Abstract
Pancreatic cancer is almost invariably associated with mutations in the KRAS gene, most commonly KRASG12D, that result in a dominant-active form of the KRAS GTPase. However, how KRAS mutations promote pancreatic carcinogenesis is not fully understood, and whether oncogenic KRAS is required for the maintenance of pancreatic cancer has not been established. To address these questions, we generated two mouse models of pancreatic tumorigenesis: mice transgenic for inducible KrasG12D, which allows for inducible, pancreas-specific, and reversible expression of the oncogenic KrasG12D, with or without inactivation of one allele of the tumor suppressor gene p53. Here, we report that, early in tumorigenesis, induction of oncogenic KrasG12D reversibly altered normal epithelial differentiation following tissue damage, leading to precancerous lesions. Inactivation of KrasG12D in established precursor lesions and during progression to cancer led to regression of the lesions, indicating that KrasG12D was required for tumor cell survival. Strikingly, during all stages of carcinogenesis, KrasG12D upregulated Hedgehog signaling, inflammatory pathways, and several pathways known to mediate paracrine interactions between epithelial cells and their surrounding microenvironment, thus promoting formation and maintenance of the fibroinflammatory stroma that plays a pivotal role in pancreatic cancer. Our data establish that epithelial KrasG12D influences multiple cell types to drive pancreatic tumorigenesis and is essential for tumor maintenance. They also strongly support the notion that inhibiting KrasG12D, or its downstream effectors, could provide a new approach for the treatment of pancreatic cancer.
Introduction
Pancreatic ductal adenocarcinoma (PDA), the most common form of pancreatic cancer, has among the worst prognoses of all human malignancies. Annually, the number of victims of the disease approaches the number of new diagnoses, and the average survival from diagnosis is less than 6 months (1, 2). Surgery is the best option for the minority of patients who present with localized disease at the time of diagnosis (about 20% of the total), but even those patients often experience local or metastatic recurrence (3, 4). Therefore, there is a dire need for new therapeutic approaches that are likely to be based on a better understanding of the biology of this disease.
The KRAS oncogene is frequently mutated in human malignancies such as colon, lung, and ovarian cancer. In pancreatic cancer, mutations in KRAS are found in more than 90% of patient samples (5, 6). The most frequent mutation is the constitutively active KRASG12D allele (herein referred to as Kras*) (for review, see refs. 7, 8). Interestingly, KRAS mutations are frequently detected in the most common precursor lesion to pancreatic cancer, pancreatic intraepithelial neoplasia (PanIN), indicating a potential role early in the disease (9). Mouse studies have provided compelling evidence that oncogenic Kras* is required for the formation of PanINs (10, 11). However, how Kras* contributes to PanIN progression and PDA maintenance has not been addressed due to the lack of a suitable in vivo model. The role of oncogenic Kras* in tumor maintenance has been addressed in lung adenocarcinoma, where Kras* is required for tumor cell survival, even in advanced stages of the disease, and in the presence of additional genetic alterations such as loss of tumor suppressor genes (12). In addition, a subset of pancreatic cancer cell lines require Kras* activity for growth and survival (13, 14). However, a mouse model for study of Kras* dependency in pancreatic cancer has so far not been developed.
Here, we describe two new mouse models of pancreatic tumorigenesis defined by tissue-specific, temporally regulated, and reversible expression of Kras*, with or without inactivation of one allele of the tumor suppressor gene p53, that we have named inducible Kras* (iKras*) and iKras*-p53+/–, respectively. We use these new models to address the role of Kras* at several key stages during pancreatic carcinogenesis: PanIN initiation, established PanIN maintenance, and the development and maintenance of invasive PDA.
Results
The iKras* mouse model resembles the well-established KC (p48-Cre;LSL-KrasG12D) model and mimics the progression of the human disease.
We used three genetically modified mouse strains to generate triple transgenic p48-Cre;R26-rtTa-IRES-EGFP;TetO-KrasG12D mice, referred to as iKras* mice. The p48-Cre, or Ptf1a-Cre, allele (15) drives Cre expression mostly in a pancreas-specific manner, thus recombining a stop cassette preceding the reverse tetracycline transactivator (rtTa) IRES-EGFP cassette in the Rosa26 (R26) locus (16). Thus, rtTa and EGFP are expressed in the pancreatic epithelium during embryogenesis and into adulthood, even in cell types that downregulate p48 expression in the adult, such as islet and ductal cells (see Cre lineage tracing and EGFP expression in Supplemental Figure 1, A and B; supplemental material available online with this article; doi: 10.1172/JCI59227DS1). Once expressed, rtTa is inactive unless doxycycline (doxy) is administered in the animals’ drinking water (Figure 1A). Activation of rtTa leads to mutant Kras* expression from the TetO-KrasG12D (12) allele, and its activation can be reversed by doxy withdrawal (Figure 1A), leading to a system that allows for organ-specific, temporally regulated, and reversible expression of Kras*.
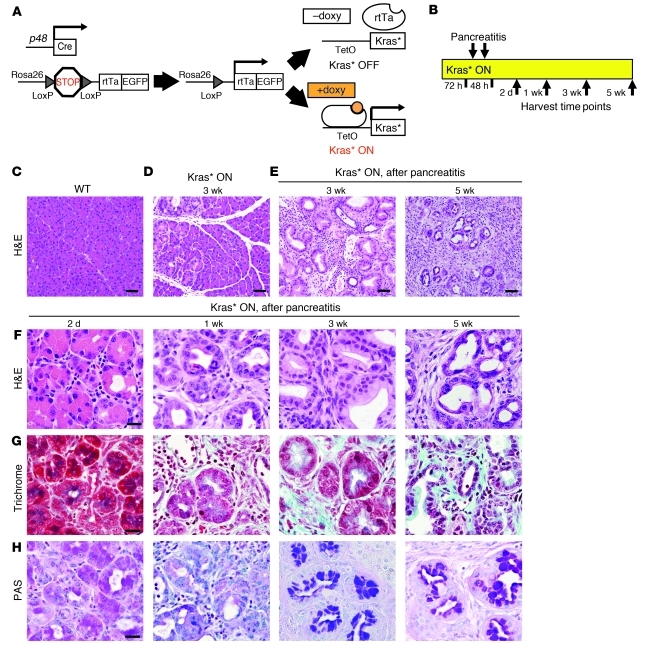
Figure 1. The iKras* mouse model of pancreatic tumorigenesis.
(A) Genetic makeup of the iKras* model: p48-Cre;R26-rtTa-IRES-EGFP;TetO-KrasG12D. (B) Experimental design. Kras* expression was induced with doxy for 72 hours before 2 consecutive days of intraperitoneal cerulein injections to induce pancreatitis and neoplasia. n = 3–5 mice per time point. (C) H&E staining of wild-type murine pancreas. Scale bar: 50: μm. (D) H&E staining of iKras* murine pancreas 3 weeks after doxy induction of Kras*. Scale bar: 50 μm. (E) H&E staining of iKras* murine pancreas 3 and 5 weeks after induction of Kras* and cerulein injections. Scale bars: 50 μm. (F) H&E staining of iKras* of iKras* murine pancreas 2 days, 1 week, 3 weeks, and 5 weeks after induction of Kras* and cerulein injections. Scale bar: 20 μm. (G) Gomori trichrome staining for interstitial collagen 2 days, 1 week, 3 weeks, and 5 weeks after induction of Kras* and cerulein injections. Scale bar: 20 μm. (H) PAS staining for mucin accumulation 2 days, 1 week, 3 weeks, and 5 weeks after induction of Kras* and cerulein injections. Scale bar: 20 μm.
We first conducted a series of experiments to compare our iKras* mouse with the well-established p48-Cre;LSL-KrasG12D model (herein referred to as KC) (10, 11, 17). The expression of Kras* is differentially regulated in the two models, since the oncogene is expressed from the endogenous Kras locus in KC mice and from an artificial transgene in iKras* mice. Moreover, in the KC model Kras* is activated during embryogenesis. In iKras* mice, we chose to express Kras* in adult mice (4–6 weeks of age), and tissues were harvested after 72 hours, 1 week, 3 weeks, 5 weeks, 18 weeks, and 23 weeks (Supplemental Figure 1C and data not shown). Doxy administration in control animals did not result in any detectable pancreatic phenotype (Supplemental Figure 1D; compare with wild-type in Figure 1C). The iKras* pancreata appeared completely normal up to 1 week following Kras* induction. However, at 3 weeks we observed rare areas of acinar-ductal metaplasia (ADM) and low-grade PanINs in 1 of 3 mice (Figure 1D and Supplemental Figure 1D). At 5 weeks of age, 2 of 3 mice had areas of ADM and low-grade PanIN formation (data not shown). Unequivocal PanINs, surrounded by areas of fibrosis and embedded in the acinar parenchyma, were observed after 18 weeks on doxy (Supplemental Figure 1D), and by 23 weeks, large areas of the pancreatic parenchyma were substituted with PanIN lesions of different grade, with frank adenocarcinoma being observed in 1 of 2 animals (Supplemental Figure 1D).
Previous reports have shown that induction of chronic or acute pancreatitis acts synergistically with oncogenic Kras* in driving carcinogenesis (18–20). Therefore, in a second set of experiments, we induced acute pancreatitis in adult mice by injecting them with the cholecystokinin analog cerulein. We used age-matched wild-type KC and iKras* mice with or without doxy. In the absence of doxy (Kras* OFF), cerulein treatment in iKras* mice led to pancreatitis-specific changes such as ADM and infiltration of inflammatory cells; however, the damage completely resolved within 3 weeks (Supplemental Figure 2, A and B) as in wild-type animals (Supplemental Figure 2C). By contrast, recovery from pancreatitis was impaired in KC mice as well as in iKras* mice treated with doxy starting 72 hours prior to the induction of pancreatitis and maintained with doxy for the duration of the experiment (Kras* ON; Figure 1B). At 2 days and at 1 week after induction of pancreatitis, both iKras* and KC mice pancreata presented with ADM (Figure 1F and Supplemental Figure 2D) with progressive accumulation of fibrotic stroma (Figure 1G), but no evidence of the intracellular mucin accumulation that characterizes PanIN lesions (Figure 1H and Supplemental Figure 2D). At 3 weeks after pancreatitis induction, in both iKras* and KC mice, the whole pancreatic parenchyma was replaced by ductal structures (Figure 1, E and F, and Supplemental Figure 2D) surrounded by collagen-rich stroma (Figure 1G). The epithelial cells showed intracellular mucin accumulation (Figure 1H and Supplemental Figure 2D) and strong positive staining of membrane claudin-18, a marker that specifically differentiates PanIN and PDA from reactive ducts in human and mouse (Supplemental Figure 2E and refs. 21, 22); thus, we identify them as PanINs and refer to them as such herein. Our classification of these morphologic lesions as PanINs is consistent with previous publications using the cerulein model (19, 20) and activation of oncogenic Kras* in adult animals (23). As expected, the MAPK/ERK pathway was active in PanINs, as determined by strong nuclear and cytoplasmic phospho-ERK1/2 staining (Supplemental Figure 2F), thus demonstrating that Kras* was biologically active in the epithelial cells. In both models, abundant proliferating cells were present both in the epithelial compartment and in the stroma (Supplemental Figure 2G). The results of this first set of experiments show that the iKras* mouse model closely recapitulates the kinetics of PanIN formation and progression of the well-established KC model, both with and without induction of pancreatitis.
In order to determine the effect of expressing oncogenic Kras* for a longer time period, we harvested pancreata of iKras* mice 5 weeks after induction of pancreatitis and found that the parenchyma was replaced by low- and high-grade PanIN lesions (Figure 1, E–H) interspersed with areas of carcinoma in situ (PanIN3). In a series of aging experiments, the iKras* mice were kept on doxy for up to 17 weeks after induction of pancreatitis without developing invasive disease. This finding was consistent with those observed in a cohort of KC mice for the same period of time and support the notion that oncogenic Kras* inefficiently drives PDA (10, 11).
Oncogenic Kras is required for PanIN maintenance.
We took advantage of the reversibility of Kras* expression in the iKras* model to address whether Kras* activity is continuously required during pancreatic carcinogenesis. For this purpose, we kept iKras* mice on doxy for 23 weeks, until the pancreatic parenchyma was largely composed of PanIN lesions, fibroinflammatory stroma, and interspersed acini (Supplemental Figure 1E). Then the mice were placed on doxy-free water, and tissues were harvested after 2 weeks. The pancreata in those animals appeared fibrotic, atrophic, and largely populated by acini with some scattered, focal areas of ADM (Supplemental Figure 1, E and F), with little evidence of inflammation or presence of PanIN lesions. Thus, these animals showed reversion of PanINs, indicating that Kras* is indeed required for PanIN maintenance.
In order to study PanIN regression in more detail, we elected to use acute pancreatitis induction with cerulein, as previously described, to trigger consistent, tissue-wide PanIN formation, thus eliminating variability in PanIN onset among individual mice. Kras* expression was induced by doxy administration in adult mice (3–5 weeks old). The mice were continuously kept on doxy (Kras* ON) starting 72 hours before induction of acute pancreatitis and for 3 weeks following the cerulein treatment. Then we inactivated oncogenic Kras* expression by returning the animals to normal water (Kras* OFF) and harvested their pancreata at subsequent time points as indicated in the scheme in Figure 2A. Transgene regulation was confirmed by quantitative RT-PCR (qRT-PCR) analysis for Kras* (Figure 2B). We also directly measured the levels of Ras-GTP, the active form of the protein, by Ras-GTP pull-down assays. We used pancreata extracted from wild-type mice and KC mice 3 weeks after pancreatitis induction and iKras* mice after pancreatitis induction with Kras* ON for 3 weeks and Kras* OFF for 2 days, 3 days, and 2 weeks. Figure 2C shows a representative assay: Ras-GTP levels were significantly increased in the pancreatic epithelial compartment of KC and iKras* mice compared with wild-type mice. Ras-GTP levels were normalized to total Ras expression (Figure 2D) and to expression of E-cadherin (Figure 2E), a pan-epithelial marker that allowed us to correct for the possible confounding presence of differing amounts of tumor stroma in the individual samples. The Ras-GTP levels in KC and iKras* mice were comparable (KC/WT ratio, 6.6 ± 2.1; iKras*/WT, 5.9 ± 1.6). In iKras* mice, Ras-GTP was rapidly downregulated upon doxy withdrawal (iKras* OFF 3 days/WT, 1.2 ± 0.8; iKras* OFF 2 weeks/WT, 0.7 ± 0.8). Phospho-ERK1/2 levels were also comparable in KC and iKras* mice, but were rapidly downregulated in the latter following Kras* inactivation (Figure 2, C and G). Our results are consistent with the previously published analysis of the TetO-KrasG12D transgene in the lung (12), where mRNA expression levels from the transgene were comparable to those of the endogenous locus, and likely explain the similar phenotype in the KC and iKras* models.
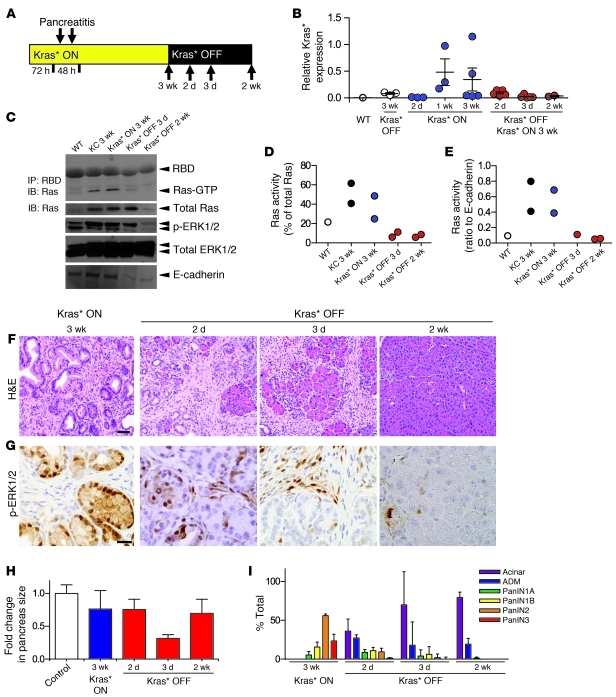
Figure 2. Kras* inactivation in mucinous ADM/early PanINs.
(A) Experimental design. Kras* expression was kept ON for 3 weeks following acute pancreatitis; then, Kras* was turned OFF; tissues were harvested at the indicated time points (arrows). n = 3–5 mice/time point. (B) Kras* expression by qRT-PCR. Each point is an individual mouse. Data represent mean ± SEM. (C) Representative Western blot showing Ras protein activity (Ras-GTP) measured by Raf1-RBD pull-down assay; as well as blots showing levels of phospho-ERK1/2, total ERK1/2, and E-cadherin. (D) Ras protein activity normalized to total Ras. (E) Ras protein activity normalized to the epithelial marker E-cadherin. (F) Histology of the pancreas at the indicated time points. Scale bar: 50 μm. (G) Activation of the MAP/ERK kinase pathway measured by phospho-ERK1/2 immunohistochemistry. Scale bar: 20 μm. (H) Quantification of the change in pancreas size. Data represent mean ± SEM. (I) Quantification of lesions at the indicated time points. Data represent mean ± SEM.
Analysis of the tissues harvested 2 and 3 days after doxy removal revealed widespread replacement of PanINs with ADM interspersed with pancreatic acini (Figure 2F), and PAS staining was progressively reduced (Supplemental Figure 3A). Strikingly, by 2 weeks after doxy removal, the pancreas was of normal size (Figure 2H) and appearance and was largely populated by acinar clusters (Figure 2, F and I). We proceeded to determine whether the tissue had recovered its normal characteristics also in terms of gene expression and activity. Analysis of phospho-ERK1/2 levels, as a readout of the MAPK/ERK pathway, revealed it to be rapidly inactivated in the epithelial cells upon doxy withdrawal (Figure 2G). Intriguingly, we observed a surge in MAPK/ERK activity in the stroma during the remodeling process. The expression of other PanIN markers, such as the ductal markers CK19 and mucin 1 (Muc1), was also rapidly downregulated upon Kras* inactivation (Supplemental Figure 3, B and C).
In order to determine whether the requirement for Kras* activity changes over time, we performed another series of experiments in which iKras* mice were kept on doxy for 5 weeks following induction of pancreatitis. At this time point, when the pancreas was largely composed of PanIN lesions surrounded by active stroma, we returned the mice to regular water, thus inactivating Kras* (Figure 3A). We collected tissues at several time points following removal of doxy (see scheme in Figure 3A). Ras-GTP expression decreased upon doxy withdrawal (Figure 3B), although the downregulation was slower than observed when doxy was removed at the 3-week time point (Figure 2C). In fact, these tissues did not appear to be undergoing significant changes at 2 and 3 days following Kras* inactivation, based on both histology and expression of PanIN markers (Figure 3C). The dynamics of tissue remodeling appeared dramatically different than in the 3-week set: PAS-positive PanIN lesions persisted 2 and 3 days after Kras* inactivation (Supplemental Figure 3D). By 2 weeks after doxy withdrawal (Kras* OFF), only a small remnant of the pancreas was present (Figure 3E). We observed partial acinar cell recovery, as well as occasional pancreatic lipomatosis (a common reaction to epithelial cell death in the pancreas) (24) and areas of ADM (Figure 3, C and F). In addition, phospho-ERK1/2 and claudin-18 levels were rapidly downregulated in most epithelial cells (Figure 3, D and G), although a surge of phospho-ERK1/2 in the stroma was transiently observed. Aberrant expression of CK19 in the basolateral membrane of epithelial cells and intracellular accumulation of Muc1 were frequently observed with Kras* ON; upon Kras* inactivation, both CK19 and Muc1 gradually returned to their normal subcellular localization, with apical accumulation in ductal cells (Supplemental Figure 3, E and F). The expression pattern and level of most other markers were largely comparable to those in normal pancreas (Supplemental Figure 4, A–F). In addition, we observed β-catenin accumulation in PanIN lesions and expression of EGFR family members, consistent with previous work in humans and mice (25–29). All of these changes were reversed upon Kras* inactivation (Supplemental Figure 5, A–C). Taken together, our data indicated that while PanIN lesions could not persist once Kras* was inactivated, the repair process was not complete and left a small, fibrotic pancreas with fewer acini than expected.
Figure 3. Kras* inactivation in established PanINs.
(A) Experimental design: Kras* expression was kept ON for 5 weeks following acute pancreatitis; then, Kras* was turned OFF; tissues were harvested at the indicated time points (arrows). n = 3–5 mice/time point. (B) Representative Western blot showing Ras protein activity (Ras-GTP) measured by Raf1-RBD pull-down assay. (C) Histology of the pancreas at the indicated time points. Scale bar: 50 μm. (D) MAP/ERK pathway activation shown by phospho-ERK1/2 immunohistochemistry. Scale bar: 20 μm. (E) Quantification of the change in pancreas size. Data represent mean ± SEM. (F) Quantification of pancreatic lesions. Data represent mean ± SEM. (G) Immunohistochemistry for the PanIN marker claudin-18. Scale bar: 20 μm.
In order to determine whether complete pancreatic repair was delayed, and could be achieved over time, we harvested additional pancreata 5 weeks following Kras* inactivation. At this time point, we did not observe evidence of persisting PanINs; however, the pancreas at histological analysis appeared fibrotic (Figure 3C) and had not increased in size back to control levels (Figure 3E).
Mechanism of PanIN regression and epithelial tissue repair.
Due to the rapid and dramatic recovery of the pancreas upon inactivation of Kras* following its expression for 3 weeks, we evaluated whether this recovery might be due to death of the cells forming PanIN lesions, followed by active proliferation of residual acinar cells that had not undergone recombination within the tissues, a mechanism that has been described following pancreatic damage in the absence of Kras* (30, 31). However, staining for cleaved caspase-3 did not show any significant changes in the number of apoptotic cells over time upon Kras* inactivation (Figure 4A). Moreover, proliferation analysis using the Ki67 marker showed rare positive cells immediately following Kras* inactivation, and active proliferation of acinar cells only at later time points (Figure 4B). Proliferation of the newly formed acinar cells is likely to play a role in the later phases of tissue repair, thus explaining the increase in pancreas size between 3 days and 2 weeks after Kras* inactivation (Figure 2H). We also found that EGFP (linked to rtTa expression in the R26-rtTa transgene; see Figure 1A) was expressed in the epithelial compartment both before and after Kras* inactivation (Supplemental Figure 6A), indicating that the newly formed acini derived from cells that had expressed rtTa and EGFP, and thus Kras*.
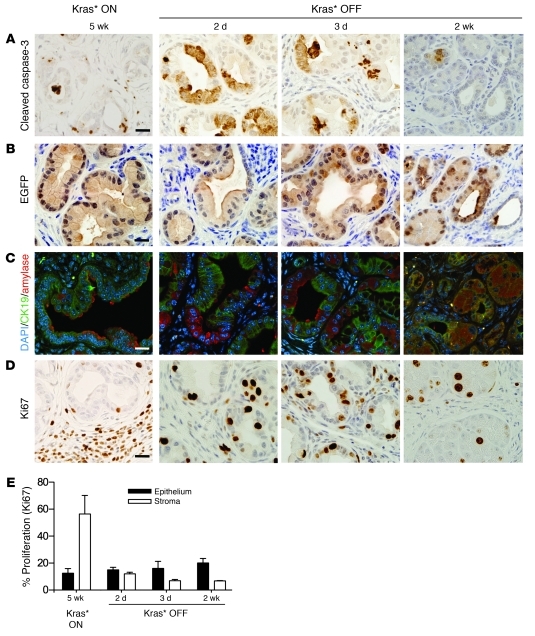
Figure 4. Mechanism of tissue recovery from early PanINs.
Kras* expression was maintained ON for 3 weeks following pancreatitis, then turned OFF for 2 days, 3 days, and 2 weeks. n = 3–5 mice/time point. (A) Apoptosis as indicated by cleaved caspase-3 immunohistochemistry. Scale bar: 20 μm. (B) Co-immunofluorescence of PanIN lesions and tissue proliferation during tissue repair: Ki67 (green), CK19 (red), and DAPI (blue). Scale bar: 20 μm. (C–F) CK19 (green), amylase (red), and DAPI (blue) co-immunofluorescence analysis of PanIN transdifferentiation in iKras* pancreas after (C) Kras* ON 3 weeks and (D) Kras* OFF 2 days, (E) 3 days, and (F) 2 weeks. Scale bar: 20 μm (G) Quantification of CK19- and amylase-positive cells at the indicated time points. Data represent mean ± SEM.
Kras* has been hypothesized to prevent ductal-to-acinar re-differentiation following pancreatitis-induced ADM, thus acting as a barrier to tissue repair and maintaining the epithelial cells in a differentiation state that is possibly more prone to neoplastic transformation (8, 19). Therefore, we investigated the differentiation status of pancreatic cells during Kras* expression and upon Kras* inactivation. PanIN lesions express the ductal marker CK19 (Figure 4C) and do not express the acinar cell marker amylase. By contrast, at 2 and 3 days after doxy withdrawal, we frequently observed cells with mixed acinar-ductal differentiation, co-expressing the ductal and PanIN marker CK19 and the acinar marker amylase (Figure 4, D and E). By 2 weeks after Kras* inactivation, these intermediate cell types were substituted by acini expressing amylase, while CK19 was confined to the ducts (Figure 4F). This may indicate that the PanIN regression occurs by reprogramming of PanIN cells into acinar cells and explain the complete recovery of the pancreas, notwithstanding its limited regenerative potential (32). Quantification of the cell types present at different time points further disproved the possibility that residual acinar cells had repopulated the pancreas: upon Kras* inactivation, the number of acinar cells increased more than 20-fold in 48 hours, an increase that could not be explained by cell division alone in any mammalian cell (Figure 4G).
In order to gain additional insight into the repair process, we addressed the expression of pancreatic progenitor markers in the PanINs and during the regression process. PanIN lesions expressed a subset of genes that are associated with pancreatic progenitors and with maintenance of an undifferentiated status (33–35) and reactivated in pancreatic cancer (10, 36), such as Sox9, Pdx1, and Hes1, a Notch signaling component and target gene (Supplemental Figure 6, B–D). The expression of those markers was maintained during the initial recovery stages, in structures with mixed acinar and ductal differentiation, and was repressed once full recovery had been achieved (Supplemental Figure 6, B–D). Taken together, our data support the hypothesis that Kras* is altering the differentiation status of pancreatic epithelial cells, derailing the repair process following pancreatitis.
We next addressed the mechanism of the recovery process in mice in which Kras* had been kept on for 5 weeks following pancreatitis. In sharp contrast to the observations at the 3-week time point, here we observed a dramatic increase of apoptotic cells (cleaved caspase-3 positive) upon Kras* inactivation (Figure 5A). This observation is consistent with the smaller size of the pancreas in these animals (Figure 3E). We then assessed the proliferation index by Ki67 immunostaining. In the presence of oncogenic Kras*, both the epithelial cells and the surrounding stroma were Ki67 positive (Figure 5, D and E). Following Kras* inactivation, the overall proliferative index was unchanged; however, the proliferation was confined to the acinar compartment. In contrast, proliferation in the stroma was rapidly downregulated. Using EGFP-based lineage tracing, we were able to determine that the vast majority of the epithelial cells in the tissue after Kras* inactivation derive from cells that had previously expressed the Kras* transgene (Figure 5B) rather than repopulation of the pancreas from acinar cells that had not undergone Cre recombination and thus never expressed the Kras* transgene. Thus, some limited re-differentiation of ductal to acinar cells might play a role in the repair process, but this was not as prominently observed as at the earlier time points (Figure 5C; compare with Figure 4, C–F). Finally, the expression of Sox9 and Pdx1 was downregulated during the repair process (Supplemental Figure 4, A and B); however, this did not occur as rapidly as seen after the earlier, 3-week time point. The limited proliferative capability of adult pancreatic cells might account for the incomplete repair process. Nevertheless, the mice at this stage appeared healthy and showed no sign of pancreatic insufficiency.
Figure 5. Extensive tissue remodeling in established PanINs following Kras* inactivation.
Kras* expression was maintained ON for 5 weeks following pancreatitis, then turned OFF for 2 days, 3 days, and 2 weeks. n = 3–5 mice/time point. (A) Cell death shown by cleaved caspase-3 immunohistochemistry. Scale bar: 20 μm. (B) Immunohistochemistry for the lineage tracer EGFP. Scale bar: 20 μm. (C) CK19 (green), amylase (red), and DAPI (blue) co-immunofluorescence. Scale bar: 20 μm. (D) Tissue proliferation shown by Ki67 immunohistochemistry. Scale bar: 20 μm. (E) Quantification of cellular proliferation (Ki67) in each tissue compartment for the indicated time points. Data represent mean ± SEM.
Interactions between the epithelial cells and their microenvironment are regulated by oncogenic Kras.
In early PanIN lesions, inactivation of Kras* is accompanied by reversal of enhanced proliferation in the stroma (Figure 4B) and complete remodeling of this compartment within 2 weeks (Supplemental Figure 7A). Although the remodeling of the stroma was not complete in tissues where Kras* had been inactivated at the 5-week time point, rapid downregulation of proliferation in the stroma was evident (Figure 5, D and E). We therefore investigated whether Kras* activity is required to maintain the extensive fibroinflammatory stroma that is a hallmark of pancreatic cancer and has been proposed to mediate its resistance to treatment (37). The pancreatic cancer stroma is largely composed of vimentin- and SMA-positive cells of mesenchymal origin. SMA is a marker of activated fibroblasts and is not expressed in quiescent pancreatic stellate cells, the resident mesenchymal cell population in the pancreas. As expected, the stroma surrounding PanIN lesions in the iKras* pancreata was SMA and vimentin positive with Kras* ON (Figure 6A and Supplemental Figure 7B). Interestingly, while vimentin expression remained unaltered following Kras* inactivation, SMA was rapidly downregulated. Loss of SMA expression preceded remodeling of the stroma, both in the 3-week and in the 5-week tissues. In the 5-week tissues, the fibrotic areas that persisted were SMA negative and non-proliferative, features consistent with scar tissue. Thus, our data indicate that Kras* activity in the epithelium is required to maintain the surrounding active stroma.
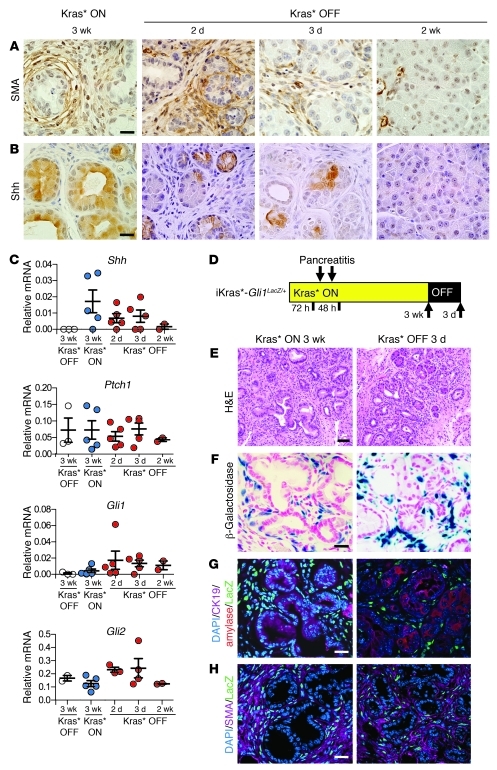
Figure 6. Oncogenic Kras* regulates interactions between epithelial cells and their microenvironment.
Kras* expression was maintained ON for 3 weeks following pancreatitis, then turned OFF for 2 days, 3 days, and 2 weeks. (A) SMA and (B) Shh ligand immunohistochemistry. Scale bar: 20 μm. (C) qRT-PCR analysis of Hedgehog signaling components Shh, Ptch1, Gli1, and Gli2. Each point represents 1 mouse. Data represent mean ± SEM. (D) Experimental design. In iKras*;Gli1LacZ/+ experimental mice, Kras* expression was kept ON for 3 weeks following pancreatitis, then turned OFF for 3 days. n = 2 mice/time point. (E) Histology. Scale bar: 50 μm. (F) β-Galactosidase staining for Gli1/LacZ expression. Scale bar: 20 μm. (G) CK19 (purple), amylase (red), Gli1/LacZ (green), and DAPI (blue) co-immunofluorescence. Scale bar: 20 μm. (H) SMA (purple), Gli1/LacZ (green), and DAPI (blue) co-immunofluorescence. Scale bar: 20 μm.
One of the key signaling pathways mediating epithelial-mesenchymal interactions in pancreatic cancer is Hedgehog signaling. The Hedgehog signaling pathway is deregulated during the onset of pancreatic cancer (38–40), since, unlike normal pancreatic epithelium, PanIN and pancreatic cancer cells secrete the Hedgehog ligands Shh and Ihh. The Hedgehog ligands act in a paracrine manner to activate signaling in the stroma (41), and activation of the pathway has been linked to stroma expansion and induction of its tumor-promoting ability (37, 42).
In iKras* tissues, Shh expression was induced upon Kras* activation, and its expression was dependent on continuous Kras activity (Figure 6, B and C). We also quantified, by qRT-PCR, the expression of Shh, Ptch1, and Gli1 — Hedgehog pathway components and targets of the pathway — and Gli2, which together with Gli1 mediates the transcriptional output of the Hedgehog pathway. Shh and Gli2 expression was strictly Kras* dependent, while Ptch1 did not significantly change, and Gli1 persisted after Kras* inactivation (Figure 6C). In order to determine which cell compartment had active Hedgehog signaling, we generated iKras*;Gli1LacZ/+ mice, in which one copy of the Gli1 gene was replaced by LacZ, and analyzed their pancreata 3 weeks after inducing pancreatitis and Kras* expression (Figure 6D), when PanINs were prevalent (Figure 6E). In these animals, the fibroblasts surrounding the epithelial lesions were LacZ positive (Figure 6F). LacZ expression overlapped with SMA-positive cells, but not with cells expressing the epithelial markers CK19 or amylase (Figure 6, G and H). We also collected tissues in iKras*;Gli1LacZ/+ mice 3 days after Kras* inactivation, at a stage when SMA expression is downregulated, and found that Gli1 was still highly expressed in the stroma surrounding the acinar-ductal clusters (Figure 6, F–H). Thus, Gli1 expression persists following fibroblast inactivation, indicating that although Hedgehog signaling is one of the pathways that mediate the paracrine interactions between the epithelial cells and the stroma, it is not the only component of a likely complex regulatory mechanism.
In addition to fibroblasts, the pancreatic cancer stroma is rich in inflammatory cells. This is not surprising, as several inflammatory cytokines are upregulated in PanIN lesions. We therefore investigated the expression of mediators of inflammatory pathways that have been shown to be important for pancreatic cancer tumorigenesis. Cox2 is overexpressed in PDA, and forced overexpression in mice has been shown to be sufficient to induce pancreatic dysplasia (43, 44). More recently, IL-6 and its downstream effector phospho-Stat3 have been shown to be important not only during the initial stages of pancreatic cancer development, but also in advanced disease (20, 45, 46). As previously observed in other mouse models of PDA (10) and in human tumors, Cox2, IL-6, and phospho-Stat3 were expressed in the PanIN lesions of iKras* mice, and their expression was reduced upon Kras* inactivation (Supplemental Figure 4E and Supplemental Figure 7, C–E). In addition, MMP7, a matrix metalloproteinase that is expressed in human (47) and mouse pancreatic cancer (10), was expressed in the PanIN lesions of the iKras* mice (Supplemental Figure 4F and Supplemental Figure 7F) but downregulated upon Kras* inactivation.
Taken together, these data show that fibroblast activation, inflammatory cell infiltration, and production of enzymes that might remodel the extracellular matrix are regulated by Kras* both during the initiation of pancreatic carcinogenesis and once PanIN lesions are established, thus highlighting the essential role of oncogenic Kras is the maintenance of the stroma.
Role of oncogenic Kras in PDA.
As previously stated, PanIN lesions in iKras* animals do not progress to invasive PDA, at least in the time frame considered. In the KC model, development of PDA has been shown to require not only Kras* mutation, but also inactivation of at least one tumor suppressor gene (11, 48, 49). The iKras* mouse was crossed with p53-null mice (50) in order to obtain iKras*-p53+/– mice, in which one allele of p53 is inactivated while the other is present in its wild-type form. Following the experimental design described above, PanIN formation was induced in iKras*-p53+/– mice by activation of Kras* expression with doxy, followed by induction of acute pancreatitis. In a first cohort of animals, the tissue was harvested after 5 weeks (Figure 7A). The pancreata of iKras*-p53+/– mice presented with PanINs (Figure 7B), dilated ducts with the presence of intracellular mucins, and extensive fibroinflammatory stroma (data not shown). Ras pathway activation in the tissues was verified by phospho-ERK1/2 staining (Figure 7C), and both epithelium and stroma were found to be highly proliferative (Figure 7D). In a subset of the animals in which Kras* had been expressed for 5 weeks, doxy was removed to inactivate Kras* and the pancreata were harvested 2 weeks later. At dissection, the pancreata appeared as a small, translucent remnant of tissue, without the characteristic fibrotic appearance characteristic of pancreata bearing PanIN lesions. Histological analysis verified the absence of PanIN lesions within 2 weeks after Kras* inactivation (Figure 7B). However, the pancreata of iKras*-p53+/– mice did not return to their normal morphology and histology following Kras* inactivation. Rather, the tissue was characterized by normal acini interspersed with dilated ducts and ADM and surrounded by fibrosis and occasional lipomatosis (Figure 7B), but with a minimal inflammatory infiltrate. Phospho-ERK1/2 was inactivated in the epithelium, but still detectable in the stroma following Kras* inactivation (Figure 7C). The proliferation index was dramatically reduced following removal of doxy (Figure 7D), with only acinar cells staining positive for Ki67 and no staining in ductal structures or in the fibrotic tissue.
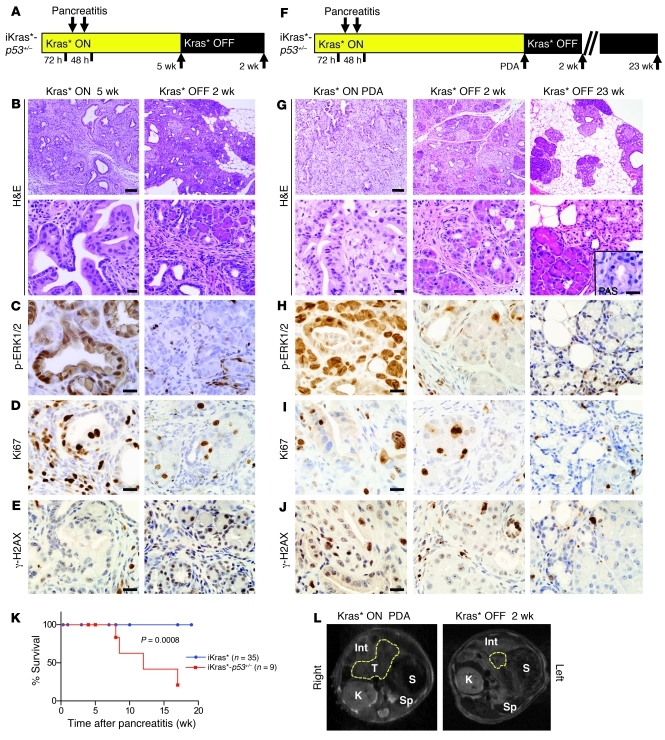
Figure 7. iKras*-p53+/– model and the effect of Kras* inactivation.
Experimental design. Kras* expression was maintained ON for 5 weeks (A) or until the mice developed frank PDA (F) before being turned OFF for 2 weeks or 23 weeks. (B and G) Histology of the pancreata at indicated time points. Scale bar: 100 μm (top row) and 20 μm (bottom row). Inset: PAS staining. Scale bar: 20 μm. (C and H) Phospho-ERK1/2 immunohistochemistry. Scale bar: 20 μm. (D and I) Ki67 immunohistochemistry. Scale bar: 20 μm. (E and J) Analysis of genomic instability in iKras*-p53+/– mice by γ-H2AX immunohistochemistry. Scale bar: 20 μm. (K) Kaplan-Meier survival curve. Log-rank statistical analysis yielded a P value of 0.0008. (L) In vivo imaging of tumor regression in one iKras*-p53+/– animal using MRI. Total animals imaged, n = 4. T, tumor, outlined in yellow; S, stomach; Sp, spleen; K, kidney; Int, intestine.
Since no bona fide PDA was observed in the 5-week cohort, a second cohort of animals was reserved for an aging experiment (Figure 7F). Between 8 and 18 weeks after Kras* activation, the animals in this iKras*-p53+/– cohort (n = 9) died or needed to be euthanized due to weight loss and a deteriorating clinical condition, in accordance with the animal protocol guidelines, while all iKras* controls (n = 35) included in this experiment reached this age without clinical sign of disease (Figure 7K). The difference in survival between the two cohorts was determined to be highly significant, with a P value of 0.0008 in a log-rank test. At necropsy, the pancreata of these animals demonstrated invasive adenocarcinoma, in some cases accompanied by hemorrhagic ascites, with admixed poorly differentiated and well-differentiated areas, and duodenal invasion (Figure 7G). The tumors presented with rare PAS positivity and extensive collagen deposition (data not shown), strong phospho-ERK1/2 positivity, and proliferation in both the epithelial and stromal compartments (Figure 7, H and I).
In some of the sick animals, Kras* was inactivated by interruption of doxy administration. The animals in this cohort that were removed from doxy returned to good health; in contrast, the animals that were kept on doxy succumbed to PDA (Figure 7K). At dissection (2 weeks following Kras* inactivation), the pancreata appeared indistinguishable from those of the 5-week cohort described above: they were atrophic, with acini interspersed by residual fibrosis (Figure 7G), and showed no or little PAS staining (data not shown). MAPK activation, measured as phospho-ERK1/2 level, was only rarely observed in epithelial or stromal cells (Figure 7H), and proliferation in the stroma was almost completely abrogated, while some of the acini remained Ki67 positive (Figure 7I). In order to confirm the tumor regression within the same animal over time, we performed MRI on iKras*-p53+/– animals (n = 4) before (when showing clinical signs of disease) and after Kras* inactivation. In the presence of active Kras*, we observed a tumor mass in the head of the pancreas or, occasionally, in the pancreas tail. Upon Kras* inactivation, the pancreatic mass regressed, leaving a small pancreatic remnant (Figure 7L, mass in the head of the pancreas and regression; and Supplemental Figure 8A, mass in the tail of the pancreas and regression).
Together, our data indicate that Kras* is required for cancer maintenance, suggesting that Kras* and/or its downstream effectors are potential therapeutic targets in this disease. In order to determine whether Kras*-independent cells had persisted that could lead to cancer recurrence, iKras*-p53+/– mice that had been on doxy for 10 weeks were taken off doxy and observed over time. Mice that were kept off doxy (Kras* OFF) for 23 weeks were healthy, with no evidence of relapse. We harvested the tissues and confirmed that there was no residual or recurring disease; a few mucinous ducts present in the pancreas were PAS negative (Figure 7G, inset) and were identified as ectopic Brunner glands.
Analysis of tumors from iKras*-p53+/– mice showed loss of expression of the wild-type allele of p53 (Supplemental Figure 8, B and C). Nuclear accumulation of a phosphorylated form of histone 2A (γ-H2AX) reflects oncogenic stress that can be associated with genomic instability (51) and is a common feature of human and mouse PDA (52). Analysis of tissues from iKras*-p53+/– mice revealed nuclear expression of γ-H2AX, both in the epithelial cells and in the stroma (Figure 7, E and J). Upon Kras* inactivation, γ-H2AX expression was strongly reduced and, when present, confined to the cytoplasm, most likely in dying cells, where the integrity of the nuclear membrane was compromised (Figure 7, E and J). In order to determine whether oncogenic stress was associated with genomic instability in our samples, we performed DNA fingerprinting (53, 54) to detect genetic alterations in the tumors compared with matched genomic DNA from the same mouse. Our results indicate the presence of genomic instability in the tumors (Supplemental Figure 8D). Interestingly, some of the tumor-specific bands became undetectable upon Kras* inactivation, indicating that the cells carrying those genetic alterations did not persist. Taken together, our data indicate that Kras* activity is still required for tumor maintenance in pancreatic cells that have accumulated genetic damage and have lost expression of tumor suppressor genes.
Discussion
A new approach to modeling Kras inhibition in pancreatic cancer.
Mouse models are widely used to study the events that lead to cancer formation and have represented a powerful tool for new discoveries (17, 55). Since most cancer patients are diagnosed with advanced disease, models that mimic Kras* inhibition in the advanced stages of cancer are particularly relevant to the human condition. In particular, a key point is to determine which oncogenes are important during the initiation and progression phases of cancer versus cancer maintenance. The term “oncogene addiction” has been used to describe the need for cancer cells to maintain the activity of oncogenes even after additional genetic and epigenetic events have occurred (56). The concept has been validated in an elegant model of pancreatic islet tumors driven by myc overexpression, where oncogene inactivation leads to tumor regression (57). Oncogenic addiction for Kras* has been studied in lung adenocarcinoma (12), where tumors remain Kras* dependent even in the presence of other genetic alterations. However, whether those findings could be extended to PDA, a tumor type characterized by activation of several signaling pathways (5) and by the extensive accumulation of desmoplastic stroma, had so far not been addressed. Expression of mutant Kras* from its endogenous locus using the LoxP-STOP-LoxP-KrasG12D allele (58) results in stepwise PanIN formation that closely mimics the human disease (10, 11, 59). Additional models have used Cre-mediated removal of a stop cassette to activate Kras* expression specifically in the pancreas but not from the endogenous locus (60). A limitation of those models is that oncogenic Kras*, once activated, is constitutively expressed. Different subtypes of pancreatic cancer have recently been defined in human patients (14), with potentially differential dependence on oncogenic Kras*, thus highlighting the need for a suitable model of reversible Kras* expression in the pancreas. Previous attempts at generating a reversible form of Kras* expression in the pancreas using the tTa/TetO system have been limited by the lack of a suitable driver for the tTa transcription factor.
Here we used an approach that allows conditional, Cre-mediated activation of rtTa expression and were thus able to use pancreas-specific Cre lines to induce its expression. We opted for the broadly expressed p48-Cre line, since previous studies in mouse models have shown that PanINs can arise from acini (18, 23), ducts (22), and even islets during conditions of tissue damage (61). Since Cre recombination is irreversible, our approach leads to expression of rtTa in most pancreatic epithelial cells. The approach allows us to express Kras* in the pancreatic epithelium in a temporally and spatially regulated manner and, more importantly, in a reversible manner. It has been recently successfully used in a model of basal cell carcinoma (62) and could easily be adapted to study other oncogenes in other organs, and should be of broad interest to scientists interested in tumor maintenance. The iKras* mouse develops PanINs, and, when crossed with mice with p53 loss of function, develops PDA that resembles the human disease and mimics the previously published LSL-KrasG12D/+;LSL-Trp53R172H/+;Pdx-1-Cre model of pancreatic cancer (48).
Oncogenic Kras prevents tissue repair following acute pancreatitis.
Acute pancreatitis can be induced in mice by injection with the cholecystokinin analog cerulein (19) and is characterized, at the tissue level, by infiltration of inflammatory cells and edema, as well as by ADM, defined as the replacement of acinar cells with duct-like structures. Wild-type animals rapidly recover from acute pancreatitis; the pancreatic parenchyma returns to its normal architecture, and the inflammatory cell infiltration and edema subside within 1–3 weeks after treatment. By contrast, it has been observed that in Kras* mutant animals, recovery is completely prevented (19), and acute pancreatitis is rapidly followed by more severe ADM surrounded by fibrosis and, by 3 weeks after the treatment, by extensive mucinous ADM/early PanINs. It has been hypothesized that Kras* directly prevents tissue repair, but so far this has not been demonstrated experimentally (8, 19). In addition, the de-differentiation of acinar cells to duct-like cells has been hypothesized to be a necessary step for PanIN formation, at least for PanINs of acinar origin (8, 19). We have taken advantage of the reversibility of Kras* expression in our model to investigate the role of this oncogene during the earliest stages of pancreatic carcinogenesis. Our results show that oncogenic Kras* prevented the repair process by maintaining the ductal differentiation of acinar cells; this process was initially fully reversible upon Kras* inactivation. An additional finding was that expression of Kras* in the epithelium was responsible for formation and maintenance of the fibrotic stroma that accompanies PanIN formation and is prevalent in PDA. The expansion of a pro-tumor stroma is one of the “hallmarks of cancer” (63), and in pancreatic cancer, the extensive desmoplastic stroma has been shown to contribute to this tumor’s chemoresistance (37).
Kras is required at all stages of pancreatic carcinogenesis.
In a last set of experiments, we have analyzed iKras*-p53+/– mice, in which activation of oncogenic Kras* is accompanied by loss of one allele of the p53 tumor suppressor (and by loss of expression from the other allele). The purpose of this set of experiments was to address the important question of the role of this oncogene in tumor maintenance, a question of great biologic and therapeutic importance. The role of Kras in advanced pancreatic cancer has in the past been addressed in established pancreatic cancer cell lines. Interestingly, two independent groups have found that pancreatic cancer cell lines can be subdivided into “Kras*-dependent” and “Kras*-independent” subsets (13, 14). Pancreatic cancer, however, is characterized by extensive stroma, which has been shown to alter the cellular response to treatment (37). Moreover, the established cell lines do not allow us to study the biology of the precursor lesions of pancreatic cancer. Kras* inactivation in iKras*-p53+/– mice with PanINs or adenocarcinoma results in tumor regression, thus indicating that the tumor cells have become “addicted” to Kras* expression and activity. Due to the limited regenerative capability of the adult pancreas, complete repair is not achieved upon Kras* inactivation; however, even when we let the animals age for several months, we did not observe any tumor recurrence.
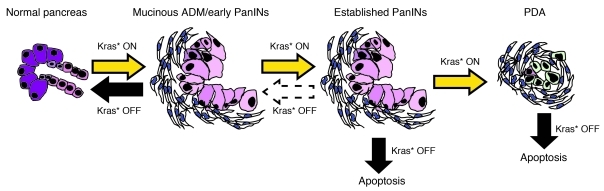
In summary, we have used our iKras* mice to study the effect of Kras* inhibition at different stages of pancreatic carcinogenesis. In addition, we have used a model of pancreatitis-induced PanIN formation to synchronize the appearance of the lesions, thus facilitating quantification and analysis of the data. In our model, activation of Kras* followed by induction of acute pancreatitis leads to pancreas-wide PanIN formation within 3 weeks and high-grade PanINs (including carcinoma in situ) by 5 weeks. In the presence of one loss-of-function allele of p53, the mice showed tissue-wide PanIN3 by 5 weeks and invasive adenocarcinoma by 8 to 18 weeks. Our findings are summarized in Figure 8: inactivation of Kras* at the early PanIN stage led to rapid and complete tissue recovery. At the mechanistic level, the recovery was accompanied by re-differentiation of PanIN cells into acinar cells and by remodeling of the stroma. However, once high-grade PanINs had formed, Kras* inactivation was linked to massive cell death, indicating that those cells had become addicted to the continuous expression of oncogenic Kras*. Finally, Kras* inhibition also led to regression of invasive adenocarcinoma. However, in both cases partial recovery of pancreatic acini was accompanied by persistence of metaplastic areas surrounded by fibrotic scar tissue. These findings differ from what was observed in lung adenocarcinoma upon Kras* inactivation (12). In that case, Kras* inactivation led to full regression, similar to what we observed when Kras* was inactivated in low-grade PanIN lesions (Figure 2). Taken together, our results indicate that targeting Kras* is likely to have a profound effect on pancreatic cancer. These data support the potential utility of targeting Kras* or its downstream signaling pathways as a therapeutic approach in patients with pancreatic cancer.
Figure 8. Proposed model for the role of oncogenic Kras in the initiation and maintenance of PanINs and PDA.
Initial oncogenic Kras activation leads to pancreatic dysplasia. When Kras is inactivated at the early time points, the pancreatic tissue reverts back to its original state. However, when dysplasia is advanced, or if frank PDA is present, turning off Kras will induce apoptosis in the dysplastic epithelium, and the remodeling of the pancreatic parenchyma is incomplete even after an extended period of time.
Methods
Mice.
Animals were housed in specific pathogen–free facilities of the University of Michigan Comprehensive Cancer Center. p48Cre (Ptf1a-Cre) mice (15) (provided by Christopher V. Wright, Vanderbilt University, Nashville, Tennessee, USA) were intercrossed with TRE-KrasG12D (The Jackson Laboratory, stock #004735) (12) and R26-rtTa (The Jackson Laboratory, stock #005670) (16) mice to create iKras* triple mutants: p48-Cre;R26-rtTa-IRES-EGFP;TetO-KrasG12D. iKras* mice were also crossed with p53-null mice to create iKras*-p53+/– quadruple mutants: p48-Cre;TRE-KrasG12D;R26-rtTa;p53+/– (The Jackson Laboratory, stock #002101) or with Gli1LacZ/LacZ (The Jackson Laboratory, stock #008211) (64) mice to generate iKras*;Gli1LacZ/+ mice. Combinations of single or double mutant littermates were used as controls. LSL-KrasG12D mice were provided by David Tuveson (Cambridge Research Institute, Cambridge, United Kingdom) and were bred with p48-Cre mice to generate KC double transgenics. A Kaplan-Meier survival curve was created to represent animals that had to be euthanized, according to the animal protocol, or died during the time of the experiments. Statistical significance was established with a log-rank test, carried out using GraphPad Prism version 5.00 for Windows (GraphPad Software). A P value less than 0.05 was considered significant, and a P value less than 0.01 was considered highly significant.
Doxy treatment.
Doxy was administered in the drinking water at a concentration of 0.2 g/l in a solution of 5% sucrose and replaced every 3–4 days.
Induction of pancreatitis.
Mice were subjected to two series of 8 hourly intraperitoneal injections of cerulein (Sigma-Aldrich) at a concentration of 75 μg/kg over a 48-hour period, as previously described (19). Littermate controls were injected in parallel with the experimental animals.
Immunohistochemistry and immunofluorescence.
Pancreatic tissues from both experimental and control mice were fixed overnight in 10% neutral buffered formalin, embedded in paraffin, and sectioned. Embedding and sectioning was performed by the University of Michigan Cancer Center Histopathology Core. H&E, PAS, Gomori trichrome, and immunohistochemistry stainings were performed as previously described (65). For a list of the antibodies used, see Supplemental Table 1. β-Galactosidase staining was performed on tissues fixed in 4% PFA and embedded in OCT for cryosectioning. Tissues were equilibrated in rinse buffer (100 mM sodium phosphate pH 7.3, 2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% NP-40 w/v) for 10 minutes, then incubated overnight in X-gal stain solution (rinse buffer plus 1 mg/ml X-gal, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide). Samples were post-fixed in 10% neutral buffered formalin and counterstained with FastRed (Vector Laboratories). Images were taken with an Olympus BX-51 microscope, Olympus DP71 digital camera, and DP Controller software. For immunofluorescence, secondary antibodies labeled with FITC, Texas red, and Alexa Fluor (Invitrogen) were used. Cell nuclei were counterstained with DAPI (Invitrogen). The immunofluorescence images were acquired using an Olympus IX-71 confocal microscope and FluoView FV500/IX software.
qRT-PCR.
Tissue for RNA extraction was prepared through overnight incubation in RNAlater-ICE (Ambion) at –20°C, then isolated using RNeasy Protect (QIAGEN) according to the manufacturer’s instructions. Reverse transcription reactions were conducted using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Samples for qRT-PCR were prepared with 1× SYBR Green PCR Master Mix (Applied Biosystems) and various primers (sequences in Supplemental Table 2). All primers were optimized for amplification under reaction conditions as follows: 95°C 10 minutes, followed by 40 cycles of 95°C 15 seconds and 60°C 1 minute. Melt curve analysis was performed for all samples after completion of the amplification protocol. Gapdh was used as the housekeeping gene expression control.
Histopathological analysis.
Histopathological analysis was performed as previously described. A minimum of 50 total acinar or ductal cluster were counted from at least 3 independent animals for each group (66). Five randomly selected, non-overlapping high-power images (×20 objective) were taken for each slide. Each cluster was classified as acinar, PanIN1A, -1B, -2, or -3, or PDA based on the classification consensus (67).
Quantification of pancreas size.
Images of control and iKras* H&E sections of pancreatic tissue were taken with a Leica MZFLIII dissection microscope and Olympus DP72 camera. Pancreatic tissue area was determined with Image Pro Plus v4 software (MediaCybernetics). Averages of the area of the iKras* pancreata per time point were normalized to control averages to determine fold change.
Proliferation analysis.
Three randomly selected, non-overlapping high-power images (×20 objective) were taken from Ki67-stained slides from 2–3 independent animals for each group. Nuclei positive for Ki67 were counted as actively proliferating cells. Epithelial and stromal compartments for each image were counted separately, and data were expressed as percentage of total counted nuclei for each compartment. Errors bars represent SEM.
Active Ras pull-down assay.
Pull-down of active Ras was performed using an Active Ras pull-down kit (Pierce). Protein bands were visualized on Kodak Biomax XAR film for 15 seconds to 3 minutes. Activity levels were normalized to total Ras as well as E-cadherin levels using ImageJ software ( http://rsbweb.nih.gov/ij/).
Western blot analysis.
Tissues were homogenized in RIPA buffer (Sigma-Aldrich, R0278) and protease inhibitor (Sigma-Aldrich, P8340). Equal amounts of protein were electrophoresed in 12% SDS-PAGE gels, transferred to PVDF membrane (Bio-Rad). Membranes were blocked with milk, and primary antibody incubations were performed at room temperature for 2 hours (phospho-ERK1/2, ERK1/2, and E-cadherin 1:1,000 dilution). Secondary antibody HRP-conjugated anti-rabbit (1:5,000) was used and detected with Supersignal West Pico substrate (Thermo Scientific). Protein bands were visualized on Kodak Biomax XAR film.
MRI.
Mice were anesthetized with 1%–2% isoflurane/air, and body temperature was maintained by blowing warm air through the bore of the magnet using an Air-Therm (World Precision Instruments). MRI scanning was performed using a 7T Agilent Direct Drive system with a quadrature rat head volume coil (M2M). Mice were placed supine in the coil, taped below the thoracic cavity on the bed to reduce respiratory motion. A subcutaneous catheter was placed into skin of the neck for delivery of contrast agent. Two T1-weighted images were acquired before and 10 minutes after a subcutaneous bolus injection of Gd-DTPA (gadopentetate dimeglumine, Bayer HealthCare) at a dose of 0.7 mmol/kg using a spin-echo sequence, with fat saturation and the following parameters: repetition time (TR)/echo time (TE) = 757/15 ms, field of view (FOV) = 25 × 25 mm2, matrix size = 128 × 128, slice thickness = 1 mm, number of slices = 25, no gap, interleaved, and 4 dummy scans. T2-weighted images were acquired using a fast spin echo multi-slice sequence with TR/TE: 4,000/30 ms, 8 echo trains, 4 averages, 2 dummy scans, and the same slice package as the T1-weighted sequence. Using in-house software, the tumor boundary was manually defined on each slice and then integrated across slices to measure the volume.
Random amplified polymorphic DNA analysis (RAPD).
Genomic DNA was isolated from tumor and normal tissues from each animal using a QIAGEN DNeasy Blood and Tissue Kit per the manufacturer’s instructions. Inter-simple sequence repeat (inter-SSR) PCR was performed as described previously (68) using (CA)8RY, (CA)8RG, and (AAC)6Y primers. PCR products were separated with 2.5% agarose gels and visualized with SYBR Safe DNA gel stain (Invitrogen).
Study approval.
All animal protocols were approved by the University of Michigan University Committee on Use and Care of Animals (UCUCA).
Supplementary Material
Acknowledgments
We would like to thank Diane Simeone, Anj Dlugosz, Jörg Zeller, Ben Allen, and Mats Ljungman (University of Michigan, Ann Arbor) for scientific discussion and critical reading of the manuscript. We thank Jimmy Hogan, Ace Josifoki, and Beth Skendrovich for help with immunostaining and histology. The p48-Cre (Ptf1a-Cre) mouse was a gift from Chris Wright (Vanderbilt University). The CK19 antibody (Troma III) was obtained from the Iowa Developmental Hybridoma Bank. The Hes1 antibody was a gift from Ben Stanger (University of Pennsylvania, Philadelphia, Pennsylvania, USA). Work in the Pasca di Magliano laboratory is supported by the University of Michigan Biological Scholar Program, NCI 1R01CA151588-01, GI SPORE P50 CA 13810, a Pancreatic Cancer Action Network (PanCan)/American Association for Cancer Research (AACR) Career Development grant, and the Michigan Gastrointestinal Peptide Research Center (P30 DK 034933-25). M.A. Collins is supported by a University of Michigan Program in Cellular and Molecular Biology training grant (NIH T32 GM07315) and by a University of Michigan Center for Organogenesis training grant (5-T32-HD007515). F. Bednar is supported by the American College of Surgeons Resident Research Scholarship and by NIH T32 HD007505.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(2):639–653. doi:10.1172/JCI59227.
References
- 1. National Cancer Institute. Surveillance, Epidemiology and End Results (SEER) Program web site. http://seer.cancer.gov . Accessed December 7, 2011.
- 2. American Cancer Society. Cancer Facts and Figures website. http://www.cancer.org/Research/CancerFactsFigures . Accessed December 7, 2011.
- 3.Hishinuma S, Ogata Y, Tomikawa M, Ozawa I, Hirabayashi K, Igarashi S. Patterns of recurrence after curative resection of pancreatic cancer, based on autopsy findings. J Gastrointest Surg. 2006;10(4):511–518. doi: 10.1016/j.gassur.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 4.Katz MH, et al. Long-term survival after multidisciplinary management of resected pancreatic adenocarcinoma. Ann Surg Oncol. 2009;16(4):836–847. doi: 10.1245/s10434-008-0295-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hruban RH, et al. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol. 2001;25(5):579–586. doi: 10.1097/00000478-200105000-00003. [DOI] [PubMed] [Google Scholar]
- 7.Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20(10):1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 8.Morris JP, Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer. 2010;10(10):683–695. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klimstra DS, Longnecker DS. K-ras mutations in pancreatic ductal proliferative lesions. Am J Pathol. 1994;145(6):1547–1550. [PMC free article] [PubMed] [Google Scholar]
- 10.Hingorani SR, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4(6):437–450. doi: 10.1016/S1535-6108(03)00309-X. [DOI] [PubMed] [Google Scholar]
- 11.Aguirre AJ, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17(24):3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher GH, et al. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 2001;15(24):3249–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh A, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15(6):489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collisson EA, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17(4):500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32(1):128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 16.Belteki G, et al. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 2005;33(5):e51. doi: 10.1093/nar/gni051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olive KP, Tuveson DA. The use of targeted mouse models for preclinical testing of novel cancer therapeutics. Clin Cancer Res. 2006;12(18):5277–5287. doi: 10.1158/1078-0432.CCR-06-0436. [DOI] [PubMed] [Google Scholar]
- 18.Guerra C, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11(3):291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 19.Morris JP, Cano DA, Sekine S, Wang SC, Hebrok M. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J Clin Invest. 2010;120(2):508–520. doi: 10.1172/JCI40045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukuda A, et al. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell. 2011;19(4):441–455. doi: 10.1016/j.ccr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karanjawala ZE, et al. New markers of pancreatic cancer identified through differential gene expression analyses: claudin 18 and annexin A8. Am J Surg Pathol. 2008;32(2):188–196. doi: 10.1097/PAS.0b013e31815701f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ray KC, et al. Epithelial tissues have varying degrees of susceptibility to Kras(G12D)-initiated tumorigenesis in a mouse model. PLoS One. 2011;6(2):e16786. doi: 10.1371/journal.pone.0016786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Habbe N, et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci U S A. 2008;105(48):18913–18918. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cano DA, Sekine S, Hebrok M. Primary cilia deletion in pancreatic epithelial cells results in cyst formation and pancreatitis. Gastroenterology. 2006;131(6):1856–1869. doi: 10.1053/j.gastro.2006.10.050. [DOI] [PubMed] [Google Scholar]
- 25.Pasca di Magliano M, et al. Common activation of canonical Wnt signaling in pancreatic adenocarcinoma. PLoS ONE. 2007;2(11):e1155. doi: 10.1371/journal.pone.0001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, et al. Oncogenic function of ATDC in pancreatic cancer through Wnt pathway activation and beta-catenin stabilization. Cancer Cell. 2009;15(3):207–219. doi: 10.1016/j.ccr.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Day JD, et al. Immunohistochemical evaluation of HER-2/neu expression in pancreatic adenocarcinoma and pancreatic intraepithelial neoplasms. Hum Pathol. 1996;27(2):119–124. doi: 10.1016/S0046-8177(96)90364-0. [DOI] [PubMed] [Google Scholar]
- 28.Friess H, Berberat P, Schilling M, Kunz J, Korc M, Buchler MW. Pancreatic cancer: the potential clinical relevance of alterations in growth factors and their receptors. J Mol Med. 1996;74(1):35–42. doi: 10.1007/BF00202070. [DOI] [PubMed] [Google Scholar]
- 29.Korc M, Chandrasekar B, Yamanaka Y, Friess H, Buchier M, Beger HG. Overexpression of the epidermal growth factor receptor in human pancreatic cancer is associated with concomitant increases in the levels of epidermal growth factor and transforming growth factor alpha. J Clin Invest. 1992;90(4):1352–1360. doi: 10.1172/JCI116001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hess DA, et al. Extensive pancreas regeneration following acinar-specific disruption of Xbp1 in mice. Gastroenterology. 2011;141(4):1463–1472. doi: 10.1053/j.gastro.2011.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strobel O, et al. In vivo lineage tracing defines the role of acinar-to-ductal transdifferentiation in inflammatory ductal metaplasia. Gastroenterology. 2007;133(6):1999–2009. doi: 10.1053/j.gastro.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanger BZ, Tanaka AJ, Melton DA. Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature. 2007;445(7130):886–891. doi: 10.1038/nature05537. [DOI] [PubMed] [Google Scholar]
- 33.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci U S A. 2003;100(25):14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cano DA, Hebrok M, Zenker M. Pancreatic development and disease. Gastroenterology. 2007;132(2):745–762. doi: 10.1053/j.gastro.2006.12.054. [DOI] [PubMed] [Google Scholar]
- 35.Seymour PA, et al. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci U S A. 2007;104(6):1865–1870. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyamoto Y, et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3(6):565–576. doi: 10.1016/s1535-6108(03)00140-5. [DOI] [PubMed] [Google Scholar]
- 37.Olive KP, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324(5933):1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thayer SP, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425(6960):851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berman DM, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425(6960):846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 40.Nakashima H, et al. Nuclear factor-kappaB contributes to hedgehog signaling pathway activation through sonic hedgehog induction in pancreatic cancer. Cancer Res. 2006;66(14):7041–7049. doi: 10.1158/0008-5472.CAN-05-4588. [DOI] [PubMed] [Google Scholar]
- 41.Yauch RL, et al. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455(7211):406–410. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 42.Bailey JM, et al. Sonic hedgehog promotes desmoplasia in pancreatic cancer. Clin Cancer Res. 2008;14(19):5995–6004. doi: 10.1158/1078-0432.CCR-08-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller-Decker K, et al. Preinvasive duct-derived neoplasms in pancreas of keratin 5-promoter cyclooxygenase-2 transgenic mice. Gastroenterology. 2006;130(7):2165–2178. doi: 10.1053/j.gastro.2006.03.053. [DOI] [PubMed] [Google Scholar]
- 44.Colby JK, et al. Progressive metaplastic and dysplastic changes in mouse pancreas induced by cyclooxygenase-2 overexpression. Neoplasia. 2008;10(8):782–796. doi: 10.1593/neo.08330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lesina M, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19(4):456–469. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Corcoran RB, et al. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res. 2011;71(14):5020–5029. doi: 10.1158/0008-5472.CAN-11-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crawford HC, Scoggins CR, Washington MK, Matrisian LM, Leach SD. Matrix metalloproteinase-7 is expressed by pancreatic cancer precursors and regulates acinar-to-ductal metaplasia in exocrine pancreas. J Clin Invest. 2002;109(11):1437–1444. doi: 10.1172/JCI15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hingorani SR, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7(5):469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 49.Bardeesy N, et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci U S A. 2006;103(15):5947–5952. doi: 10.1073/pnas.0601273103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacks T, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4(1):1–7. doi: 10.1016/S0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 51.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 52.Caldwell ME, et al. Cellular features of senescence during the evolution of human and murine ductal pancreatic cancer [published online ahead of print August 22, 2011]. Oncogene. doi: 10.1038/onc.2011.350. doi:10.1038/onc.2011.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ong TM, Song B, Qian HW, Wu ZL, Whong WZ. Detection of genomic instability in lung cancer tissues by random amplified polymorphic DNA analysis. Carcinogenesis. 1998;19(1):233–235. doi: 10.1093/carcin/19.1.233. [DOI] [PubMed] [Google Scholar]
- 54.Malkhosyan S, Yasuda J, Soto JL, Sekiya T, Yokota J, Perucho M. Molecular karyotype (amplotype) of metastatic colorectal cancer by unbiased arbitrarily primed PCR DNA fingerprinting. Proc Natl Acad Sci U S A. 1998;95(17):10170–10175. doi: 10.1073/pnas.95.17.10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. 2007;7(9):645–658. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- 56.Weinstein IB. Cancer. Addiction to oncogenes — the Achilles heal of cancer. Science. 2002;297(5578):63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 57.Soucek L, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455(7213):679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15(24):3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hruban RH, et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: consensus report and recommendations. Cancer Res. 2006;66(1):95–106. doi: 10.1158/0008-5472.CAN-05-2168. [DOI] [PubMed] [Google Scholar]
- 60.Ji B, et al. Ras activity levels control the development of pancreatic diseases. Gastroenterology. 2009;137(3):1072–1082. doi: 10.1053/j.gastro.2009.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gidekel Friedlander SY, et al. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell. 2009;16(5):379–389. doi: 10.1016/j.ccr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grachtchouk M, et al. Basal cell carcinomas in mice arise from hair follicle stem cells and multiple epithelial progenitor populations. J Clin Invest. 2011;121(5):1768–1781. doi: 10.1172/JCI46307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 64.Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2010;129(20):4753–4761. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- 65.Pasca di Magliano M, Sekine S, Ermilov A, Ferris J, Dlugosz AA, Hebrok M. Hedgehog/Ras interactions regulate early stages of pancreatic cancer. Genes Dev. 2006;20(22):3161–3173. doi: 10.1101/gad.1470806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kojima K, et al. Inactivation of Smad4 accelerates Kras(G12D)-mediated pancreatic neoplasia. Cancer Res. 2007;67(17):8121–8130. doi: 10.1158/0008-5472.CAN-06-4167. [DOI] [PubMed] [Google Scholar]
- 67.Hruban RH, Rustgi AK, Brentnall TA, Tempero MA, Wright CV, Tuveson DA. Pancreatic cancer in mice and man: the Penn Workshop 2004. Cancer Res. 2006;66(1):14–17. doi: 10.1158/0008-5472.CAN-05-3914. [DOI] [PubMed] [Google Scholar]
- 68.Benavides F, et al. Application of inter-simple sequence repeat PCR to mouse models: assessment of genetic alterations in carcinogenesis. Genes Chromosomes Cancer. 2002;35(4):299–310. doi: 10.1002/gcc.10129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.