Abstract
Hereditary spastic paraplegias (HSPs) are a group of genetically heterogeneous neurodegenerative conditions. They are characterized by progressive spastic paralysis of the legs as a result of selective, length-dependent degeneration of the axons of the corticospinal tract. Mutations in 3 genes encoding proteins that work together to shape the ER into sheets and tubules — receptor accessory protein 1 (REEP1), atlastin-1 (ATL1), and spastin (SPAST) — have been found to underlie many cases of HSP in Northern Europe and North America. Applying Sanger and exome sequencing, we have now identified 3 mutations in reticulon 2 (RTN2), which encodes a member of the reticulon family of prototypic ER-shaping proteins, in families with spastic paraplegia 12 (SPG12). These autosomal dominant mutations included a complete deletion of RTN2 and a frameshift mutation predicted to produce a highly truncated protein. Wild-type reticulon 2, but not the truncated protein potentially encoded by the frameshift allele, localized to the ER. RTN2 interacted with spastin, and this interaction required a hydrophobic region in spastin that is involved in ER localization and that is predicted to form a curvature-inducing/sensing hairpin loop domain. Our results directly implicate a reticulon protein in axonopathy, show that this protein participates in a network of interactions among HSP proteins involved in ER shaping, and further support the hypothesis that abnormal ER morphogenesis is a pathogenic mechanism in HSP.
Introduction
The ER is a continuous membrane system comprising the nuclear envelope and a dynamic network of proximal sheets and peripheral tubules. Proteins of 2 classes — the reticulons and the REEP/DP1/yop1p family (referred to herein as the REEPs) — are fundamental to the generation of both sheets and tubules. These proteins share a characteristic sequence feature; in the case of the reticulons this feature is termed the reticulon homology domain (RHD) and consists of 2 long hydrophobic stretches separated by a hydrophilic sequence. Each hydrophobic stretch is thought to sit in the ER membrane as a hairpin loop. Such loop domains have been suggested to generate membrane curvature by occupying more space in the outer leaflet of the membrane than the inner in a process termed “hydrophobic wedging” (1–3). These proteins are thus critical for producing and stabilizing the high membrane curvature found at tubules and ER sheet edges (1–4).
Reticulons and REEPs are necessary and sufficient to generate the tubular ER (1, 2). However, additional factors are required for generation of a correct tubular ER network, including another class of proteins containing a hairpin membrane domain, the atlastin GTPases, which mediate homotypic fusion of ER tubules (5, 6). Proper ER morphology also depends on microtubules. An isoform of the microtubule severing ATPase spastin localizes to the ER and contains a hairpin loop domain (7). REEP1 and atlastin-1 interact with this form of spastin, probably via hairpin loop domain interactions (8–10). Expression of an ATPase-defective version of this isoform, which is unable to sever microtubules, results in profound tubulation of the ER, implicating spastin as an ER morphogen as well (8).
The HSPs are a group of clinically and genetically heterogeneous neurodegenerative conditions, in which the cardinal feature is progressive spastic paralysis of the legs. This is caused by a distal axonopathy involving the main motor pathway, the corticospinal tract (reviewed in ref. 11). The most common subtype of HSP in Northern Europe and North America is autosomal dominant, uncomplicated HSP. Strikingly, mutations in 3 genes encoding proteins involved in ER morphogenesis, receptor accessory protein 1 (REEP1), atlastin-1 (ATL1), and spastin (SPAST), have been found in this HSP subtype. This has led to the proposal that defects in an “ER morphogen complex” could be a mechanism underlying axonopathy in up to 60% of HSP cases (reviewed in ref. 12). However, to date, no disease process has been associated with mutations in the genes encoding the prototypic ER-shaping proteins, the reticulons. In this study, we show that mutations in the gene encoding reticulon 2 (RTN2) cause the spastic paraplegia 12 (SPG12) subtype of autosomal dominant HSP. We also demonstrate that the RTN2 protein interacts with spastin and that this interaction requires sequences of the spastin protein that contain the predicted hairpin membrane domain. As well as identifying a new HSP gene, to our knowledge for the first time our results directly implicate a reticulon protein in a disease process and strengthen the evidence that abnormal ER morphogenesis is involved in causing HSP.
Results
Mutation of RTN2 causes HSP.
In 2000, we mapped the SPG12 gene at chromosome 19q13 in a British family (family 28, logarithm of the odds [LOD] score, 3.72) with autosomal dominant uncomplicated HSP (13). Next, a North American family, which appeared to be linked to the SPG12 locus (family 5, LOD score, 1.91) turned out to have a REEP1 change at the SPG31 locus (14, 15). However, subsequent linkage analysis in an Italian family (family ROM-05, LOD score, 5.52), also with autosomal dominant uncomplicated HSP, narrowed this locus to an interval corresponding to a 13-Mbp physical region containing 377 coding RefSeq genes (chromosome 19:34,068,612-47,011,334) (16). We screened 196 of these genes by sequencing without finding a pathogenic mutation. Finally, we identified an insertion, c.178insC (chromosome 19:45,998,163), in the third exon of RTN2 in both the British and Italian SPG12-linked families. This change leads to a frameshift mutation, R60fs, and premature stop after 9 codons and thus is highly likely to be pathogenic (Figure 1, A and B). As whole-exome sequencing became available during the study, we performed it on both families and did not identify any other significant sequence change in the linkage region. The RTN2 mutation cosegregated with the disease phenotype in both families (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI60560DS1), and genotyping of 1,700 control subjects of mixed European descent from North America and 500 control subjects from Italy (4,400 chromosomes) did not detect it. We also identified a shared haplotype around RTN2 in both families, suggesting a possible founder effect rather than a mutational hot spot (Supplemental Table 1).
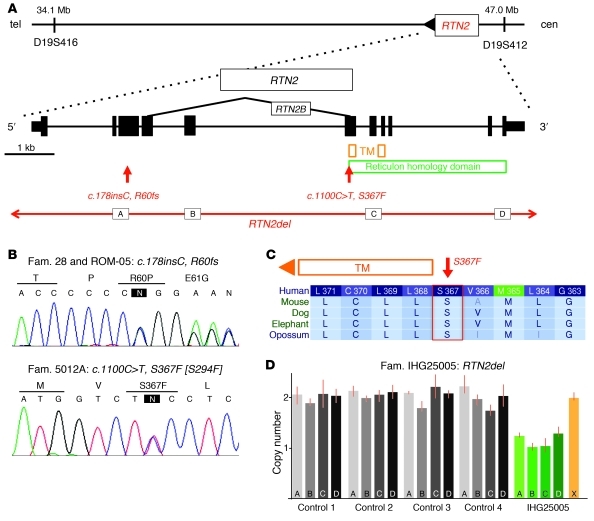
Figure 1. Identification of RTN2 as the SPG12 gene.
(A) By candidate gene screening of the linked chromosomal locus, we identified a mutation in RTN2. A schematic of the RTN2 gene with the corresponding conserved protein domains is drawn to scale. Subsequently, we identified another missense change and a gene deletion. Exons are represented by rectangles. The letters “A,” “B,” “C,” and “D” indicate the location of the copy number variation assays (see D below). TM, transmembrane domain; tel, telomere; cen, centromere. (B) Sanger sequence traces of the identified mutations. The variant nucleotide is highlighted by an “N” in a black rectangle. Fam., family. (C) The missense mutation (S367F) is highly conserved and immediately flanks the transmembrane domain. Letters in lighter font represent not fully conserved amino acids. (D) Four copy number variant assays across the gene (referred to as “A,” “B,” “C,” and “D” on the bars of the graph) identified a patient with a heterozygous loss of RTN2 (green bars). Gray bars indicate samples that had 2 copies of RTN2. The orange bar (X) indicates a control locus on a different chromosome tested in the patient with the RTN2 deletion.
We sought additional SPG12 mutations by Sanger sequencing the coding exons of RTN2 in 342 patients of mixed European descent and 160 Japanese patients with HSP, all previously excluded for mutations in the most common HSP gene, spastin (SPAST). In one patient with HSP (from family 5012A), we identified a coding sequence change c.1100C>T, S367F, chr19:45,992,745 (Figure 1, A and B). The S367 amino acid affected is highly conserved (Genomic Evolutionary Rate Profiling [GERP] score, 4.00; phastCons score, 1.00; Polyphen2, probably damaging), and the sequence change was not present in 1,700 control samples (Figure 1C and Supplemental Figure 2). We also screened 84 HSP index patients to identify copy number variants in RTN2 and identified a sporadic case (IHG25005) with a complete deletion of the gene (Figure 1D). This deletion was not present in 1,400 control individuals.
Clinical features associated with RTN2 mutations.
Clinical features of family 28 and ROM-05 have been described previously and are summarized in Table 1 (13, 16–18). The patient with the point mutation S367F developed slowly progressive pure spastic paraplegia, accompanied by mild urge incontinence at the age of 39 years. At age 54, he was still able to walk without aid for several miles but was not able to run. Neurophysiological examination revealed prolonged central motor conduction time as well as delayed sensory evoked potentials, indicating abnormality of the corticospinal tract and the dorsal column. Nerve conduction velocities were normal. His 2 children both received physical therapy during infancy, due to spasticity. Later, however, they showed normal motor development and, at the age of 30 and 32 years, did not report any motor problems. Unfortunately, they were not available for neurological examination. The individual with a RTN2 gene deletion (from family IHG25005) had onset of spasticity at an age of 36 years. The patient had typical symptoms of pure spastic paraplegia, with weakness of the legs, brisk reflexes, and a spastic gait, accompanied by urinary urgency and frequency. He was relatively severely affected, using foot orthoses and a cane to aid walking. Neurological examination also revealed diminished vibration sensation at both ankles. Median motor nerve conduction velocity was normal at 52 m/s (Table 1).
Table 1 .
RTN2 mutation summary and clinical characteristics
Wild-type but not truncated RTN2 localizes to the ER.
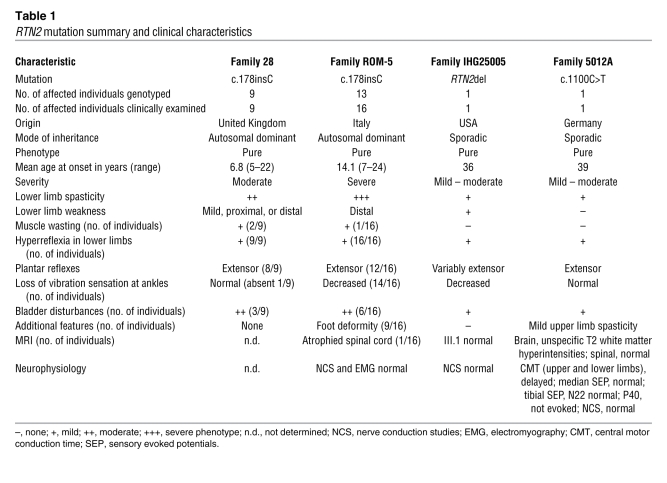
We considered the molecular pathological mechanisms of the RTN2 mutations identified. We began by examining the subcellular localization of RTN2. For cell-based experiments, we chose the RTN2B isoform, which appears to be preferentially expressed in brain (19). Consistent with the ER localization described for other reticulons, we found that epitope-tagged RTN2B tightly colocalized with ER markers (Figure 2A). In cells strongly expressing the reticulon protein punctate structures were observed (Figure 2A).
Figure 2. Wild-type, but not truncated RTN2B, localizes to the ER.
HEK293T cells were cotransfected with GFP-tagged RTN2B constructs and pmKate2-ER, which expresses a fluorescent protein fused to an ER-targeting signal and the ER retention signal KDEL. Images show cells transfected with (A) RTN2B-WT, (B) RTN2B-R60f, or (C) RTN2B-S294F. Scale bars: 10 μm.
The R60fs mutation is predicted to encode a severely truncated protein that lacks most of the protein sequence, including the RHD. However, such transcripts can be subject to nonsense-mediated mRNA decay, and so we first examined whether any mutant mRNA was expressed. RT-PCR and sequencing demonstrated that the mutated transcript was present in peripheral blood lymphocytes from an affected patient (Supplemental Figure 3). We therefore examined the cellular expression pattern of the truncated protein predicted for this transcript. When expressed in HEK293 cells, the truncated protein was diffusely present in the cytosol as well as the nucleus and showed no specific colocalization with ER markers. ER morphology was normal in these cells, and none of the punctate structures present after wild-type RTN2B overexpression were observed (Figure 2B).
We also examined the effect of the S367F (S294F in the RTN2B isoform that we studied) sequence change on the ER. The affected amino acid directly flanks the first hairpin loop domain in the RHD and could potentially interfere with function of this domain (Figure 1, A and C). However, colocalization with the ER was preserved in cells overexpressing RTN2B-S294F, and we could not identify any significant qualitative or quantitative differences in ER morphology in these cells (Figure 2C and Supplemental Figure 4). As with wild-type RTN2B, punctate structures appeared in cells strongly expressing the S294F protein (Figure 2C).
RTN2 interacts with spastin.
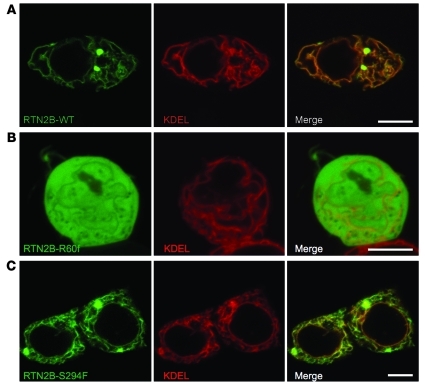
A network of interactions has been detected among the 3 previously known ER morphogens that are HSP proteins: spastin interacts with atlastin-1, and REEP1 interacts with both atlastin-1 and spastin. In addition, as well as the interactions among reticulons and the REEP family, 2 reticulons, RTN3 and RTN4, are known to interact with atlastin-1, while RTN1 interacts with spastin (Supplemental Figure 5 and refs. 2, 5, 8–10, 20). All of these proteins have at least one long membrane domain predicted to form a hairpin loop, and these appear to be crucial for the interactions. In order to demonstrate that RTN2 is part of this interacting network, we examined interactions between RTN2B and spastin. We chose spastin as an example because mutations in the gene encoding it are the most common cause of HSP (11). Spastin is a microtubule severing ATPase with 2 main cellular isoforms: a full-length form (M1 spastin) and a short form (M87 spastin) that lacks the N-terminal 86 amino acids of the full-length form (21). Importantly, spastin’s predicted hairpin membrane domain is only present in the M1 isoform, and only this isoform localizes to the ER (7). We carried out coimmunoprecipitation experiments between epitope-tagged RTN2B and M1 or M87 spastin. We found that M1 spastin, but not M87 spastin, coimmunoprecipitated RTN2B (Figure 3, A and B, and Supplemental Figure 6). Consistent with this, we found strong colocalization of M1 spastin, but not M87 spastin, with RTN2B (Figure 3C and Supplemental Figure 7). This colocalization was often strongest in, but not limited to, the punctate structures induced by RTN2B expression. Cellular expression of ATPase-defective M1 spastin redistributes the ER onto abnormally thickened and elongated microtubule bundles (7, 8, 22, 23), and we found that RTN2B was also redistributed onto these bundles (Figure 3D). Interestingly, the microtubule bundles tended to run through the punctate structures induced by RTN2B expression (Figure 3D). As well as causing no effect on ER morphology, the S294F amino acid change did not affect the ability of RTN2 to interact with M1 spastin (Figure 3A and Supplemental Figure 8).
Figure 3. RTN2B interacts with spastin.
(A) HeLa cells were transfected with the constructs indicated and immunoprecipitated with spastin86-340 antibody or preimmune serum (PI), and then the immunoprecipitates were immunoblotted with the antibodies shown. Syntaxin-17 is an ER membrane protein used to demonstrate specificity of the immunoprecipitation between spastin and RTN2. The M87A mutation of spastin prevents transcription of M87 spastin from the M1 transcript but does not alter function of the M1 form. (B) Quantitation of immunoblot band strength in 3 experiments in which HeLa cells were cotransfected with Myc-M1 spastin M87A and RTN2B-V5 and then immunoprecipitated with spastin86-340 antibody or preimmune serum. Immunoprecipitates were immunoblotted with antibodies against the V5-tag or syntaxin-17, and the relevant band density was quantified. P values were calculated using 2-tailed paired t tests. Values plotted are mean ± SEM. (C and D) Confocal micrographs of representative HeLa cells cotransfected with RTN2B-V5 and either Myc-M1 spastin or ATPase-defective Myc-M1 spastin K388R. Some smaller colocalized structures are indicated with arrowheads in the higher-magnification boxes. The color of the words in each black and white image correlates with the color of that image in the corresponding color panel. Scale bars: 10 μm.
Discussion
In this study we have identified RTN2 as the gene mutated in the SPG12 subtype of HSP. We identified a frameshift mutation shared by the 2 previously known SPG12-linked families as well as a whole gene deletion in another patient. In addition, we identified only a single missense change in a large cohort of unrelated index patients, indicating that RTN2 mutations are a rare cause of HSP. The clinical features in these families with RTN2 mutations were typical of a pure, early- to late-onset HSP and so are broadly similar to those seen with other ER morphogen complex genes mutated in HSP.
Our finding of a whole gene deletion of RTN2 in a patient with HSP suggests that haploinsufficiency can cause the disease. Consistent with this, the truncated protein predicted for the frameshift transcript lacked the RHD and did not localize to the ER. Thus, this mutant is also likely to act via a haploinsufficiency mechanism, although we cannot formally exclude an additional toxic gain of function of the truncated protein in the cytosol or nucleus. The S367F sequence change results in substitution of a highly conserved polar amino acid adjacent to a hairpin membrane domain for one with a bulky hydrophobic side chain. In addition, the underlying causative DNA mutation was not found in a large number of control chromosomes and so is likely to be pathogenic. Although we could not detect any functional effect of this mutation, it could still have subtle effects on dynamic aspects of ER morphogenesis that are only apparent in extreme situations, such as in the highly polarized corticospinal tract neurons. Alternatively, it might specifically affect interactions with complex members other than spastin. However, we cannot completely rule out the possibility that it is a rare polymorphism.
The 4 mammalian reticulon genes (RTN1–RTN4) encode at least 11 protein isoforms (24). These proteins share little sequence homology, except at the C-terminal RHD, which mediates a number of verified interactions between the reticulons and other ER morphogens, including M1 spastin and the atlastins (2, 5, 8–10, 20). We have extended this interaction network by demonstrating that spastin and RTN2 interact and that the interaction requires spastin sequences that contain the predicted hairpin loop domain. RTN2 therefore participates in the network of hairpin loop-containing ER morphogens that includes REEP1, atlastin-1, and M1 spastin.
In summary, for the first time to our knowledge we show that mutations in a member of the prototypic family of ER morphogens, the reticulons, cause disease and that haploinsufficiency of RTN2 is a likely molecular pathological mechanism for axonopathy. Our results demonstrate that RTN2 participates in the network of HSP proteins involved in ER shaping and provide further direct evidence for abnormal ER morphogenesis as a potential pathogenic mechanism in HSP. These findings will act as a platform for future studies aimed at determining how abnormal ER morphogenesis causes axonal degeneration.
Methods
Patients and families.
The study included patients and family members from previously described SPG12 families (13, 16). In addition, we included 342 HSP probands of mixed European ethnicity and 160 Japanese HSP samples. Of the samples from individuals of mixed European descent, 252 (74%) represented autosomal dominant patients, 80 (23%) represented sporadic cases or those that had no family history available, and 10 (3%) represented individuals that had a recessive mode of inheritance. Based upon clinical and electrophysiological data, 267 (78%) patients of mixed European descent had a pure, uncomplicated type of HSP. All HSP samples had been previously excluded for spastin mutations. In addition, 2,200 samples from clinically healthy individuals of mixed European descent were screened for the c.178insC variant, and 1,700 samples were screened for the missense mutation. Written informed consent was taken from each study participant prior to the study. We also received genotype data from Yao-Shan Fan (University of Miami, Miami, Florida, USA) on RTN2 deletions in clinical control individuals. These were based on CGH array analysis.
Study approval.
The research was performed according to protocols reviewed and approved by local institutional review boards (IRBs): University of Miami Miller School of Medicine IRB committee; Cambridge Local Research Ethics Committee, Cambridge, United Kingdom; IRCCS Santa Lucia Ethics Committee, Rome, Italy.
Sanger sequencing and whole-exome sequencing.
Sanger sequencing was performed according to established standard protocols on an Applied Biosystems (ABI) 3730 capillary instrument. Exons and flanking intronic sequences were amplified on ABI Veriti 96-well Fast Thermal Cyclers using a touchdown protocol. Sequence traces were analyzed with Sequencher 4.10 software (Gene Codes Corporation). Exome sequencing was carried out, applying the SureSelect Human All Exon 50Mb Kit (Agilent), following the manufacturer’s standard protocol. Enriched DNA was subjected to standard sample preparation for the Hiseq2000 instrument (Illumina). Sequence reads were aligned with the MAQ software. Variants were called with MAQ, applying the standard parameters. All variants were submitted to SeattleSeq for further categorization into coding, noncoding, and novel single nucleotide polymorphisms. Resulting data were converged, filtered, and ranked by GERP conservation scores.
Genotyping assays.
In controls, the RTN2 mutations were confirmed and screened for using the ABI TaqMan technology. Sequences of the primers and probes were as follows: RTN2-ex3_F, 5′-CCACGTCGCAGGACTGG-3′; RTN2-ex3_R, 5′ CAAAGGCGATGTAGGAGAAGGT-3′; VIC reporter, 5′-CACCCCCCGGGAGC-3′; FAM reporter, 5′-CCCCCCCGGGAGC-3′. Likewise, we designed a similar custom assay for the point mutation on exon 6: RTN2-ex6_F, 5′-CGTCAGGAGTGGTCTTCACA-3′; RTN2-ex6_R, 5′-CACGGACACGATGCTAAAGTG-3′; VIC reporter, 5′-AGAGGAGGGAGACCAT-3′; FAM reporter, 5′-AGAGGAGGAAGACCAT-3′. Standard protocols were applied, and plates were scanned on an ABI 7900HT Real-Time PCR System.
Copy number variant analysis.
Four predesigned ABI TaqMan copy number assays, with RNase P reference assay as endogenous control, were chosen to cover the RTN2 gene (hs01270067, hs02017504, hs01153465, and hs00213975). A total of 84 HSP samples were screened for all 4 assays and normalized against a control panel of 3 samples. Reactions were run on an ABI 7900HT Real-Time PCR System in quadruplets. Data were analyzed using SDS 2.3 and ABI CopyCaller software. We also ran each sample against a probe on chromosome 12 in the gene KIF5A.
Constructs.
The gene encoding human RTN2B (RefSeq accession no. NM_206900.1; http://www.ncbi.nlm.nih.gov/RefSeq/) was PCR amplified from a pCMV6-XL vector (Origene). Purified PCR products were TA topo cloned into pcDNA3.1/NT-GFP and pcDNA3.1/V5-His (Invitrogen). cDNA encoding M1 or M87 spastin was cloned into the pIRES vector. Mutagenesis was performed with the QuikChange II Mutagenesis Kit (Agilent) for the point mutations C.1100C>T and c.178insC, and site-directed mutagenesis was used to generate the construct encoding M87A M1 spastin. All clones were confirmed for sequence fidelity using Sanger sequencing.
Antibodies.
Spastin86-340 rabbit polyclonal antibody was described previously (7). Anti-spastin mouse monoclonal 6C6 was from Sigma-Aldrich. Mouse monoclonal anti-V5 antibody was from AbD Serotec. The syntaxin 17 antibody was a gift of Andrew Peden (University of Cambridge).
Cell culture and imaging.
Human embryonal kidney HEK293T cells and HeLa cells were cultured in Dulbecco’s medium supplemented with 10% fetal bovine serum and antibiotics. HEK293 cells were plated to reach approximately 70% confluency when plasmids were transfected using X-tremeGENE 9 DNA transfection reagent (Roche), while HeLa cells were transfected with HeLaMonster Transit reagent (Mirus), as described by the manufacturers. After 24 to 48 hours of transfection, cells were processed for confocal microscopy as described previously (7). Live cell imaging was performed with a Zeiss LSM710 point scanning confocal microscope, using a plan-apochromat ×40 objective with a numerical aperture of 1.30. Z-stacks of cells overexpressing both RTN2B-GFP and pmKate2-ER (Evrogen) were performed using Zen2009 and analyzed with ImageJ ( http://rsbweb.nih.gov/ij/). Images were first filtered by applying a band pass filter of 3 to 40 pixels in Fourier space using the Fast Hartley Transform in which a subsequent background subtraction was performed using rolling ball radius of 1. The filtered images were then made into a maximum z-projection and then binarized using the mean threshold function. The binarized image of the ER was then skeletonized and analyzed using the plug-in binary connectivity from the morphology package produced by G. Landini ( http://www.dentistry.bham.ac.uk/landinig/software/software.html).
Coimmunoprecipitation.
Constructs were transfected into HeLa cells. After 48 hours, cells were washed in PBS and lysed in 1 ml ice-cold lysis buffer (300 mM NaCl, 50 mM Tris HCl, 5 mM EDTA) containing 1% Triton Tx-100 and protease inhibitors (Roche). Lysate was spun at 10,000 g for 10 minutes, and supernatant was retained, before being precleared by incubation with 100 μl of a 50% slurry of protein A-Sepharose beads (Sigma-Aldrich) in lysis buffer, and rotated for 1 hour at 4°C. Samples were spun to remove beads and divided into immunoprecipitates and control fractions. Ten μl spastin86-340 antibody was added to the immunoprecipitate fraction, and an equivalent volume of preimmune serum was added to the control fraction. Samples were incubated and rotated for 1 hour at 4°C; 100 μl of a 50% slurry of protein A-Sepharose beads was added to each sample; and then the samples were rotated for 2 hours at 4°C. Protein A-Sepharose beads were isolated by brief centrifugation and then washed 3 times for 5 minutes in lysis buffer at room temperature. Beads were resuspended in 3× SDS-PAGE sample buffer containing 10% β-mercaptoethanol. SDS-PAGE and immunoblotting were then carried out. Immunoblot band density was quantified on scanned films using Quantity One software (Bio-Rad).
Statistics.
Statistical analyses for coimmunoprecipitation immunoblot band density comparisons were carried out in GraphPad Prism version 5.00 for Windows (GraphPad Software). A 2-tailed Student’s t test was applied, and a P value of less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
We are thankful to the family members studied and to their support for our research. This study was supported by grants from the NIH (grant no. R01NS072248 and R01NS054132 to S. Züchner), the Hereditary Spastic Paraplegia Foundation (to S. Züchner), the Wellcome Trust (grant no. 082381 and 079895 to E. Reid and Cambridge Institute for Medical Research, respectively), the Comitato Telethon Fondazione Onlus, Italy (grant no. GGP10121 to A. Orlacchio), the Italian Ministero dell’Istruzione, dell’Università e della Ricerca (PRIN 2008 grant no. 020903052 to A. Orlacchio), the Università di Roma “Tor Vergata” (FAA 2008 grant no. 020903023 to A. Orlacchio), and the European Union (E-RARE grant no. 01GM0807 to L. Schöls).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(2):538–544. doi:10.1172/JCI60560.
References
- 1.Collins RN. How the ER stays in shape. Cell. 2006;124(3):464–466. doi: 10.1016/j.cell.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 2.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124(3):573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 3.Hu J, et al. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319(5867):1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- 4.Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. Mechanisms determining the morphology of the peripheral ER. Cell. 2010;143(5):774–788. doi: 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu J, et al. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138(3):549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orso G, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460(7258):978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- 7.Connell JW, Lindon C, Luzio JP, Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 2009;10(1):42–56. doi: 10.1111/j.1600-0854.2008.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanderson CM, et al. Spastin and atlastin, two proteins mutated in autosomal-dominant hereditary spastic paraplegia, are binding partners. Hum Mol Genet. 2006;15(2):307–318. doi: 10.1093/hmg/ddi447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans K, Keller C, Pavur K, Glasgow K, Conn B, Lauring B. Interaction of two hereditary spastic paraplegia gene products, spastin and atlastin, suggests a common pathway for axonal maintenance. Proc Natl Acad Sci U S A. 2006;103(28):10666–10671. doi: 10.1073/pnas.0510863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park SH, Zhu PP, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest. 2010;120(4):1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reid E, Rugarli E. Hereditary spastic paraplegias. In: Beaudet AL, Vogelstein B, Kinzler KW, Antonarakis SE, Ballabio A, eds.The Online Molecular and Metabolic Basis of Inherited Diseases . New York, New York, USA: McGraw Hill; 2010:1–65. [Google Scholar]
- 12.Blackstone C, O’Kane CJ, Reid E. Hereditary spastic paraplegias: membrane traffic and the motor pathway. Nat Rev Neurosci. 2011;12(1):31–42. doi: 10.1038/nrn2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reid E, Dearlove AM, Osborn O, Rogers MT, Rubinsztein DC. A locus for autosomal dominant “pure” hereditary spastic paraplegia maps to chromosome 19q13. Am J Hum Genet. 2000;66(2):728–732. doi: 10.1086/302783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashley-Koch A, et al. Fine mapping and genetic heterogeneity in the pure form of autosomal dominant familial spastic paraplegia. Neurogenetics. 2001;3(2):91–97. doi: 10.1007/s100480000098. [DOI] [PubMed] [Google Scholar]
- 15.Züchner S, et al. Mutations in the novel mitochondrial protein REEP1 cause hereditary spastic paraplegia type 31. Am J Hum Genet. 2006;79(2):365–369. doi: 10.1086/505361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orlacchio A, et al. Clinical and genetic study of a large Italian family linked to SPG12 locus. . Neurology. 2002;59(9):1395–1401. doi: 10.1212/01.wnl.0000031423.43482.19. [DOI] [PubMed] [Google Scholar]
- 17.Reid E, Grayson C, Rubinsztein DC, Rogers MT, Rubinsztein JS. Subclinical cognitive impairment in autosomal dominant “pure” hereditary spastic paraplegia. J Med Genet. 1999;36(10):797–798. doi: 10.1136/jmg.36.10.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reid E, Dearlove AM, Whiteford ML, Rhodes M, Rubinsztein DC. Autosomal dominant spastic paraplegia: refined SPG8 locus and additional genetic heterogeneity. . Neurology. 1999;53(8):1844–1849. doi: 10.1212/wnl.53.8.1844. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Vidensky S, Ruggiero AM, Maier S, Sitte HH, Rothstein JD. Reticulon RTN2B regulates trafficking and function of neuronal glutamate transporter EAAC1. J Biol Chem. 2008;283(10):6561–6571. doi: 10.1074/jbc.M708096200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mannan AU, et al. Spastin, the most commonly mutated protein in hereditary spastic paraplegia interacts with Reticulon 1 an endoplasmic reticulum protein. Neurogenetics. 2006;7(2):93–103. doi: 10.1007/s10048-006-0034-4. [DOI] [PubMed] [Google Scholar]
- 21.Claudiani P, Riano E, Errico A, Andolfi G, Rugarli EI. Spastin subcellular localization is regulated through usage of different translation start sites and active export from the nucleus. Exp Cell Res. 2005;309(2):358–369. doi: 10.1016/j.yexcr.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 22.Errico A, Ballabio A, Rugarli EI. Spastin, the protein mutated in autosomal dominant hereditary spastic paraplegia, is involved in microtubule dynamics. Hum Mol Genet. 2002;11(2):153–163. doi: 10.1093/hmg/11.2.153. [DOI] [PubMed] [Google Scholar]
- 23.Orlacchio A, et al. Hereditary spastic paraplegia: clinical genetic study of 15 families. Arch Neurol. 2004;61(6):849–855. doi: 10.1001/archneur.61.6.849. [DOI] [PubMed] [Google Scholar]
- 24.Yang YS, Strittmatter SM. The reticulons: a family of proteins with diverse functions. Genome Biol. 2007;8(12):234. doi: 10.1186/gb-2007-8-12-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.