top 30 differentially expressed probe sets (based on magnitude of change) are summarized in a heat map (Figure 1D). Among the most highly induced genes were 2 closely related chemokine receptors, CXCR4 and CXCR7, which increased 16.5 and 7.1 fold, respectively. In addition, we discovered increased expression (2.0 fold) of the common cognate ligand for both of these receptors, SDF-1. Taken together, these data highlighted a CXCR4/CXCR7/SDF-1 pathway that was perturbed by Stx in the microvascular endothelium.
Stx alter the CXCR4/CXCR7/SDF-1 axis.
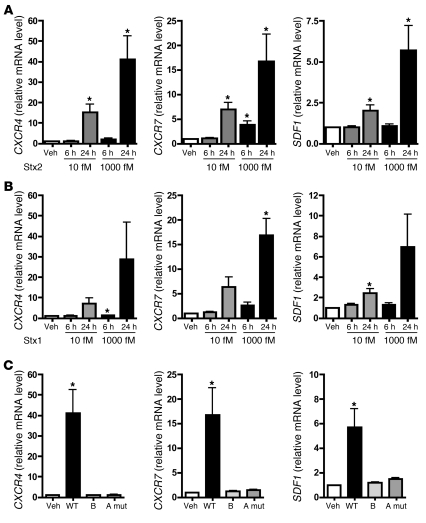
In addition to its critical roles in blood vessel development, CXCR4/SDF-1 signaling is important in revascularization of ischemic tissues and maintenance of the vasculature (16, 18, 24, 25). A disrupted CXCR4/SDF-1 axis has also been described in a variety of thrombotic and renal disorders (19–21, 26). Less is currently known about the biology of CXCR7, although it has been shown to be important in cardiac development and biology (27, 28). Since HUS is a microangiopathic disorder, at least in humans, we were interested in further examining the involvement of the CXCR4/CXCR7/SDF-1 pathway in Stx pathogenesis. We independently confirmed the microarray results using qRT-PCR and found time- and concentration-dependent increases in all 3 genes (Figure 2A). These findings were recapitulated in HMVECs treated with another Stx variant, Stx1 (Figure 2B), which shares 58% identity in amino acid sequence with Stx2 (29). SDF-1 is the sole known ligand for CXCR4 to date, while CXCR7 binds to SDF-1 as well as to interferon-inducible T cell α chemoattractant (I-TAC) (30). Although I-TAC was represented by 2 independent probe sets on the HG-U133A arrays, its expression was not significantly altered by Stx2 treatment (10 fM, 24 hours), and these findings were confirmed by qRT-PCR (data not shown).
Figure 2. Stx activates the CXCR4/CXCR7/SDF-1 pathway in microvascular endothelium by a mechanism requiring enzymatic activity of the Stx A subunit.
HMVECs were treated with the indicated concentrations of (A) Stx2 or (B) Stx1 for 6 or 24 hours, and mRNA levels were quantitated by qRT-PCR. (C) HMVECs were treated with 1,000 fM wild-type Stx2 (WT), Stx1 B subunit alone, or Stx2 holotoxin consisting of a mutant A subunit and wild-type B subunit (A mut). mRNA levels were measured by qRT-PCR. In all cases, data are normalized for GAPDH and 18S and are shown relative to vehicle-treated cells. Similar trends were observed whether or not data were normalized to housekeeping genes. Basal levels of CXCR4, CXCR7, and SDF1 were 1.53 ± 0.43 × 104, 7.40 ± 2.20 × 104, and 2.21 ± 0.69 × 104 copies/μg of input RNA, respectively. The mean ± SEM of at least 4 independent experiments is shown. *P < 0.05 vs. vehicle-treated cells.
Changes in gene expression require a catalytically active Stx A subunit.
Stx consist of pentameric receptor-binding B subunits covalently linked to an enzymatic A subunit, which depurinates eukaryotic 28S rRNA and inhibits protein synthesis (Figure 1, A and B, and ref. 10). To determine the basis for differential gene expression, we tested whether the B subunit alone was sufficient to upregulate CXCR4, CXCR7, and SDF1. Whereas 24-hour treatment of HMVECs with 1,000 fM wild-type Stx2 substantially increased expression of these genes, equimolar concentrations of the Stx1 B subunit, as expected, had no effect on protein synthesis (Figure 1B) and did not change mRNA levels (Figure 2C). This indicated that binding of the toxin to its receptor, Gb3, was not sufficient to induce expression of these genes. Similarly, when cells were treated with an inactive Stx2 holotoxin mutant containing 2 amino acid substitutions within the A subunit (E166Q, R169L) (31, 32), mRNA levels were not changed (Figure 2C). Taken together, these studies indicate that the A subunit must be present and catalytically active to elicit changes in gene expression.
Stx increases steady-state CXCR4 mRNA by transcriptional as well as posttranscriptional mechanisms.
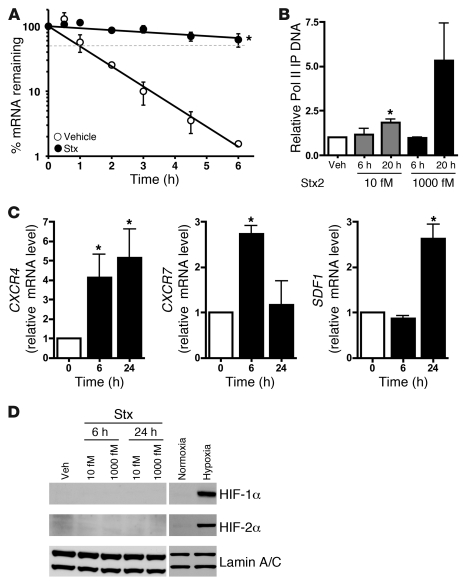
We and others have shown that Stx increases levels of mRNAs via transcript stabilization, without transcriptional activation (11, 33–35). These observations compel study of the role of ribosome and mRNA interactions, especially because related ribosome-inactivating proteins, such as ricin, have comparable effects on gene expression (11). Like ET1, CXCR4 is a short-lived mRNA with a half-life of 1.18 ± 0.09 hours (Figure 3A). Upon Stx treatment (1,000 fM, 20 hours), its mRNA half-life increased to 19.2 ± 7.8 hours (Figure 3A). Although we observed a dramatic posttranscriptional effect, a role for transcription could not be excluded. We therefore assessed the levels of RNA polymerase II associated with the CXCR4 promoter in Stx-treated HMVECs by ChIP. A concentration- and time-dependent increase in polymerase II recruited to the CXCR4 locus was observed (Figure 3B). Hence, even though transcriptional effects were modest relative to those of posttranscriptional mechanisms, a combination of both nuclear events and posttranscriptional mechanisms contributed to the marked mRNA induction of CXCR4 by Stx. This could explain why CXCR4 was among the most highly differentially expressed mRNA transcripts in our microarray studies (Figure 1D).
Figure 3. Stx enhances CXCR4 expression by both transcriptional and posttranscriptional mechanisms.
(A) HMVECs treated with vehicle or 1,000 fM Stx2 for 20 hours were subjected to transcription arrest with 10 μg/ml actinomycin D, and RNA was harvested at various times thereafter. Remaining CXCR4 mRNA was determined using qRT-PCR and normalized to 18S. The mean ± SEM of 3 independent experiments is shown. Half-lives were calculated using exponential regression to be 1.18 ± 0.09 hours in vehicle-treated cells and 19.2 ± 7.8 hours in Stx-treated cells. (B) HMVECs were exposed to 1,000 fM Stx2 for 20 hours, followed by ChIP using anti-RNA polymerase II (Pol II) antibodies. qRT-PCR for CXCR4 was then used to determine the amount of immunoprecipitated DNA (IP DNA). The mean ± SEM of at least 4 independent experiments is shown. (C) HMVECs were maintained under normoxic (21% O2) or hypoxic (<1% O2) conditions for the indicated times. qRT-PCR was used to determine mRNA levels. Data are normalized for 18S levels and are shown relative to normoxic cells. The mean ± SEM of 3 independent experiments is shown. Similar trends were observed whether or not data are normalized to housekeeping genes. (D) Western blot analyses were performed for HIF-1α and HIF-2α in HMVECs treated with 10 fM or 1,000 fM Stx for 6 or 24 hours. Lamin A/C was used as a loading control. Negative control and positive control were normoxic and hypoxic HMVECs. *P < 0.05 vs. vehicle or normoxia.
The CXCR4/CXCR7/SDF-1 pathway is regulated specifically by Stx through mechanisms distinct from those of other classic endothelial stimuli.
TNF-α is an important proinflammatory cytokine and potent endothelial cell activator, which has been suggested to mediate many of the in vivo effects of Stx. To determine whether the CXCR4/CXCR7/SDF-1 pathway responds nonspecifically when the endothelium is activated, or whether this response is specific to Stx, HMVECs were treated with TNF-α (10 ng/ml), and mRNA levels were measured using qRT-PCR (Supplemental Figure 3). Similar to that with Stx treatment, TNF-α increased CXCR7 expression, but, in contrast to that with Stx treatment, CXCR4 and SDF1 levels were dramatically reduced by TNF-α. These data indicate that Stx-mediated stimulation of the CXCR4/CXCR7/SDF-1 pathway in microvascular endothelium clearly differs from expression patterns produced by another classic endothelial activator.
An additional, biologically important vascular stimulus is hypoxia. Similar to the Stx-induced response, hypoxia increased expression of CXCR4, CXCR7, and SDF1 mRNAs (Figure 3C). However, Stx induced upregulation of all 3 components of the pathway by 24 hours, while hypoxia increased these receptor messages by 6 hours, followed by a delayed increase in their ligand at 24 hours. CXCR4 and SDF-1 expression is increased in a variety of hypoxic tissues and cells, and this pair is a target of HIF (16, 36, 37). CXCR7 has been shown to increase in lung, but not other tissues, during hypoxia (38). The similarity of the effects of Stx and hypoxia on the CXCR4/CXCR7/SDF-1 pathway, and our observation that CXCR4 is partly induced by transcription by Stx, prompted us to examine the potential involvement of HIF in Stx biology. Neither HIF-1α nor HIF-2α accumulated in Stx-treated cells as they did in hypoxic HMVECs (1 % O2, 24 hours) (Figure 3D). Hence, HIF appears not to have a direct transcriptional effect on this pathway in the context of Stx, and this discordance suggests that Stx and hypoxia target the pathway via distinct mechanisms.
Stx enhances association of CXCR4, CXCR7, and SDF-1 transcripts with cellular polyribosomes.
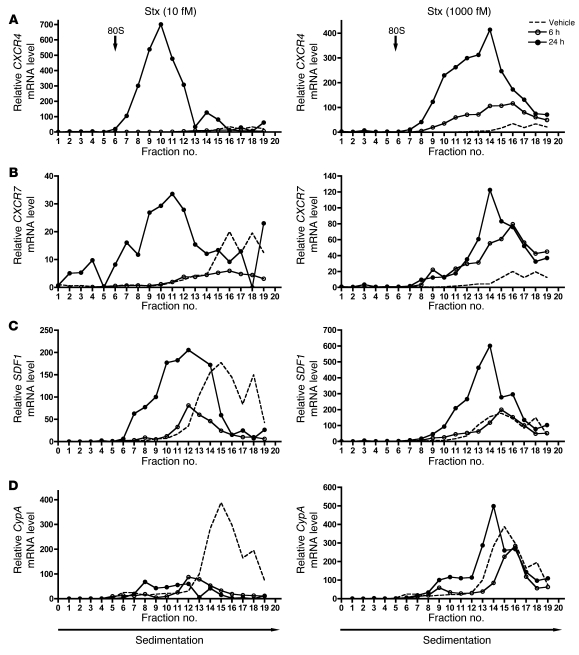
Because Stx in sufficient concentrations inactivates protein synthesis, it was important to address translation of CXCR4, CXCR7, and SDF-1 in addition to steady-state mRNA levels. We assessed whether the individual transcripts were being loaded onto ribosomes for translation by fractionating cellular ribosomes and their associated RNAs from Stx- and vehicle-treated HMVECs. We also used a synthetic exogenous mRNA transcript to control for mRNA recovery and first-strand cDNA synthesis efficiency for rigorous quantification of total cellular RNA and ribosome fraction RNA content. CXCR4 and CXCR7 transcripts accumulated in polyribosome fractions (heavier fractions) after 24-hour treatment with 10 fM and 1,000 fM Stx, compared with those in vehicle-treated cells (Figure 4, A and B). The most dramatic effect was the increase in CXCR4 mRNA associated with polyribosomes after addition of 10 fM Stx for 24 hours. This effect was also evident for both CXCR4 and SDF1. Importantly, there was increased abundance of these transcripts in the polyribosome fractions after treatment with only 10 fM Stx for 24 hours, a concentration at which there is only minimal inhibition of global protein synthesis (Figure 1A). A key finding is that, even in the absence of a significant increase in total levels of CXCR4 (1.00-fold change vs. vehicle) and CXCR7 (2.06-fold change vs. vehicle) mRNA after 6-hour treatment with 1,000 fM Stx (Supplemental Figure 4, B and 4C), there was increased association of these transcripts with the polyribosome fractions (Figure 4, A and B), suggesting increased translational efficiency for these particular transcripts. Increased endothelial cell surface expression of CXCR4 was also observed by flow cytometry (Supplemental Figure 4A). The observed translational effects of Stx on SDF1 were less pronounced than those on the SDF-1 receptors (Figure 4C) but were consistent with a previous report of enhanced SDF-1 release from microvascular endothelial cells exposed to Stx (39). The translational changes observed for these Stx target genes are specific and clearly different from other genes, such as cyclophilin A, which appears to fall off the ribosomes in the presence of only 10 fM Stx (Figure 4D). These findings indicate that Stx, a bacterial derived exotoxin that catalytically modifies the 28S ribosomal RNA of the large 60S ribosome subunit, has transcript-specific effects on the functional interaction between mRNA transcripts and the translating ribosome.
Figure 4. Stx-treated cells exhibit increased CXCR4, CXCR7, and SDF1 transcripts associated with cellular polyribosomes.
HMVECs were treated with vehicle or the indicated concentrations of Stx for 6 or 24 hours. Cellular ribosomes were fractionated based on density into 20 fractions, and RNA was purified from each fraction. mRNA levels were quantitated by qRT-PCR, normalized for luciferase mRNA, and shown relative to vehicle fraction no. 1. 80S refers to 80S ribosome. (A) CXCR4, (B) CXCR7, (C) SDF1, and (D) cyclophilin A. A representative experiment is shown.
Functional role of the CXCR4/CXCR7/SDF-1 pathway in Stx-mediated endothelial injury.
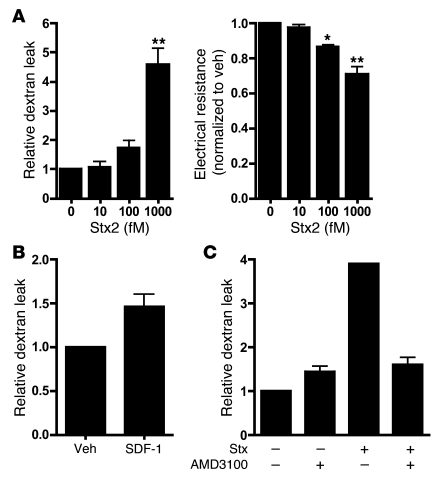
To explore the involvement of an activated CXCR4/CXCR7/SDF-1 pathway in Stx-mediated endothelial injury, we examined endothelial monolayer integrity using in vitro permeability assays. Dextran leak and reduced electrical resistance were evident across monolayers treated with Stx (Figure 5A). As previously shown (40), SDF-1α induces modest increases in endothelial permeability (Figure 5B). Inhibition of SDF-1/CXCR4 signaling using the CXCR4 antagonist AMD3100/plerixafor reduced permeability of Stx-treated HMVECs (1,000 fM Stx, 24 hours, n = 3; Figure 5C), thus implicating a role for this pathway in Stx-mediated endothelial damage.
Figure 5. Functional effects of Stx and the CXCR4/CXCR7/SDF-1 axis on endothelial permeability.
(A) Confluent endothelial monolayers were grown on transwell filters and treated with the indicated concentrations of Stx. Stx treatment causes increased dextran leak and decreased electrical resistance across the monolayer. The mean ± SEM of at least 3 independent experiments is shown. (B) Treatment of HMVECs with SDF-1 (50 ng/ml, 5 hours) induces dextran leak. The mean ± SEM of at least 3 independent experiments is shown. (C) Stx-induced HMVEC permeability is decreased by treatment of AMD3100/plerixafor. A representative experiment of 3 is shown (mean ± SD). *P < 0.05, **P < 0.001 vs. vehicle-treated cells.
Stx causes kidney dysfunction in an animal model of pathogenesis.
Based on our in vitro findings, we hypothesized that the CXCR4/CXCR7/SDF-1 pathway plays a role in Stx-mediated pathogenesis in vivo. To test this hypothesis, we established and characterized a mouse model of Stx exposure. Importantly, murine in vivo Stx models imperfectly recapitulate human HUS. Motto et al. have previously developed mice deficient in Adamts13, the vWF protease (41). When Adamts13 was ablated via gene targeting in the widely used inbred lab strain, C57BL/6, no phenotype was observed; however, knockout of this gene in the wild-derived Mus musculus castaneus (CAST/Ei) strain rendered these mice extremely susceptible to Stx-induced thrombotic microangiopathy. It was demonstrated that C57BL/6 mice express a truncated form of ADAMTS13 due to an insertional element within the genetic locus and are already hypomorphic for this protease (42). Interestingly, however, these mice do not develop overt thrombotic microangiopathy, and genetic modifiers that protect these inbred strains from such diseases have been mapped (41, 43). Consistent with this, castaneus mice are naturally more susceptible to thrombotic diseases. For these reasons, we opted to use CAST/Ei mice for our in vivo studies. Male and female CAST/Ei mice received a single i.v. injection of various concentrations of purified Stx2, and survival was monitored for a period of 10 days. While both Stx1 and Stx2 affected the CXCR4/CXCR7/SDF-1 pathway, our studies focused specifically on Stx2, as this is the isoform associated with the most severe forms of human disease (7, 8). As expected, increasing concentrations of Stx resulted in increased mortality, with a LD50 approximating 2,400 pg/g. In contrast, we noted that neither the Stx2 A subunit mutant (10 times the molar LD50) nor the Stx1 B subunit alone (50 times the LD50) were lethal over a 10-day observation interval (Figure 6A). Of note, animals treated with wild-type Stx2 exhibited significant weight loss (86.1% ± 1.9% of original weight at day 0; mean ± SEM; P < 0.05 vs. weight at day 0) within 4 days of challenge.
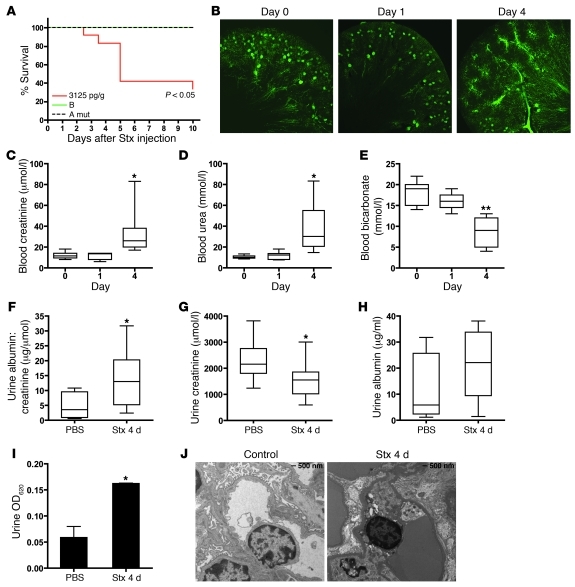
Figure 6. Stx challenge causes impaired renal function and mortality in vivo.
(A) CAST/Ei mice were injected i.v. with wild-type Stx2 (3,125 pg/g), Stx2 A subunit mutant (10 times LD50), or Stx1 B subunit (50 times LD50), and survival was monitored for 10 days. (B) Mice were injected with Stx2 (2,400 pg/g) and, at the indicated times, animals were perfused with fluorescent microbeads. A representative confocal image of a 100-μm section is shown (original magnification, ×5). Plasma collected from Stx-treated animals shows elevated (C) creatinine and (D) urea and reduced (E) bicarbonate levels by 4 days after Stx injection. Mice injected with Stx had (F) significantly higher urine albumin/creatinine ratios by 4 days after Stx injection, (G) lower urine creatinine, and (H) slightly elevated urine albumin. Horizontal bars indicate the median values, boxes indicate 25th to 75th percentiles, and whiskers indicate the minimum and maximum values. (I) Mice injected with Stx or PBS were injected i.v. with 0.1% Evans Blue dye. Urine samples were collected 1 hour later, and the amount of dye leak was measured spectrophotometrically. (J) Transmission electron micrographs of glomeruli from control and Stx-treated animals. *P < 0.05, **P < 0.001 vs. day 0 or PBS control.
Numerous measures used to evaluate kidney function were indicative of renal impairment. Fluorescence microangiography (fluorescent microbead size averaging 0.02 m) showed normal microvascular architecture within the renal cortex and medulla in control animals and those treated with Stx for 24 hours (Figure 6B). However, 4 days after Stx injection, cortical perfusion of the functional vasculature was severely affected, with some mice demonstrating marked changes in glomerular microperfusion (Figure 6B). At the same time, blood creatinine and urea were increased in Stx-treated animals (Figure 6, C and D). Decreased bicarbonate levels were also observed (Figure 6E). Urinalysis provided further evidence of kidney dysfunction. Urine albumin/creatinine ratios were significantly higher in Stx-treated animals (Figure 6F) as a result of significantly lower urine creatinine (Figure 6G) and elevated albumin leak (Figure 6H). Additionally, mice injected i.v. with Evans Blue dye, which binds to albumin and serves as a marker of albumin leak from the vasculature (44), showed release of Evans Blue/albumin complexes into the urine (Figure 6I). Consistent with these parameters of disruption of the glomerular permselectivity barrier by Stx, transmission electron microscopic examination of the glomerular microvasculature demonstrated partial effacement of podocyte foot processes and glomerular endothelial cell ultrastructural alterations, specifically swelling and vacuolization of the endothelial cells (Figure 6J). The fall in urine creatinine concentrations, together with the observed elevated plasma creatinine levels, are indicative of Stx-induced acute renal failure in this mouse model. Animals injected with either mutant form of the toxin showed blood creatinine and urea levels within the normal range for this strain (data not shown).
Effects of Stx on hematology and gastrointestinal biology.
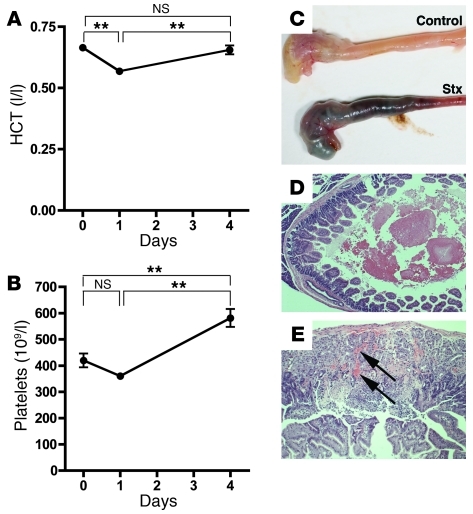
Thrombocytopenia and hemolytic anemia are prominent pathophysiological features of HUS in patients. The majority of previously described mouse models do not report anemia, and development of thrombocytopenia in one model was attributed to cotreatment with endotoxin (45). Hematologic analysis in our model showed a modest but significant decrease in hematocrit levels by 24 hours after Stx injection, which then increased to basal levels by day 4 (Figure 7A). As we previously noted (41) in the outbred CAST/Ei strain, fragmented red blood cells were not pronounced in peripheral blood smears (data not shown). Platelet levels were not significantly altered by 24 hours after toxin injection; however, they were substantially increased above basal levels by day 4 (Figure 7B), as in mice that do not develop thrombocytopenia following Stx (41).
Figure 7. Effects of Stx on animal hematology and gastrointestinal biology.
(A) CAST/Ei mice were injected i.v. with Stx2 (2,400 pg/g) on day 0. Blood samples were taken via saphenous vein at the indicated times. (A) Hematocrit (HCT) and (B) platelet levels are shown (n = 13 per time point). (C–E) Approximately 60% of mice develop gastrointestinal hemorrhage. (C) Gross images of stomach and small intestine from PBS- and Stx-treated mice. (D) H&E staining shows intraluminal hemorrhage (original magnification, ×50) and (E) intramucosal hemorrhage (arrows; original magnification, ×100). **P < 0.001.
E. coli O157:H7 cause bloody diarrhea and gut injury that normally precedes HUS. Previous studies have demonstrated expression of Gb3 on human gut epithelial cells (46). Additionally, Stx has previously been shown to induce cecal or colonic hemorrhagic lesions in rabbits (47), and both purified Stx and culture filtrate from O157:H7 produced hemorrhagic lesions in a rat model (48). Purified Stx damages the stomach and intestinal microvasculature, causing microinfarctions and both intramucosal and lumenal hemorrhagic lesions, including frank intraluminal blood content with upper gastrointestinal hemorrhage, in approximately 60% of treated mice (Figure 7, C–E).
Stx regulation of the CXCR4/CXCR7/SDF-1 pathway in vivo.
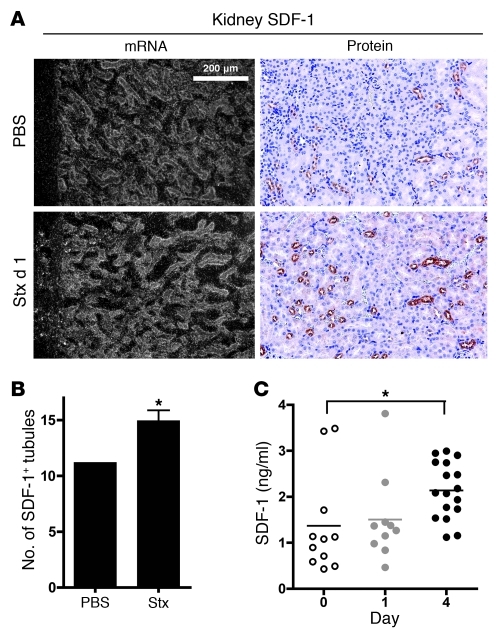
To determine whether Stx alters the CXCR4/CXCR7/SDF-1 axis in vivo, mice were injected with Stx, and expression of these messages was surveyed across tissues by in situ cRNA hybridization (Figure 8A and Supplemental Figure 6). Consistent with previous studies, CXCR4 and SDF-1 were highly expressed in the bone marrow (Supplemental Figure 5), and CXCR7 was expressed in the hearts of control animals (Supplemental Figure 6). Perturbation of the pathway at the mRNA level was evident as early as 24 hours after Stx injection. In particular, CXCR4 expression was increased in the thymus (Supplemental Figure 6). In contrast, CXCR7 levels decreased in cardiac tissue by 24 hours after Stx and remained low at day 4 (Supplemental Figure 6 and data not shown). In situ hybridization revealed increases in SDF1 mRNA levels in the kidney and liver by 1 day after toxin treatment (Figure 8A and Supplemental Figure 6), and immunohistochemistry demonstrated increased SDF-1 protein in the kidney of toxin-treated animals (Figure 8, A and B). A significant increase in the secreted SDF-1 peptide was observed in the blood of treated animals by day 4 (median, 2.1 ng/ml; interquartile range [IQR], 1.6–2.7 ng/ml) compared with that at day 0 (median, 1.1 ng/ml; IQR, 0.6–1.7 ng/ml) (Figure 8C). It is possible that decreased expression of CXCR7, which is a scavenging receptor (49, 50), contributed to increased blood levels. It is of great interest that our in vitro mechanistic studies demonstrated enhanced translation of the mRNA transcripts for CXCR4, CXCR7, SDF1 after Stx treatment.
Figure 8. Stx challenge causes dysregulation of SDF-1 in vivo.
(A) SDF-1 in situ hybridization and immunohistochemistry demonstrate increased SDF-1 production in the renal cortical tubules (original magnification, ×160). (B) The number of tubules staining positive for SDF-1 by immunohistochemistry were quantified in 6 nonoverlapping cortical fields (magnification, ×100). (C) Animals were injected with Stx2 (2,400 pg/g) on day 0, and SDF-1α levels were measured in blood plasma at the indicated times. Horizontal bars indicate the mean, and each symbol represents an individual animal. *P < 0.01.
CXCR4 inhibition improves animal survival.
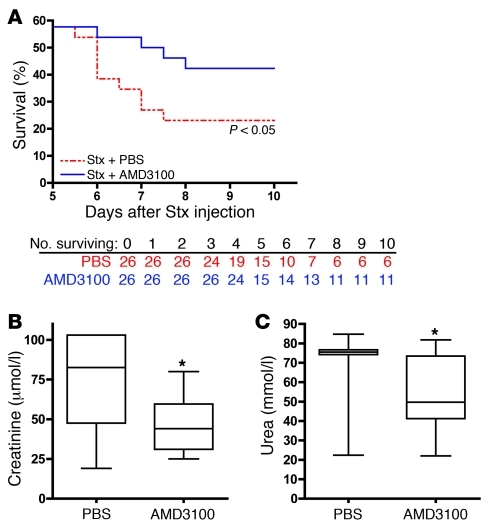
Given the multifaceted nature of the CXCR4/CXCR7/SDF-1 pathway, from proangiogenic functions for tissue repair (12, 13, 51) to its pathological roles in renal disease and inflammation (19–21), we examined its role in Stx pathogenesis in our in vivo model. We determined the effects of the CXCR4 antagonist, AMD3100/plerixafor, which is currently approved for stem cell mobilization in non-Hodgkin’s lymphoma and multiple myeloma and represents an attractive option avenue for patients (52). In conjunction with AMD3100/plerixafor administration, animals received 200–300 μl saline. We chose to replace fluids, because animals treated with Stx lost weight compared with untreated animals, and their hematocrit levels increased significantly between days 1 and 4 after Stx injection (Figure 7A). In addition, we observed albumin leak from the microvasculature in the kidney (Figure 6, G and H). Administration of AMD3100/plerixafor beginning 1 day after Stx challenge improved animal survival over the course of 10 days (Figure 9A). Survival between days 0 and 5 was identical in both the AMD3100/plerixafor and PBS groups, with 57.7% survival (15 out of 26 mice in each group). Beyond day 5, animal survival in the group receiving AMD3100/plerixafor improved significantly compared with that in the group receiving PBS (P < 0.05). 42.3% of mice receiving AMD3100/plerixafor survived to day 10, compared with only 23.1% of mice receiving PBS control injections. These data suggest a key role for the CXCR4/CXCR7/SDF-1 pathway in Stx pathogenesis and demonstrate that AMD3100/plerixafor treatment after Stx exposure enhances survival of Stx-treated animals.
Figure 9. Blockade of CXCR4/SDF-1 interaction by AMD3100 improves renal function and promotes animal survival.
(A) CAST/Ei mice were injected with Stx2 (2,400 pg/g) on day 0, followed by daily administration of AMD3100 (10 μg/g) or an equal volume of PBS beginning 1 day after Stx. Survival was monitored for 10 days (n = 26 per treatment group). (B and C) CAST/Ei mice were injected with Stx2 (625 pg/g) on day 0, followed by daily injections of AMD3100 (10 μg/g) or an equivalent volume of PBS vehicle. Plasma was collected 4 days after Stx injection and analyzed for (B) creatinine and (C) urea (n = 10–26 animals per group). Horizontal bars indicate the median values, boxes indicate 25th to 75th percentiles, and whiskers indicate the minimum and maximum values. *P < 0.05.
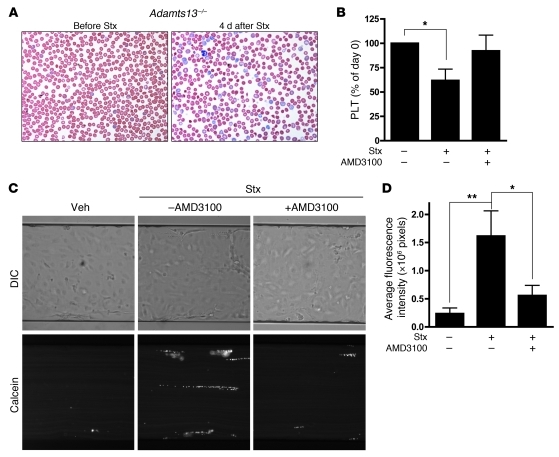
Kidney function was also restored in part after AMD3100/plerixafor treatment, as demonstrated by reductions in both blood creatinine and urea levels (Figure 9, B and C). Because common inbred lab mouse strains (45) and the wild-derived castaneus strain described here do not naturally develop remarkable anemia or thrombocytopenia when treated with purified Stx, we evaluated the effects of AMD3100/plerixafor on hematologic parameters in an Adamts13-deficient castaneus mouse model of thrombotic microangiopathy described elsewhere (41). Nolasco et al. have shown that high concentrations of Stx can induce ultra-large vWF multimer secretion from endothelial cells in culture and delayed multimer cleavage by ADAMTS13, which, the authors propose, leads to enhanced platelet aggregation (53). As previously reported (41), Adamts13–/– mice treated with Stx developed features consistent with thrombotic microangiopathy, including hemolytic anemia, with red blood cell fragments and marked reticulocytosis evident in a peripheral blood smear (Figure 10A), and thrombocytopenia (Figure 10B). Importantly, platelet counts were restored to normal levels with AMD3100/plerixafor treatment, while saline administration alone had no effect (Figure 10B). Despite the effect on platelets, no beneficial effect of the drug was observed with respect to hematocrit levels (data not shown). To further investigate the mechanism by which this may occur, platelet adhesion to endothelial cells under fluid flow conditions was measured in vitro after Stx treatment in the presence and absence of AMD3100/plerixafor. Toxin-treated endothelial cells exhibited enhanced platelet adhesion, and inhibition of CXCR4/SDF-1 interaction significantly reduced platelet strings attached to the endothelial monolayer (Figure 10, C and D). Other thrombotic abnormalities observed in HMVECs include increased tissue factor (Tf) (Supplemental Figure 7, A and B) and decreased tissue factor pathway inhibitor (TFPI) mRNA, protein levels, and biochemical activity (Supplemental Figure 7, C and D).
Figure 10. Blockade of CXCR4/SDF-1 interaction by AMD3100 restores platelet levels in vivo and inhibits platelet adhesion to endothelial cells under flow conditions in vitro.
(A) Blood smears from Adamts13–/– mice injected with Stx2 (625 pg/g) show evidence of red blood cell fragments 4 days after Stx exposure, with high numbers of reticulocytes (original magnification, ×150). (B) Adamts13–/– mice were injected with Stx2 (625 pg/g) on day 0, followed by daily injections of AMD3100 (10 μg/g) or PBS. Blood was analyzed for platelet levels prior to Stx injection and again at day 4 after toxin injection in both groups (n = 9–21 animals per group). *P < 0.05 vs. control animals that received neither Stx nor AMD3100. (C) HMVECs were grown to confluence in microfluidic channels and were either left untreated or were treated with 10 fM Stx2 (24 hours), after which calcein-labeled platelets were perfused at constant shear rates of 2.5 dyn/cm2 over the endothelial monolayer. Preincubation of HMVECs with AMD3100 (10 μM) significantly inhibited platelet adhesion. Images are representative of 5 independent experiments (original magnification, ×15). (D) Quantification of platelet adhesion to endothelial cells. Data are expressed as mean ± SEM from 5 independent experiments. *P < 0.05; **P < 0.01.
SDF-1 levels in patients with gastrointestinal E.
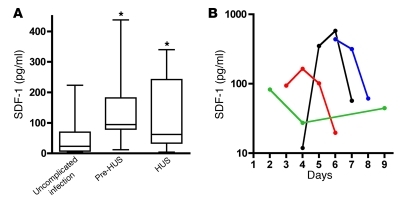
coli O157:H7 infection. To examine whether the CXCR4/CXCR7/SDF-1 axis is altered in patients with Stx-induced disease, SDF-1 levels were measured in plasma collected from children younger than 10 years of age with documented gastrointestinal E. coli O157:H7 infection (54, 55). The samples were obtained after a similar numbers of days of illness in 2 groups of children with infection: those with uncomplicated infection and those in whom HUS subsequently developed. The hematocrit, platelet count, and serum creatinine concentration were normal and similar in these 2 groups, as described previously (55). Thus, hemolysis, quantitative platelet abnormalities, and renal insufficiency had yet to evolve in the children in whom HUS subsequently developed. Circulating SDF-1 levels were compared in children whose infection resolved without further complication and in those children who subsequently developed HUS (Figure 11A). Despite the absence of clinically evident microangiopathic changes or renal insufficiency, the children in whom HUS subsequently developed had a 4-fold increase in median plasma levels of SDF-1 in the days preceding onset of HUS (median, 94.2 pg/ml, IQR, 79.3–181.6 pg/ml vs. median, 23.0 pg/ml, IQR, 7.3–69.46 pg/ml in individuals who were later diagnosed with the disease compared with children with uncomplicated infection, respectively). Median SDF-1 levels during HUS (median, 62.1 pg/ml; IQR, 33.69–242.2 pg/ml) remained significantly higher compared with those in individuals with uncomplicated infection. Importantly, the observed increases in blood SDF-1 levels preceded development of HUS, suggesting that aberrant regulation of the pathway, either as a cause or a consequence of endothelial injury, contributes to progression of the disease. Serial measurements were available for some patients and showed that SDF-1 levels peaked prior to development of HUS and subsequently decreased (Figure 11B).
Figure 11. Circulating SDF-1 levels in HUS patients.
SDF-1 levels were measured in plasma samples from individuals infected with E. coli O157:H7. (A) Infected samples obtained from patients with uncomplicated O157:H7 infection within 4 days after the onset of diarrhea (n = 36). Horizontal bars indicate the median values, boxes indicate 25th to 75th percentiles, and whiskers indicate the minimum and maximum values. Pre-HUS, samples collected prior to HUS diagnosis in those individuals who later progressed to HUS (n = 7); HUS, samples collected on the day of HUS diagnosis (n = 22). *P < 0.05. (B) SDF-1 levels over the course of E. coli O157:H7–mediated illness. Each line represents 1 patient, and the last data point for each patient was obtained on the day of HUS diagnosis.
Discussion
Stx-mediated HUS is a clinically important microangiopathy that may follow infection with enterohemorrhagic E. coli, particularly serotype O157:H7 and the newer O104:H4 enteroaggregative strain, which has acquired Stx expression. We have established an in vitro system to study Stx-mediated injury to cells that are relevant to Stx-mediated HUS and demonstrated the importance of the CXCR4/CXCR7/SDF-1 axis in eukaryotic injury. An understanding of the effects of Stx on its target vascular cells is critical to development of efficient intervention strategies. To this end, we established an in vitro model using primary microvascular endothelial cells. In many studies to date, HUVECs have been the endothelial model of choice to study effects of Stx, despite the fact that they express quantitatively less of the toxin receptor (Gb3) on their surface and, therefore, exhibit less sensitivity to the toxin (22, 23). Moreover, published reports have shown fundamental differences in the gene expression patterns of endothelial cells isolated from different vascular beds, in particular macrovascular- and microvascular-derived endothelium (56, 57). The microvascular localization of Stx pathology therefore emphasizes the relevance of our microvascular cell model. In addition, the exquisite sensitivity of microvascular endothelium allowed us to focus specifically on the effects of Stx, whereas other cells, such as HUVECs, have usually required sensitization with cytokine pretreatment (58, 59).
In the current study, we expanded our previous work, demonstrating that Stx upregulated ET1 mRNA that occurred at Stx concentrations that had minimal effects on global cellular protein synthesis (11). Specifically, we now more comprehensively characterized the gene expression profile of HMVECs after treatment with 10 fM Stx2, which produces only approximately 10% inhibition of protein synthesis. It is interesting that this low concentration of Stx had such potent and reproducible effects on endothelial phenotype. Previous expression profiling studies have been performed using Stx in HUVECs and in vivo (45, 60). We identified many of the same genes in our work; however, modulation of the CXCR4/CXCR7/SDF-1 pathway was not described in either previous study. This might be attributable to the cell model used in the former study, or source of tissue studied, or LPS cotreatment in the latter study. Interestingly, Guessous et al. observed elevated SDF-1α protein levels in conditioned media from HMVECs treated with Stx (39).
Our in vitro and in vivo data indicate that A subunit activity is indispensable for disruption of the CXCR4/CXCR7/SDF-1 pathway and for Stx-mediated mortality. First, studies with the mutant holotoxin and the receptor-binding B subunit demonstrated that the toxin must possess a functional A subunit in order to regulate expression of the CXCR4/CXCR7/SDF-1 pathway both in vitro and in vivo. Second, though our studies do not exclude important biological roles of the B subunit (61), our in vivo studies using the B subunit demonstrated 100% animal survival.
Most of the genes whose expression was affected were upregulated. We and others have previously shown that Stx increases particular mRNAs by increasing the half-life of the transcript (11, 33–35). We have also observed Stx-induced ET1 and ICAM1 mRNA expression via transcript stabilization but observed no change in nuclear run-off signal (ref. 11 and P.A. Marsden, unpublished observations). CXCR4, SDF-1, CXCR7, ICAM-1, and ET-1 share commonalities of basal mRNA lability, enhanced association with polyribosomes after Stx addition, and stabilization of mRNA transcripts after Stx treatment (ref. 11 and our unpublished observations). It is of great interest, therefore, that CXCR4 transcription is also enhanced by Stx, explaining the marked increases in CXCR4 mRNA steady-state levels in Stx-treated HMVECs. However, nuclear events play a smaller part in the overall increase in steady-state mRNA, as the slight induction in transcription of CXCR4 with 10 fM Stx cannot explain the substantial increase in transcript levels at that concentration.
The stark differences between the regulation of the CXCR4/CXCR7/SDF-1 pathway by Stx and TNF-α suggest that the effect of Stx on endothelial gene regulation is a function of this particular stimulus. CXCR4 and SDF-1 are well-known transcriptional targets of HIF, and as we showed, CXCR7 expression is also augmented in endothelium during hypoxia. Therefore, we considered the possibility that Stx is a hypoxia mimetic, possibly via stabilization of HIF-α subunits. However, HIF-1α and HIF-2α levels were not affected by Stx in HMVECs. Notably, while a subset of hypoxia-regulated genes were upregulated in our studies (e.g., VEGF, CXCL1), most did not change (e.g., GLUT1, GLUT3, VEGFR2, TIE2, ANGPTL4). This dichotomy provides further evidence that Stx acts via a mechanism distinct from the cellular response to hypoxic stress.
It is key mechanistically that, even without changes in mRNA levels, we observed substantial increases in polyribosome loading of specific target transcripts only 6 hours after Stx addition and with very low concentrations of toxin. Moreover, the magnitude of change in polyribosome association induced by Stx far outweighed the changes in mRNA steady-state levels (up to 700 fold vs. 40 fold, respectively). Comparison analysis demonstrates that only select mRNAs are abundantly associated with polyribosomes during Stx exposure. For instance, cyclophilin A mRNA transcripts exhibited decreased polyribosome association in Stx-treated endothelial cells. These findings highlight that changes in mRNA levels alone do not explain the effects and that specific translational regulation must be taken into consideration, especially in the case of a bacterial toxin that modifies ribosome activity. Together, these findings suggest the intriguing possibility that low levels of ribosome damage by small quantities of Stx may modulate selected cell signaling pathways that change ribosome function. Future mechanistic studies will be needed to gain greater understanding of how these putative signaling pathways modify the cell’s expression profile and phenotype and whether activation of the CXCR4/CXCR7/SDF-1 pathway is common to other ribosome inhibiting proteins.
To assess the biological relevance of our in vitro findings, we established an in vivo model of Stx toxicity. Kidney impairment is evident in this model, but hemoconcentration and gastrointestinal vascular damage also figure prominently. Our studies also implicated the CXCR4/CXCR7/SDF-1 pathway as a mediator of Stx pathology in a murine model. The pattern of tissue damage in our mouse model coincided with tissues in which the CXCR4/CXCR7/SDF-1 pathway has previously been demonstrated to play critical roles, including the gastrointestinal vasculature (12, 13) and kidneys (14, 19, 20). Interestingly, overexpression of CXCR4 or SDF-1 in the glomerulus causes kidney disease and induces proteinuria (19, 20). In our studies, the Stx-induced albuminuria and hemoconcentration were consistent with the observed in vitro endothelial permeability caused by Stx and SDF-1, which was reduced in our in vitro studies by inhibition of the pathway using AMD3100/plerixafor.
The aberrant regulation of the complex and biologically important CXCR4/CXCR7/SDF-1 axis offers new insight into the pathogenesis of HUS. We acknowledge the limitation of our studies, in that the murine model is not a robust clinical surrogate for Stx-induced HUS. However, advances in understanding the complexities of studying procoagulant pathways in murine models has led to the utilization of the outbred CAST/Ei strain, especially when deficient in ADAMTS13. We found that CXCR4 was expressed in lymphoid organs, and its expression was increased in thymus after Stx exposure. CXCR4 is expressed on a variety of cells, including T- and B-thymocytes, and its altered expression here might indicate an immune response to the toxin. CXCR7 has recently been described as a scavenger receptor that effectively creates and maintains a SDF-1 gradient to support CXCR4 signaling functions (49) by binding SDF-1 and targeting the chemokine for degradation (50). Stx decreases the amount of CXCR7 in the heart and, therefore, may increase the amount of free SDF-1 available to signal through CXCR4. Interestingly, while CXCR4/SDF-1 is essential in a variety of organ systems, CXCR7 is particularly important in proper development and function of the heart (27, 28), and CXCR7 present in cardiac tissue has previously been shown to sequester SDF-1 (50). To date, transgenic overexpression of SDF-1 in the heart has not been studied. However, adenovirus-mediated overexpression of CXCR4 in cardiac tissue increased SDF-1 levels and has been reported to enhance ischemia/reperfusion injury (62). The CXCR4/SDF-1 axis is emerging as a key pathway in vascular and other tissue biology, and, thus, a tight balance is necessary to maintain homeostasis. The increase in free SDF-1 might tip the balance toward CXCR4 signaling on cells expressing the receptor and contribute to the endothelial dysfunction characteristic of Stx-mediated disease. It is important to note that our in vivo studies specifically addressed mRNA levels, but as we have shown in vitro, Stx pathobiology can involve profound changes in translational efficiency of CXCR4, CXCR7, and SDF1 transcripts, and this mechanism may also be important in vivo in tissues in which changes in mRNA levels are not observed.
We addressed the in vivo role of the CXCR4/CXCR7/SDF-1 pathway using the CXCR4 inhibitor AMD3100/plerixafor. This bicylam compound is a specific antagonist of CXCR4 and does not appear to inhibit CXCR7 function (63). AMD3100/plerixafor increases CXCR7 affinity for SDF-1 and functions as a CXCR7 agonist (64). When AMD3100/plerixafor therapy was initiated in a clinically relevant setting, namely 1 day after Stx challenge, we observed a beneficial effect on overall survival (Figure 9A). Importantly, CXCR4 inhibition is beneficial to mice already exposed to the toxin. This has important clinical implications for patients who visit the hospital only after becoming ill from enteric infection with E. coli O157:H7.
Our data also demonstrate involvement of the CXCR4/CXCR7/SDF-1 axis in pediatric HUS cases, using plasma from a prospective cohort study of patients with E. coli O157:H7 infection (54, 55). Children in this phase of infection have prothrombotic abnormalities, which appear before, and plausibly contribute to, HUS. We demonstrated marked elevation in blood SDF-1 before the onset of HUS and azotemia, which could contribute to the pathogenesis of HUS. SDF-1 has been shown to enhance platelet activation by low levels of ADP (65) or thrombin (66), leading to increased aggregation (67) through CXCR4 platelet surface expression. Platelet thrombi are a characteristic feature of HUS. Our data indicate that Stx augment platelet adhesion in an in vitro flow model and that AMD3100/plerixafor could inhibit this. Moreover, we found increased and decreased activity of Tf and TFPI, respectively. Therefore, increased thrombin generation observed in HUS (55), in combination with increased SDF-1 levels, may be an important mechanism involved in Stx pathogenesis. The relationship between increased tissue factor, decreased tissue factor pathway inhibitor, and the effects of thrombin and SDF-1 on platelet aggregation in the in vivo setting warrants further study. SDF-1 is also stored in and released by activated platelets (68) and could exacerbate thrombosis in later stages of disease. Our in vitro studies demonstrate that AMD3100/plerixafor inhibited formation of platelet strings on the endothelium and may explain why Adamts13–/– mice receiving the drug displayed normal platelet levels after Stx challenge compared with those of animals that did not receive the drug.
Taken together, these studies identify Stx-induced activation of the CXCR4/CXCR7/SDF-1 pathway in vitro and in vivo, demonstrate that this pathway contributes to Stx-mediated pathogenesis, and present targets which we believe to be novel for therapeutic intervention. We have found that the median SDF-1 level decreases in overt HUS. SDF-1 binding to CXCR4 induces permanent internalization of the receptor from the cell surface, so that by the time HUS ensues, SDF-1 might be cleared from the circulation by CXCR4 or its other scavenger receptor, CXCR7. Alternatively, platelet decreases during HUS may account for the fall in median SDF-1 levels. Little overlap exists between the IQRs of SDF-1 measurements in children with uncomplicated infection and those who later developed HUS. Further studies are therefore warranted to determine the potential value of plasma SDF-1 level as a predictive indicator of complications and progression to HUS after infection with E. coli O157:H7.
In summary, aberrant activation of the CXCR4/CXCR7/SDF-1 pathway in Stx-mediated pathogenesis could provide novel insight into the molecular mechanisms underlying HUS. Specifically, CXCR4 inhibition improves renal function and, ultimately, survival in a murine model, and circulating SDF-1 in O157:H7-infected patients provides complementary data in support of an important role for this axis in the development of HUS.
Methods
Reagents
[methyl-3H]thymidine (70–90 Ci/mmol), [5,6-3H]uridine (30–50 Ci/mmol), and l-[3,4,5-3H(N)]leucine (>140 Ci/mmol) were purchased from NEN-Dupont. l-[methyl-3H]methionine (70–85 Ci/mmol) was purchased from Amersham Biosciences. TNF-α was purchased from Sigma-Aldrich. cDNA I.M.A.G.E. clones were obtained from ATCC or Invitrogen.
Toxin purification
Wild-type Stx2 holotoxin and Stx2 holotoxin mutant (E167Q and R170L substitutions in the A subunit) were extracted from recombinant E. coli strains using a combination of 10 μg/ml lysozyme and 0.1 mg/ml colimycin in PBS in the presence of 5 mM EDTA and 0.1 g/l PMSF (Sigma-Aldrich). The extract was cleared by centrifugation, dialyzed against 50 mM acetate buffer (pH 5), resolved on DEAE Sepharose Fast Flow (Amersham Biosciences), and eluted using a NaCl gradient 0 M to 0.5 M in 50 mM acetate buffer. Fractions containing Stx were precipitated with 80% ammonium sulfate, backwashed with 0.8 M ammonium sulfate in 50 mM acetate buffer (pH 5), and diluted to 0.6 M ammonium sulfate in 50 mM acetate buffer (pH 5). Fractions were then loaded on Phenyl Sepharose HP (Amersham Biosciences), eluted with a decreasing salt gradient, dialyzed against 50 mM ammonium bicarbonate, lyophilized for storage, and resuspended in PBS. Stx1 B subunit was purified as previously described (69). Contaminating lipopolysaccharide was removed by incubating with Affi-Prep polymyxin B resin overnight (Bio-Rad) at 4°C. Endotoxin content was tested by Limulus Amebocyte Lysate assay (Associates of Cape Cod Inc.) and determined to be less than 1 pg endotoxin/μg Stx.
Cell culture
Human dermal neonatal microvascular endothelial cells (HMVECs) were purchased from Lonza, cultured on 0.2% gelatin, and maintained in EGM-2MV growth medium (Lonza). Studies were performed on HMVECs at passages 5 and 6. HUVECs were maintained in M199 (Invitrogen) containing 20% fetal bovine serum (Hyclone), 17 units/ml heparin, and 0.05 mg/ml endothelial mitogen (Biomedical Technologies Inc.), as previously described (70), and used at passages 3 and 4. Confluent monolayers were treated with the specified concentrations of Stx or TNF-α (10 ng/ml) for the indicated times before harvesting. For hypoxia studies, HMVECs were treated as previously described (71). Total cellular RNA was purified by CsCl ultracentrifugation, as previously described (72).
Metabolic assays
Metabolite incorporation assays with radiolabeled thymidine, uridine, and leucine were used to determine the dose-dependent effects of Stx on global DNA, RNA, and protein synthesis, as described previously (11). HMVECs were seeded at a density of 40,000 cells per well in gelatin-coated 24-well plates. Two days after reaching confluence, cells were treated with varying concentrations of Stx2 for 24 hours. 1 μCi/ml [3H]thymidine, [3H]uridine, [3H]leucine, or [3H]methionine was added to each well 1 hour before harvest. After labeling, the cells were quickly washed with ice-cold PBS to terminate isotope uptake, and incorporated radioactivity was precipitated with 15% ice-cold TCA for 20 minutes. Cell monolayers were washed with water and then solubilized with 0.1 M NaOH-0.1% SDS (w/v). Aliquots from each well were counted using a scintillation counter. Each experiment was performed on triplicate wells. Data were normalized to vehicle-treated control wells.
Affymetrix GeneChip expression profiling — sample preparation, hybridization, and image acquisition
Total cellular RNA (10 μg) from Stx- and vehicle-treated (n = 6 replicates per group) HMVECs was submitted to a core microarray facility (The Centre for Applied Genomics, Hospital for Sick Children). Sample preparation, array hybridization, and image acquisition were performed according to the manufacturer’s technical documentation (Affymetrix). All samples were hybridized to Affymetrix GeneChip Human Genome U133A Arrays, housing approximately 14,500 well-characterized human genes. Raw probe-level intensities were derived from pixel-level data of the scanned images of hybridized arrays using the manufacturer’s software (Affymetrix Microarray Suite Version 5.0). Microarray data were deposited in NCBI’s Gene Expression Omnibus (accession no. GSE32710; http://www.ncbi.nlm.nih.gov/geo/) in conformity with the standards put forward by the Microarray Gene Expression Data Society ( http://www.mged.org/).
Quality control metrics.
Quality control metrics were assessed using packages made available through the Bioconductor Project implemented in the R statistical language and environment (version 2.4.0). Quality control metrics recommended by Affymetrix (average background, scale factor, percent present, and 3ι/5ι ratio of control probe sets) were computed using the simpleaffy package and conformed to the manufacturer’s guidelines. In addition, the absence of important spatial artifacts, RNA degradation plots, and the distribution of probe-level intensities across all arrays were assessed using functions made available in the affy package.
Data preprocessing and determination of differential expression.
Raw probe-level intensities were preprocessed using 13 different computational methods made available as part of the Bioconductor Project (v1.6) ( http://www.bioconductor.org) or as freely available, stand-alone software applications as outlined below. The base affy package was used for the computation of MAS5.0, RMA, dChip.PM (PM-only), and dChip.PM-MM (subtract MM) summary expression measures. Various other Bioconductor packages were used for the computation of GCRMA, PLIER, VSN, mgMOS, mmgMOS, q.FARMS (quantile normalization), and l.FARMS (loess normalization) measures. The preferred method of Choe et al. was also used (73). A stand-alone software application was used for the computation of the PDNN summary expression measure (PerfectMatch v2.3) (74). All preprocessing methods were used in their default implementations. Next, a filtering step was performed to remove probe sets called “absent” across all hybridized arrays (n = 12), as determined by the Affymetrix Detection Call algorithm implemented in the affy package. Finally, filtered probe set lists were independently submitted to a test of differential expression, Cyber-T, a regularized t test based on a Bayesian statistical framework. Probe sets were considered differentially expressed if they changed by at least 2 fold, with a Bayesian P value of 0.05 and a posterior probability of differential expression (PPDE(<p)) of 0.95. This denotes a global confidence of 95% based on the experiment-wide false-positive level.
Real-time RT-PCR
First-strand cDNA was synthesized from DNase-treated total cellular RNA, using random hexamers and SuperScript II reverse transcriptase (Invitrogen), according to the manufacturer’s protocol. SYBR Green and TaqMan Master Mixes (Applied Biosystems) were used according to manufacturer’s protocols. Quantitative real-time PCR assays were performed in triplicate using the ABI 7900HT Sequence Detection System (Applied Biosystems) with the following cycle parameters: 10 minutes at 95°C and 40 cycles of 10 seconds at 95°C and 1 minute at 60°C. Primers were designed such that amplified products crossed exon boundaries to exclude amplification of any remaining genomic DNA in the sample. Serial dilutions of reference plasmids were used to quantitate template copy number. Results were normalized to the reference genes GAPDH and 18S, both of which yielded similar results. Primers used in these experiments are listed in Supplemental Table 2. The cycles at which 1,000 copies of target amplicon passed through the cycle threshold (0.2) were 25.3 ± 0.3, 24.4 ± 0.1, 24.2 ± 0.4 for CXCR4, CXCR7, and SDF-1, respectively.
Polyribosome profiling
Polyribosome profiling was performed as previously described (71). Briefly, HMVECs were treated with vehicle or 10 fM Stx2 for 6 or 24 hours, washed in ice-cold phosphate-buffered saline containing 100 μg/ml cycloheximide (Sigma-Aldrich) and harvested in lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES (pH 7.4), 100 μg/ml cycloheximide (Sigma-Aldrich), and 1,000 U/ml RNaseOUT (Invitrogen). Cellular extracts were sheared using a 27-gauge needle, and the nuclei were pelleted by centrifugation (12,000 g, 4°C, 5 minutes). Cytoplasmic extracts were layered onto 10 ml sucrose gradients (15%–45%). After ultracentrifugation (SW Ti 41 rotor, 151,260 g, 4°C, 2 hours) (Beckman Coulter), 20 fractions were collected using a programmable density gradient fraction collector (Isco; PerkinElmer Life Sciences). RNA was extracted from each of the fractions using standard guanidinium-based protocols and resuspended in 10 μl 0.025 ng exogenously synthesized luciferase. RNA was added to each fraction prior to RNA purification in order to control for the efficiency of RNA purification and first-strand reverse transcription. 5 μl RNA from each fraction was reverse transcribed using SuperScript III (Invitrogen), followed by real-time PCR to measure mRNA levels in each fraction.
RNA polymerase II ChIP assays
ChIP was performed as previously described (75). Briefly, 5 μg anti–polymerase II antibody (N-20; Santa Cruz Biotechnology Inc.) was added to each immunoprecipitation reaction. Control immunoprecipitations containing no antibody were performed in parallel. An input sample was removed prior to immunoprecipitation. CXCR4 primers (+13 to +124 bp) were as follows: 5′-CGGCAGCAGGTAGCAAAGTGAC-3′ and 5′-GCAGGTTGAAACTGGACTTACACTG-3′. The number of copies of CXCR4 was determined in the bound chromatin, a no-antibody control, and a diluted input sample using serial dilutions of genomic DNA. Immunoprecipitated DNA was calculated by subtracting the number of copies in the no-antibody control from the bound chromatin and dividing by the input.
Determination of mRNA half-life
To study the effect of Stx on mRNA half-life, confluent HMVECs were left untreated or were treated with 1,000 fM Stx2 for 20 hours before addition of 10 μg/ml actinomycin D (Sigma-Aldrich) to arrest transcription. Total cellular RNA was extracted at 0, 5, 15, 30, 60, 120, 180, 270, and 360 minutes after the addition of actinomycin D. Levels of specific transcripts were determined by real-time RT-PCR using the primers listed above. Data were normalized to 18S rRNA. Rates of mRNA decay were determined using exponential regression.
Western blot
HMVECs were treated with Stx or vehicle, and total cellular protein collected as described previously (76). 15–20 μg total cellular protein was size fractionated on NuPAGE Novex 4–12% Bis-Tris gels (Invitrogen) and transferred onto nitrocellulose membranes using the XCell SureLock Mini-Cell and Xcell II Blot Module (Invitrogen). Mouse monoclonal anti–HIF-1α (Abcam; 1:200), rabbit polyclonal anti–HIF-2α (Novus Biologicals; 1:800), and mouse monoclonal anti-Lamin A/C (Santa Cruz Biotechnology Inc.; 1:200) were used as primary antibodies. HRP-linked rabbit anti-mouse IgG (H & L) and HRP-linked goat anti-rabbit IgG were used as secondary antibodies. Chemiluminescence signals from immunoreactive bands were detected using ECL Plus (GE Amersham).
Permeability studies
HMVECs were grown on polyester transwell filters (0.4-mm pore size; Corning Life Sciences) coated with Attachment Factor (Cascade Biologics) to confluency until the cells achieved a transendothelial electrical resistance of more than 21 cm2 (measured using Endohm-12, World Precision Instruments). The cells were treated with the indicated concentrations of Stx for 24 hours or SDF-1α (50 ng/ml) for 5 hours. Fluorescein-conjugated dextran (MW 70 kDa; Invitrogen) was then added (0.05 mg/ml) to the upper chamber of the transwell. Permeability of endothelial monolayers was measured by passage of the dextran from the upper chamber to the lower chamber, as determined by the ratio of fluorescence in the lower chamber divided by total (upper and lower chamber) fluorescence. Fluorescence was measured in a spectrofluorometer (Spectramax Gemini EM, Molecular Devices) using an excitation wavelength of 492 nm and an emission wavelength of 540 nm. In other experiments, cells were exposed to AMD3100 (10 μM) for 30 minutes prior to the addition of Stx (1,000 fM, 24 hours). Fresh AMD3100 was added to the media after 16 hours.
In vivo studies
Inbred, wild-derived CAST/Ei mice were obtained from The Jackson Laboratory. Adamts13–/– mice generated on a castaneus background, as previously described (41), were obtained by heterozygote crosses. All animals were maintained in pathogen-free facilities at the University of Toronto. Equal numbers of male and female mice between 8 to 11 weeks of age were injected i.v. via tail vein injection with the indicated concentrations of Stx2, Stx1 B subunit (12 ng/g body weight), or Stx2 holotoxin A subunit mutant (24 ng/g body weight) diluted in PBS, such that injection volumes were 10 μl/g body weight. Control mice received an injection of PBS.
Blood analysis
For hematology analysis, daily blood samples were collected in EDTA-coated tubes via saphenous vein. Alternatively, animals were anesthetized with isoflurane and exsanguinated by cardiac puncture. Blood was analyzed using a Coulter AcT diff Analyzer (Coulter Corporation). For plasma biochemistry measurements, blood was collected in heparinized tubes and centrifuged at 13,000 g for 5 minutes, and plasma was then stored at –80°C until subsequent analysis.
Urine studies
Urine was collected passively from CAST/Ei mice at various times after Stx injection. Creatinine levels were quantitated using an alkaline picrate method, and albumin levels were quantitated using an indirect competitive ELISA (Exocell Inc.). Alternatively, animals treated with vehicle or Stx (2,400 pg/g) for 4 days were injected i.v. with 100 μl 0.1% Evans Blue dye. One hour after injection, urine samples were collected, and the OD620 was determined.
CXCR4 inhibition studies
Mice were injected i.v. with 2,400 pg/g Stx2 on day 0 and received daily i.p. injections of the CXCR4 inhibitor AMD3100/plerixafor (1-1′-[1,4-phenylenebis(methylene)-bis(1,4,8,11-tetraazacyclotetradecane]) (Sigma-Aldrich) at a concentration of 10 μg/g, diluted in PBS. Control mice received daily injections of PBS. AMD3100/plerixafor administration began either 1 day prior to Stx injection or 1 day after Stx injection. Survival was monitored for a total of 10 days after toxin injection. Alternatively, beginning 1 day after Stx injection, mice received 10 μg anti-CXCR4 (Torrey Pines BioLabs) or rabbit IgG daily via i.p. injection in a volume of 250 μl PBS.
In situ cRNA hybridization
CAST/Ei mice were injected with Stx2 (2,400 pg/g) or an equivalent volume of PBS and euthanized at days 1 or 4. Animals were euthanized with CO2, and OCT was administered by oral gavage to distend the gastrointestinal tract. After being shaved completely, the animals were frozen in isopentane and stored at –80°C. 10-μm sections were fixed, hybridized, and washed as described previously (77). Slides were then dehydrated, exposed to Kodak BioMaxMR x-ray film, emulsion dipped in Kodak NTB nuclear track emulsion, and exposed in light-tight boxes at 4°C for 12 days for CXCR4 and CXCR7 and 14 days for SDF-1. A 776-nt probe within exon 2 was used to detect CXCR4, a 714-nt probe representing exon 2 and a portion of the 3′-UTR was used to detect CXCR7, and a 307-nt probe, including a portion of the 5′-UTR in addition to ORF sequence, was used to detect SDF-1. The probe used to detect SDF-1 was designed in a region common to all known murine isoforms. Control sense cRNA probes were used to detect background levels of hybridization signals.
Immunohistochemistry
Immunohistochemistry was performed with a mouse monoclonal anti-human/mouse SDF-1 antibody (clone 79018) (R&D Systems) at a dilution of 10 μg/ml, as previously described (78). After incubation with the appropriate HRP-conjugated secondary antibody, sections were labeled with Liquid Diaminobenzidine and Substrate Chromogen System (DakoCytomation), before counterstaining in Mayer’s hematoxylin. Positively stained tubules were counted in 6 nonoverlapping cortical fields at ×100 magnification.
Apoptosis and cytotoxicity assays
HMVECs were treated with 1,000 fM Stx2 for 24 hours. Where indicated, cells were pretreated with 20 μM ZVAD-FMK or vehicle for 1 hour before addition of toxin. Apoptosis was assessed by flow cytometry using the Annexin V-FITC Apoptosis Kit I, per the manufacturer’s instructions (BD Pharmingen). To determine cytotoxicity, cells were treated with Stx2 for 24 hours, and LDH release was measured using the CytoTox96 Non-Radioactive Cytotoxicity Assay according to the manufacturer’s instructions (Promega).
Platelet-endothelial adhesion studies
HMVECs were grown to confluence in gelatin-coated channels of the Bioflux microfluidic system (Fluxion Biosciences) and either left untreated or treated with 10 fM Stx2 for 24 hours. When AMD3100 was used, it was added 30 minutes prior to addition of platelets at a concentration of 10 μM. Whole blood samples were collected from healthy donors into acid citrate dextrose and centrifuged at 160 g for 15 minutes to obtain platelet-rich plasma (PRP). The PRP was centrifuged at 800 g for 10 minutes and washed twice with PBS (pH 6.3). The platelet pellet was resuspended in HEPES-Tyrode’s buffer (136 mM NaCl, 2.7 mM KCl, 0.42 mM NaH2PO4, 19 mM NaHCO3, 0.35 mM Na2HPO4, 5.5 mM glucose, 1 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, pH 7.2). Calcein-labeled platelets (2 × 107/ml) were perfused through the channels at constant shear rates of 2.5 dyn/cm2 for up to 10 minutes. Images were acquired using a Nikon Eclipse Ti inverted confocal microscope while the platelets were still under flow, and the fluorescent intensity of adhered platelets was quantified using the Bioflux 200 analysis software.
Tissue factor and TFPI protein assays
To collect membrane fractions of HMVECs, cells were washed with cold PBS, scraped in 100 μl PBS, and sonicated 3 times for 10 seconds using Vibra Cell VCX400 sonicator (Sonics&Materials Inc.) at 40% maximal power. Samples were centrifuged at 100,000 g for 60 minutes at 4°C to obtain membrane proteins in the supernatants. Tissue factor and tissue factor pathway inhibitor activities were determined using Actichrome TF and Actichrome TFPI Kits (American Diagnotica), per the manufacturer’s instructions.
SDF-1 protein measurements
CAST/Ei mice were injected with Stx2 (2,400 pg/g) and euthanized at various times thereafter. Blood was collected by cardiac puncture into EDTA-coated tubes and centrifuged to collect plasma. SDF-1α levels were quantified by ELISA (R&D Systems). The mean minimal detectable dose for the assay was 0.044 ng/ml.
Blood SDF-1 measurements in patients
A description of the patient cohort and study protocol are provided elsewhere (54, 55). Briefly, the prospective cohort study consisted of children under the age of 10, who were recruited to the study upon notification of a stool culture positive for E. coli O157:H7. Participating laboratories were located in Washington, Oregon, Idaho, Wyoming, Missouri, and Illinois. In addition, children who reported to the emergency department of the Children’s Hospital and Regional Medical Center in Seattle, Washington, USA, with acute bloody diarrhea were enrolled at the time of presentation, with appropriate consent. Data from these samples were included in the study if E. coli O157:H7 was later confirmed in a stool culture. Each child’s caregiver(s) completed a standardized questionnaire to identify when diarrhea began. Day 1 of illness was defined as the first day on which diarrhea occurred. Venous blood samples were obtained at enrollment and as clinically indicated thereafter until each child either developed and recovered from the HUS or reached the end of the period of risk for HUS (14 days after the onset of diarrhea) without further illness. Children were classified as having HUS if hemolytic anemia (hematocrit below 30%), thrombocytopenia (platelet count less than 150,000/mm3), and renal insufficiency (serum creatinine concentration exceeding the upper limit of normal for the child’s age) developed (55). Children were classified as having uncomplicated infection if fewer than 3 of the above criteria were present. Plasma derived from patient blood was collected using sodium citrate as anticoagulant and stored in aliquots at –70°C until use. All measurements were performed on blinded samples by sandwich ELISA (R&D Systems), per the manufacturer’s instructions. Samples were assigned to 1 out of 3 groups for analysis: those taken from children who had uncomplicated infection that resolved without subsequent progression to HUS, those taken on days 2 to 6 of illness from children in whom HUS later developed, and those from children during HUS on days 1–10 of illness (55). The first day of diarrhea is defined as day 1 of illness.
Fluorescence microangiography
Kidney microvascular architecture was assessed in CAST/Ei mice injected with PBS or Stx2 (2,400 pg/g) using a fluorescence microangiography technique, essentially as described previously (79). In brief, animals were anesthetized with inhaled isoflurane 2.5%. The abdominal aorta was cannulated with a 26 G plastic catheter with the tip advanced to a point just distal to the origins of the renal arteries. Whole-body perfusion/exsanguination commenced at 100–120 mmHg via the abdominal aorta with PBS (pH 7.4) to remove circulating blood, and the right internal jugular vein was simultaneously severed, allowing free flow of perfusate. 5 ml of a pre-warmed (50°C) 1% agarose–fluorescent microbead (10%) (0.02-μm diameter; Invitrogen) mixture was then infused via the 26 G catheter into the aorta to achieve whole-body microbead perfusion. After cooling the animals on ice for 10 minutes, the kidneys were harvested and immersed in neutral buffered formalin for 24 hours at 4°C. 200-μm sections were then viewed using a confocal microscope at ×5 or ×30 optical zoom (Leica TCS SL, Leica). Neurolucida software (MBF Bioscience) was used to digitally reconstruct tissue microvasculature.
Statistics
Data represent the mean ± SEM of at least 3 independent experiments, unless otherwise stated. Statistical significance was determined using the Student’s t test or ANOVA, as appropriate, with Bonferroni, Newman-Keuls, or Dunnett’s post-hoc test. Patient data was analyzed by Wilcoxon signed-rank test. P < 0.05 was considered statistically significant.
Study approval
All animal studies were performed in accordance with the guidelines of the Canadian Council on Animal Care and were approved by the University of Toronto Animal Care Committee. For patient SDF-1 measurements, approval was obtained from the Human Research Protection Office at Washington University in St. Louis, Missouri, USA. Written, informed consent was obtained from the child’s parent or guardian at time of study entry. Where appropriate, assent was also obtained from the child.
Supplementary Material
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health, especially MOP 79475, and grant PO1 HL076540-06A1 from NHLBI/NIH to P.A. Marsden. P.A. Marsden holds the Keenan Chair in Medical Research at St. Michael’s Hospital and the University of Toronto.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(2):759–776. doi:10.1172/JCI57313.
References
- 1.Karmali MA. Infection by verocytotoxin-producing Escherichia coli. Clin Microbiol Rev. 1989;2(1):15–38. doi: 10.1128/cmr.2.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bielaszewska M, et al. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect Dis. 2011;11(9):671–676. doi: 10.1016/S1473-3099(11)70165-7. [DOI] [PubMed] [Google Scholar]
- 3.Frank C, et al. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med. 2011;365(19):1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 4.Rohde H, et al. Open-source genomic analysis of Shiga-toxin-producing E. coli O104:H4. N Engl J Med. 2011;365(8):718–724. doi: 10.1056/NEJMoa1107643. [DOI] [PubMed] [Google Scholar]
- 5.Rasko DA, et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 2011;365(8):709–717. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarr PI, Gordon CA, Chandler WL. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 2005;365(9464):1073–1086. doi: 10.1016/S0140-6736(05)71144-2. [DOI] [PubMed] [Google Scholar]
- 7.Scotland SM, Willshaw GA, Smith HR, Rowe B. Properties of strains of Escherichia coli belonging to serogroup O157 with special reference to production of Vero cytotoxins VT1 and VT2. Epidemiol Infect. 1987;99(3):613–624. doi: 10.1017/S0950268800066462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bitzan M, Karch H, Altrogge H, Strehlau J, Blaker F. Hemolytic-uremic syndrome associated with a variant Shiga-like cytotoxin of Escherichia coli O111. Pediatr Infect Dis J. 1988;7(2):128–132. doi: 10.1097/00006454-198802000-00013. [DOI] [PubMed] [Google Scholar]
- 9.Lingwood CA, et al. Glycolipid binding of purified and recombinant Escherichia coli produced verotoxin in vitro. J Biol Chem. 1987;262(18):8834–8839. [PubMed] [Google Scholar]
- 10.Endo Y, Tsurugi K, Yutsudo T, Takeda Y, Ogasawara T, Igarashi K. Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. RNA N-glycosidase activity of the toxins. Eur J Biochem. 1988;171(1–2):45–50. doi: 10.1111/j.1432-1033.1988.tb13756.x. [DOI] [PubMed] [Google Scholar]
- 11.Bitzan MM, Wang Y, Lin J, Marsden PA. Verotoxin and ricin have novel effects on preproendothelin-1 expression but fail to modify nitric oxide synthase (ecNOS) expression and NO production in vascular endothelium. J Clin Invest. 1998;101(2):372–382. doi: 10.1172/JCI522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ara T, Tokoyoda K, Okamoto R, Koni PA, Nagasawa T. The role of CXCL12 in the organ-specific process of artery formation. Blood. 2005;105(8):3155–3161. doi: 10.1182/blood-2004-07-2563. [DOI] [PubMed] [Google Scholar]
- 13.Tachibana K, et al. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature. 1998;393(6685):591–594. doi: 10.1038/31261. [DOI] [PubMed] [Google Scholar]
- 14.Takabatake Y, et al. The CXCL12 (SDF-1)/CXCR4 axis is essential for the development of renal vasculature. J Am Soc Nephrol. 2009;20(8):1714–1723. doi: 10.1681/ASN.2008060640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strasser GA, Kaminker JS, Tessier-Lavigne M. Microarray analysis of retinal endothelial tip cells identifies CXCR4 as a mediator of tip cell morphology and branching. Blood. 2010;115(24):5102–5110. doi: 10.1182/blood-2009-07-230284. [DOI] [PubMed] [Google Scholar]
- 16.Ceradini DJ, et al. Progenitor cell trafficking is regulated by hypoxic gradients through HIF-1 induction of SDF-1. Nat Med. 2004;10(8):858–864. doi: 10.1038/nm1075. [DOI] [PubMed] [Google Scholar]
- 17.Jin DK, et al. Cytokine-mediated deployment of SDF-1 induces revascularization through recruitment of CXCR4+ hemangiocytes. Nat Med. 2006;12(5):557–567. doi: 10.1038/nm1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz de Almodovar C, Luttun A, Carmeliet P. An SDF-1 trap for myeloid cells stimulates angiogenesis. Cell. 2006;124(1):18–21. doi: 10.1016/j.cell.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 19.Ding M, et al. Loss of the tumor suppressor Vhlh leads to upregulation of Cxcr4 and rapidly progressive glomerulonephritis in mice. Nat Med. 2006;12(9):1081–1087. doi: 10.1038/nm1460. [DOI] [PubMed] [Google Scholar]
- 20.Sayyed SG, et al. Podocytes produce homeostatic chemokine stromal cell-derived factor-1/CXCL12, which contributes to glomerulosclerosis, podocyte loss and albuminuria in a mouse model of type 2 diabetes. Diabetologia. 2009;52(11):2445–2454. doi: 10.1007/s00125-009-1493-6. [DOI] [PubMed] [Google Scholar]
- 21.Wang A, et al. CXCR4/CXCL12 hyperexpression plays a pivotal role in the pathogenesis of lupus. . J Immunol. 2009;182(7):4448–4458. doi: 10.4049/jimmunol.0801920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obrig TG, Louise CB, Lingwood CA, Boyd B, Barley-Maloney L, Daniel TO. Endothelial heterogeneity in Shiga toxin receptors and responses. J Biol Chem. 1993;268(21):15484–15488. [PubMed] [Google Scholar]
- 23.Ohmi K, Kiyokawa N, Takeda T, Fujimoto J. Human microvascular endothelial cells are strongly sensitive to Shiga toxins. Biochem Biophys Res Commun. 1998;251(1):137–141. doi: 10.1006/bbrc.1998.9417. [DOI] [PubMed] [Google Scholar]
- 24.Walter DH, et al. Impaired CXCR4 signaling contributes to the reduced neovascularization capacity of endothelial progenitor cells from patients with coronary artery disease. Circ Res. 2005;97(11):1142–1151. doi: 10.1161/01.RES.0000193596.94936.2c. [DOI] [PubMed] [Google Scholar]
- 25.Grunewald M, et al. VEGF-induced adult neovascularization: recruitment, retention, and role of accessory cells. Cell. 2006;124(1):175–189. doi: 10.1016/j.cell.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 26.Gerli G, et al. SDF1-3′A gene polymorphism is associated with chronic myeloproliferative disease and thrombotic events. Clin Chem. 2005;51(12):2411–2414. doi: 10.1373/clinchem.2005.057802. [DOI] [PubMed] [Google Scholar]
- 27.Sierro F, et al. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc Natl Acad Sci U S A. 2007;104(37):14759–14764. doi: 10.1073/pnas.0702229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerrits H, et al. Early postnatal lethality and cardiovascular defects in CXCR7-deficient mice. Genesis. 2008;46(5):235–245. doi: 10.1002/dvg.20387. [DOI] [PubMed] [Google Scholar]
- 29.Jackson MP, Newland JW, Holmes RK, O’Brien AD. Nucleotide sequence analysis of the structural genes for Shiga-like toxin I encoded by bacteriophage 933J from Escherichia coli. Microb Pathog. 1987;2(2):147–153. doi: 10.1016/0882-4010(87)90106-9. [DOI] [PubMed] [Google Scholar]
- 30.Burns JM, et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203(9):2201–2213. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson MP, Deresiewicz RL, Calderwood SB. Mutational analysis of the Shiga toxin and Shiga-like toxin II enzymatic subunits. J Bacteriol. 1990;172(6):3346–3350. doi: 10.1128/jb.172.6.3346-3350.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohmura M, Yamasaki S, Kurazono H, Kashiwagi K, Igarashi K, Takeda Y. Characterization of non-toxic mutant toxins of Vero toxin 1 that were constructed by replacing amino acids in the A subunit. Microb Pathog. 1993;15(3):169–176. doi: 10.1006/mpat.1993.1067. [DOI] [PubMed] [Google Scholar]
- 33.Thorpe CM, Smith WE, Hurley BP, Acheson DW. Shiga toxins induce, superinduce, and stabilize a variety of C-X-C chemokine mRNAs in intestinal epithelial cells, resulting in increased chemokine expression. Infect Immun. 2001;69(10):6140–6147. doi: 10.1128/IAI.69.10.6140-6147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harrison LM, van Haaften WC, Tesh VL. Regulation of proinflammatory cytokine expression by Shiga toxin 1 and/or lipopolysaccharides in the human monocytic cell line THP-1. Infect Immun. 2004;72(5):2618–2627. doi: 10.1128/IAI.72.5.2618-2627.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrison LM, van den Hoogen C, van Haaften WC, Tesh VL. Chemokine expression in the monocytic cell line THP-1 in response to purified shiga toxin 1 and/or lipopolysaccharides. Infect Immun. 2005;73(1):403–412. doi: 10.1128/IAI.73.1.403-412.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staller P, Sulitkova J, Lisztwan J, Moch H, Oakeley EJ, Krek W. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature. 2003;425(6955):307–311. doi: 10.1038/nature01874. [DOI] [PubMed] [Google Scholar]
- 37.Zagzag D, et al. Stromal cell-derived factor-1alpha and CXCR4 expression in hemangioblastoma and clear cell-renal cell carcinoma: von Hippel-Lindau loss-of-function induces expression of a ligand and its receptor. Cancer Res. 2005;65(14):6178–6188. doi: 10.1158/0008-5472.CAN-04-4406. [DOI] [PubMed] [Google Scholar]
- 38.Costello CM, et al. Lung-selective gene responses to alveolar hypoxia: potential role for the bone morphogenetic antagonist gremlin in pulmonary hypertension. Am J Physiol Lung Cell Mol Physiol. 2008;295(2):L272–L284. doi: 10.1152/ajplung.00358.2007. [DOI] [PubMed] [Google Scholar]
- 39.Guessous F, et al. Shiga toxin 2 and lipopolysaccharide induce human microvascular endothelial cells to release chemokines and factors that stimulate platelet function. Infect Immun. 2005;73(12):8306–8316. doi: 10.1128/IAI.73.12.8306-8316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee BC, Lee TH, Avraham S, Avraham HK. Involvement of the chemokine receptor CXCR4 and its ligand stromal cell-derived factor 1alpha in breast cancer cell migration through human brain microvascular endothelial cells. Mol Cancer Res. 2004;2(6):327–338. [PubMed] [Google Scholar]
- 41.Motto DG, et al. Shigatoxin triggers thrombotic thrombocytopenic purpura in genetically susceptible ADAMTS13-deficient mice. J Clin Invest. 2005;115(10):2752–2761. doi: 10.1172/JCI26007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banno F, Kaminaka K, Soejima K, Kokame K, Miyata T. Identification of strain-specific variants of mouse Adamts13 gene encoding von Willebrand factor-cleaving protease. J Biol Chem. 2004;279(29):30896–30903. doi: 10.1074/jbc.M314184200. [DOI] [PubMed] [Google Scholar]
- 43.Shavit JA, Manichaikul A, Lemmerhirt HL, Broman KW, Ginsburg D. Modifiers of von Willebrand factor identified by natural variation in inbred strains of mice. Blood. 2009;114(26):5368–5374. doi: 10.1182/blood-2009-07-233213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sellers AL, Smith S 3rd, Marmorston J, Goodman HC. Studies on the mechanism of experimental proteinuria. J Exp Med. 1952;96(6):643–652. doi: 10.1084/jem.96.6.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keepers TR, Psotka MA, Gross LK, Obrig TG. A murine model of HUS: Shiga toxin with lipopolysaccharide mimics the renal damage and physiologic response of human disease. J Am Soc Nephrol. 2006;17(12):3404–3414. doi: 10.1681/ASN.2006050419. [DOI] [PubMed] [Google Scholar]
- 46.Jacewicz MS, Acheson DW, Mobassaleh M, Donohue-Rolfe A, Balasubramanian KA, Keusch GT. Maturational regulation of globotriaosylceramide, the Shiga-like toxin 1 receptor, in cultured human gut epithelial cells. J Clin Invest. 1995;96(3):1328–1335. doi: 10.1172/JCI118168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barrett TJ, Potter ME, Wachsmuth IK. Continuous peritoneal infusion of Shiga-like toxin II (SLT II) as a model for SLT II-induced diseases. J Infect Dis. 1989;159(4):774–777. doi: 10.1093/infdis/159.4.774. [DOI] [PubMed] [Google Scholar]
- 48.Tashiro H, et al. Verotoxin induces hemorrhagic lesions in rat small intestine. Temporal alteration of vasoactive substances. Dig Dis Sci. 1994;39(6):1230–1238. doi: 10.1007/BF02093788. [DOI] [PubMed] [Google Scholar]
- 49.Boldajipour B, et al. Control of chemokine-guided cell migration by ligand sequestration. Cell. 2008;132(3):463–473. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- 50.Naumann U, et al. CXCR7 functions as a scavenger for CXCL12 and CXCL11. PLoS One. 2010;5(2):e9175. doi: 10.1371/journal.pone.0009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ceradini DJ, Gurtner GC. Homing to hypoxia: HIF-1 as a mediator of progenitor cell recruitment to injured tissue. Trends Cardiovasc Med. 2005;15(2):57–63. doi: 10.1016/j.tcm.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 52.[No authors listed]. Plerixafor: AMD 3100, AMD3100, JM 3100, SDZ SID 791. Drugs R D. 2007;8(2):113–119. doi: 10.2165/00126839-200708020-00006. [DOI] [PubMed] [Google Scholar]
- 53.Nolasco LH, et al. Hemolytic uremic syndrome-associated Shiga toxins promote endothelial-cell secretion and impair ADAMTS13 cleavage of unusually large von Willebrand factor multimers. Blood. 2005;106(13):4199–4209. doi: 10.1182/blood-2005-05-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong CS, Jelacic S, Habeeb RL, Watkins SL, Tarr PI. The risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N Engl J Med. 2000;342(26):1930–1936. doi: 10.1056/NEJM200006293422601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chandler WL, et al. Prothrombotic coagulation abnormalities preceding the hemolytic-uremic syndrome. N Engl J Med. 2002;346(1):23–32. doi: 10.1056/NEJMoa011033. [DOI] [PubMed] [Google Scholar]
- 56.Ho M, et al. Identification of endothelial cell genes by combined database mining and microarray analysis. Physiol Genomics. 2003;13(3):249–262. doi: 10.1152/physiolgenomics.00186.2002. [DOI] [PubMed] [Google Scholar]
- 57.Chi JT, et al. Endothelial cell diversity revealed by global expression profiling. Proc Natl Acad Sci U S A. 2003;100(19):10623–10628. doi: 10.1073/pnas.1434429100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaye SA, Louise CB, Boyd B, Lingwood CA, Obrig TG. Shiga toxin-associated hemolytic uremic syndrome: interleukin-1 beta enhancement of Shiga toxin cytotoxicity toward human vascular endothelial cells in vitro. Infect Immun. 1993;61(9):3886–3891. doi: 10.1128/iai.61.9.3886-3891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Louise CB, Obrig TG. Shiga toxin-associated hemolytic-uremic syndrome: combined cytotoxic effects of Shiga toxin, interleukin-1 beta, and tumor necrosis factor alpha on human vascular endothelial cells in vitro. Infect Immun. 1991;59(11):4173–4179. doi: 10.1128/iai.59.11.4173-4179.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matussek A, et al. Molecular and functional analysis of Shiga toxin-induced response patterns in human vascular endothelial cells. Blood. 2003;102(4):1323–1332. doi: 10.1182/blood-2002-10-3301. [DOI] [PubMed] [Google Scholar]
- 61.Huang J, Motto DG, Bundle DR, Sadler JE. Shiga toxin B subunits induce VWF secretion by human endothelial cells and thrombotic microangiopathy in ADAMTS13-deficient mice. Blood. 2010;116(18):3653–3659. doi: 10.1182/blood-2010-02-271957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen J, et al. Effects of CXCR4 gene transfer on cardiac function after ischemia-reperfusion injury. Am J Pathol. 2010;176(4):1705–1715. doi: 10.2353/ajpath.2010.090451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mazzinghi B, et al. Essential but differential role for CXCR4 and CXCR7 in the therapeutic homing of human renal progenitor cells. J Exp Med. 2008;205(2):479–490. doi: 10.1084/jem.20071903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalatskaya I, Berchiche YA, Gravel S, Limberg BJ, Rosenbaum JS, Heveker N. AMD3100 is a CXCR7 ligand with allosteric agonist properties. Mol Pharmacol. 2009;75(5):1240–1247. doi: 10.1124/mol.108.053389. [DOI] [PubMed] [Google Scholar]
- 65.Kowalska MA, et al. Stromal cell-derived factor-1 and macrophage-derived chemokine:2 chemokines that activate platelets. Blood. 2000;96(1):50–57. [PubMed] [Google Scholar]
- 66.Panicot-Dubois L, Thomas GM, Furie BC, Furie B, Lombardo D, Dubois C. Bile salt-dependent lipase interacts with platelet CXCR4 and modulates thrombus formation in mice and humans. J Clin Invest. 2007;117(12):3708–3719. doi: 10.1172/JCI32655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gear AR, Suttitanamongkol S, Viisoreanu D, Polanowska-Grabowska RK, Raha S, Camerini D. Adenosine diphosphate strongly potentiates the ability of the chemokines MDC, TARC, and SDF-1 to stimulate platelet function. Blood. 2001;97(4):937–945. doi: 10.1182/blood.V97.4.937. [DOI] [PubMed] [Google Scholar]
- 68.Massberg S, et al. Platelets secrete stromal cell-derived factor 1alpha and recruit bone marrow-derived progenitor cells to arterial thrombi in vivo. J Exp Med. 2006;203(5):1221–1233. doi: 10.1084/jem.20051772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramotar K, Boyd B, Tyrrell G, Gariepy J, Lingwood C, Brunton J. Characterization of Shiga-like toxin I B subunit purified from overproducing clones of the SLT-I B cistron. Biochem J. 1990;272(3):805–811. doi: 10.1042/bj2720805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marsden PA, et al. Molecular cloning and characterization of human endothelial nitric oxide synthase. FEBS Lett. 1992;307(3):287–293. doi: 10.1016/0014-5793(92)80697-F. [DOI] [PubMed] [Google Scholar]
- 71.Fish JE, et al. Hypoxia-inducible expression of a natural cis-antisense transcript inhibits endothelial nitric-oxide synthase. J Biol Chem. 2007;282(21):15652–15666. doi: 10.1074/jbc.M608318200. [DOI] [PubMed] [Google Scholar]
- 72.Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 73.Choe SE, Boutros M, Michelson AM, Church GM, Halfon MS. Preferred analysis methods for Affymetrix GeneChips revealed by a wholly defined control dataset. Genome Biol. 2005;6(2):R16. doi: 10.1186/gb-2005-6-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang L, Miles MF, Aldape KD. A model of molecular interactions on short oligonucleotide microarrays. Nat Biotechnol. 2003;21(7):818–821. doi: 10.1038/nbt836. [DOI] [PubMed] [Google Scholar]
- 75.Fish JE, et al. The expression of endothelial nitric-oxide synthase is controlled by a cell-specific histone code. J Biol Chem. 2005;280(26):24824–24838. doi: 10.1074/jbc.M502115200. [DOI] [PubMed] [Google Scholar]
- 76.Wang Y, Goligorsky MS, Lin M, Wilcox JN, Marsden PA. A novel, testis-specific mRNA transcript encoding an NH2-terminal truncated nitric-oxide synthase. J Biol Chem. 1997;272(17):11392–11401. doi: 10.1074/jbc.272.17.11392. [DOI] [PubMed] [Google Scholar]
- 77.Robb GB, et al. Post-transcriptional regulation of endothelial nitric-oxide synthase by an overlapping antisense mRNA transcript. J Biol Chem. 2004;279(36):37982–37996. doi: 10.1074/jbc.M400271200. [DOI] [PubMed] [Google Scholar]
- 78.Advani A, et al. The (Pro)renin receptor: site-specific and functional linkage to the vacuolar H+-ATPase in the kidney. Hypertension. 2009;54(2):261–269. doi: 10.1161/HYPERTENSIONAHA.109.128645. [DOI] [PubMed] [Google Scholar]
- 79.Dutly AE, Kugathasan L, Trogadis JE, Keshavjee SH, Stewart DJ, Courtman DW. Fluorescent microangiography (FMA): an improved tool to visualize the pulmonary microvasculature. Lab Invest. 2006;86(4):409–416. doi: 10.1038/labinvest.3700399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.