Whole-genome sequencing identifies a novel germline variant in the oncogene MITF, which is associated with the development of melanoma.
Supplementary information
The online version of this article (doi:10.1038/nature10630) contains supplementary material, which is available to authorized users.
Subject terms: Rare variants, Genetic predisposition to disease, Sumoylation, Melanoma
MITF, a melanoma predisposition gene
Two papers in this issue of Nature demonstrate that missense substitutions in the gene encoding for microphthalmia-associated transcription factor (MITF) are associated with susceptibility to melanoma and renal cell carcinoma. Functional analysis shows that the variant has impaired sumoylation that leads to differential regulation of several MITF targets, and promotes tumour cell clonogenicity, migration and invasion.
Supplementary information
The online version of this article (doi:10.1038/nature10630) contains supplementary material, which is available to authorized users.
Abstract
So far, two genes associated with familial melanoma have been identified, accounting for a minority of genetic risk in families. Mutations in CDKN2A account for approximately 40% of familial cases1, and predisposing mutations in CDK4 have been reported in a very small number of melanoma kindreds2. Here we report the whole-genome sequencing of probands from several melanoma families, which we performed in order to identify other genes associated with familial melanoma. We identify one individual carrying a novel germline variant (coding DNA sequence c.G1075A; protein sequence p.E318K; rs149617956) in the melanoma-lineage-specific oncogene microphthalmia-associated transcription factor (MITF). Although the variant co-segregated with melanoma in some but not all cases in the family, linkage analysis of 31 families subsequently identified to carry the variant generated a log of odds (lod) score of 2.7 under a dominant model, indicating E318K as a possible intermediate risk variant. Consistent with this, the E318K variant was significantly associated with melanoma in a large Australian case–control sample. Likewise, it was similarly associated in an independent case–control sample from the United Kingdom. In the Australian sample, the variant allele was significantly over-represented in cases with a family history of melanoma, multiple primary melanomas, or both. The variant allele was also associated with increased naevus count and non-blue eye colour. Functional analysis of E318K showed that MITF encoded by the variant allele had impaired sumoylation and differentially regulated several MITF targets. These data indicate that MITF is a melanoma-predisposition gene and highlight the utility of whole-genome sequencing to identify novel rare variants associated with disease susceptibility.
Supplementary information
The online version of this article (doi:10.1038/nature10630) contains supplementary material, which is available to authorized users.
Main
Cutaneous malignant melanoma is predominantly a disease of fair-skinned individuals. Aetiology is complex, with environmental (mainly ultraviolet radiation exposure) and genetic factors affecting disease risk. Phenotypic risk factors, which are largely heritable, include pigmentation (fair skin, blue or green eyes, blonde or red hair), sun sensitivity, an inability to tan3,4,5,6, high number of melanocytic naevi7,8, or the presence of clinically atypical naevi7. Candidate-gene studies and genome-wide association studies (GWAS) for melanoma and these melanoma-associated phenotypes have identified several variants associated with melanoma risk in the general population9,10,11,12,13. Family studies, on the other hand, have identified only two high-penetrance melanoma genes, CDKN2A (ref. 1) and CDK4 (ref. 2), accounting for a minority of genetic risk in melanoma families.
As part of a larger sequencing effort to identify novel melanoma risk genes, we sequenced the genome of an affected individual from an eight-case melanoma family negative for alterations in CDKN2A or CDK4 (Fig. 1, FAM1) using a nanoarray-based short-read sequencing-by-ligation strategy14. From among the 410 novel variants predicted to affect protein structure, we prioritized for follow-up a single nucleotide polymorphism (SNP) resulting in a glutamic acid to lysine substitution in MITF (E318K, codon numbering based on the melanocyte-specific MITF-M isoform; c.G1075A, NCBI accession NM_000248.3; p.E318K, NCBI accession NP_000239.1; rs149617956). Although linkage15 and GWAS studies9,10 have not provided evidence implicating MITF in either predisposition to melanoma or the melanoma-associated phenotypes of pigmentation and naevogenesis11,12,16,17,18,19, MITF is known to regulate a broad repertoire of genes whose functions in melanocytes range from development, differentiation, survival, cell-cycle regulation and pigment production. MITF is somatically amplified20,21 or mutated22 in a subset of melanomas, and strongly overexpressed in others20, making it an attractive candidate despite the lack of prior evidence for involvement in germline risk.
Figure 1. Co-segregation analysis of the MITF E318K variant in the family in which it was identified.
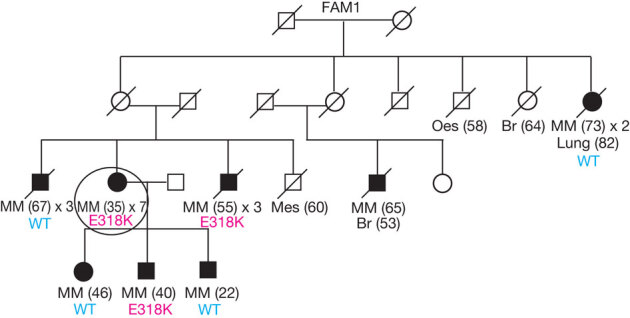
The pedigree shows individuals that have had melanoma (shaded circles or boxes), with the age of first melanoma diagnosis indicated in brackets and the number of melanomas that have occurred in the individual so far (for example, × 2 indicates two primary melanomas). If the number of melanomas is not stated, the individual has had a single melanoma. A diagonal line through the symbol indicates that the person is deceased. The genotype for the MITF E318K variant for individuals with an available DNA sample for testing is annotated ‘E318K’ if a carrier or wild type ‘WT’. Other cancer types are also indicated with the age of first diagnosis indicated in brackets if known. Br, breast; Mes, mesothelioma; MM, melanoma; Oes, oesophagus. The individual circled in Family 1 (FAM1) is the melanoma case in which the MITF E318K variant was discovered through whole-genome sequencing. See Supplementary Fig. 3 for pedigrees of all other families identified as carrying E318K.
We evaluated whether MITF E318K is a high-penetrance melanoma susceptibility variant in Family 1 by genotyping the remaining affected individuals available for study. The MITF variant allele was found in 3/7 melanoma cases assessed in this family (Fig. 1), consistent with it being a medium-penetrance melanoma risk variant. To assess further this possibility, we genotyped two large Australian melanoma case–control samples for MITF E318K. The variant was found in 14/1,953 controls (carrier frequency = 0.0072) and thus represents a rare population variant (Table 1). We observed a significantly higher frequency (34/2,059) in cases (carrier frequency = 0.0165) than controls (Fisher exact P = 0.008, odds ratio (OR) 2.33, 95% confidence interval (CI) 1.21–4.70), indicating that the variant correlates with increased melanoma risk in the general population. The effect size for E318K is larger than those reported for variants from melanoma GWAS9,10 and similar to that observed for red-hair-colour-associated variants of the melanocortin 1 receptor (MC1R) gene (OR for most populations ∼2.4)23. Among cases, the MITF E318K variant was enriched in those with multiple primary melanomas (OR 4.22, 95% CI 1.52–10.91), a family history of melanoma (OR 2.95, 95% CI 1.23–6.92), or both (OR 8.37, 95% CI 2.58–23.80), but not in cases with earlier age of onset (comparing diagnosis before age 40 versus after 40 years) (Table 2).
Table 1.
MITF E318K association with melanoma
| Population | Group | No. of individuals with variant | No. of individuals without variant | Variant carrier frequency | OR | 95% CI | One-sided exact P | Two-sided exact P |
|---|---|---|---|---|---|---|---|---|
| Australia | Cases | 34 | 2,025 | 0.0165 | 2.33 | 1.21–4.70 | 0.0045 | 0.0083 |
| Controls | 14 | 1,939 | 0.0072 | |||||
| UK | Cases | 34 | 1,895 | 0.0176 | 2.09 | 1.14–3.94 | 0.0074 | 0.0115 |
| Controls | 18 | 2,097 | 0.0085 | |||||
| Australia + UK | Cases | 68 | 3,920 | 0.0171 | 2.19 | 1.41–3.45 | 0.0001 | 0.0003 |
| Controls | 32 | 4,036 | 0.0079 |
It should be noted that the reported allele frequencies for MITF E318K in the population-based samples are without removing individuals with CDKN2A or CDK4 mutations, as screening for these genes was not routinely performed.
Table 2.
Association of MITF E318K with melanoma-associated variables
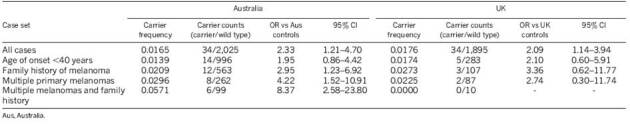
We replicated these findings in two independent population-based case–control samples from the United Kingdom. In the combined UK sample, the variant allele frequency was also significantly higher in cases (carrier frequency = 0.0176) than controls (carrier frequency = 0.0085, P = 0.012, OR 2.09, 95% CI 1.14–3.94, Table 1). The association with melanoma in the pooled UK and Australian data was highly significant (combined P = 0.0003, OR 2.19, 95% CI 1.41–3.45). In the UK cases there were also trends towards family history, earlier age of onset, and the occurrence of more than one primary melanoma in variant carriers (Table 2).
To extend assessment of the MITF variant in melanoma-prone families, we screened for E318K in 182 UK families with at least two melanoma cases and 88 Australian families with at least three cases, all of which are negative for mutations in CDKN2A or CDK4. Six families (2.2%) were found to carry the variant. In the UK, E318K was enriched in the more melanoma-dense families; 4/54 (7.4%) families with at least three melanoma cases versus 1/128 (0.8%) families with two melanoma cases (Fisher’s exact P = 0.013). We subsequently evaluated whether MITF E318K co-segregated with melanoma in these as well as additional multiple-case families identified from the case–control sample. In total, we identified 31 unrelated cases carrying MITF E318K from Australia and the UK with at least one first- or second-degree relative diagnosed with melanoma (listed in Supplementary Table 1; Supplementary Fig. 3), 22 of which had DNA available from additional affected family members for genotyping. In 9/31 families (five three-case and four two-case families) the variant was found in all affected individuals (Supplementary Fig. 3a; non-segregating families shown in Supplementary Fig. 3b), whereas in 12 additional families, the variant co-segregated with melanoma in the available cases, but DNA from all cases was not available for screening (Supplementary Fig. 3c). To test more formally for linkage of melanoma with MITF E318K in these families, we calculated a lod score of 2.7 under a dominant model, again consistent with an incompletely penetrant medium risk variant.
To assess whether the MITF variant is related to known melanoma-associated risk phenotypes of pigmentation and naevus count, we tested for association both in cases and controls from the Australian and British populations. The MITF variant allele is significantly associated with increased naevus count (combined P = 0.002, OR 2.54, 95% CI 1.42–4.55; Supplementary Table 2) and non-blue eye colour (combined P = 0.018, OR 2.01, 95% CI 1.11–3.81; Supplementary Table 3). It was not associated with skin colour, hair colour, or freckling (Supplementary Table 4). Reassessing the case–control analysis accounting for naevus count and eye colour gave a slightly reduced effect size for association of the variant with melanoma (OR 1.82, 95% CI 0.85–3.92), suggesting that the risk of melanoma attributable to MITF E318K may be mediated at least in part via one or both of these phenotypes, but that there is a substantial residual risk conferred by the variant through an as yet undetermined mechanism.
We next sought to evaluate whether the E318K mutation alters MITF function. The E318K variant is located within one of two IKXE consensus sites on MITF previously shown to be post-translationally modified by the addition of the small ubiquitin-like-modifier SUMO24. Mutation of the residue to which SUMO is covalently attached in this motif (K316R) has previously been shown to abrogate MITF sumoylation and significantly increase MITF transcriptional activity in vitro24,25. We thus hypothesized that E318K would similarly alter sumoylation and transcriptional activity of MITF. To test this we constructed a cDNA encoding His-tagged MITF carrying the E318K mutation. We evaluated the effects of E318K on sumoylation in comparison to the wild type and previously characterized synthetic mutations of the two known MITF sumoylation sites (K316R and K182R) by co-transfecting with haemagglutinin (HA)-tagged SUMO1 in COS-7 cells (Fig. 2a). Wild-type MITF shows two SUMO1-modified forms, whereas MITF mutants K182R or K316R each show only one modified form (Fig. 2a). Similar to the synthetic K316R and K182R mutants, E318K abrogates sumoylation, resulting in complete loss of the doubly sumoylated form of MITF and reducing the mono-sumoylated form. When the second site is mutated (K182R) simultaneously with E318K, MITF sumoylation is completely abolished. Immunoprecipitation of endogenously expressed MITF E318K from melanoma cells homozygous for E318K (NAE) when transfected with SUMO similarly revealed only bands corresponding to mono- and non-sumoylated isoforms of MITF on western blot (Fig. 2b).
Figure 2. E318K prevents MITF sumoylation and results in differential expression of MITF target genes.
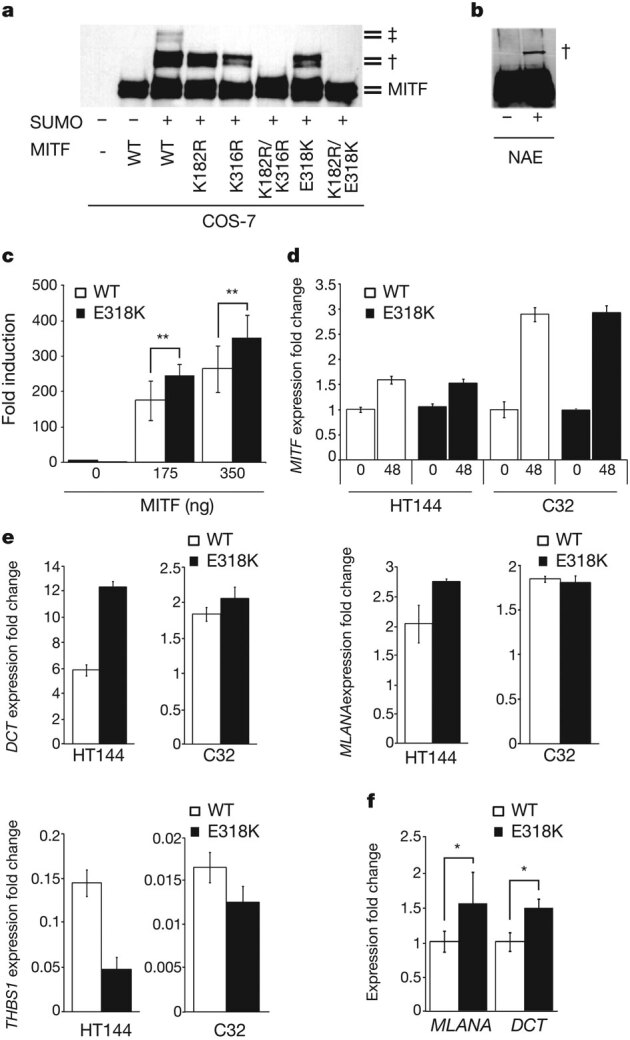
a, His-tagged wild-type MITF or the indicated single or double point mutants were co-transfected with HA–SUMO1 in COS-7 cells or b, HA–SUMO was transfected alone into homozygous mutant E318K MITF melanoma cells (NAE). Single- and double-sumoylated forms of MITF are indicated by a dagger and double dagger, respectively. The doublet bands are caused by MAPK-mediated phosphorylation at serine 73 (ref. 30). c, UACC62 human melanoma cells were transfected with TRPM1-promoter constructs with indicated amounts of expression vector encoding wild-type or mutant forms of MITF. Fold induction is shown as the ratio to the average of no MITF transfection (0 ng). Data are mean ± s.d. of at least four independent experiments. d, Expression of MITF in two melanoma cell lines (HT144 and C32) engineered to inducibly express wild-type (WT) or mutant (E318K) MITF after treatment with tetracycline for 48 h (48), as determined by qRT–PCR. Performed in triplicate, error bars depict s.d. e, Expression of MITF target genes DCT (top left), MLANA (top right) and THBS1 (bottom left) determined by qRT–PCR in melanoma cell lines 48 h after induction of wild-type or E318K MITF. Gene expression is normalized to GAPDH and shown as fold change compared to uninduced cells. Performed in triplicate, error bars denote s.d. f, qRT–PCR analysis of total RNA isolated from UACC62 human melanoma cells, which were transfected with expression vector encoding wild-type or mutant forms of MITF. The expression level of each target gene was normalized to MITF mRNA. Fold induction is shown as the ratio to each mRNA expression with wild-type MITF. Data are mean ± s.d. of at least three independent experiments. *P < 0.05, **P < 0.01.
We then looked for differences between mutant and wild-type MITF transcriptional activity using a reporter construct containing the MITF-responsive TRPM1 promoter25. At two concentrations tested, the E318K mutant exhibited 1.34–1.40 fold induction of the TRPM1 luciferase reporter relative to wild-type MITF (Fig. 2c). This fold induction is similar to that observed previously on multiple MITF target promoters using single or double artificial sumoylation-site MITF mutants24,25 and suggests that the E318K variant found in melanoma changes the transcriptional potency of MITF. To study this in greater detail, we determined the effect of the E318K mutation on global MITF target gene transcription. We developed a tetracycline-inducible system for expression of wild-type MITF or the E318K variant in melanoma cell lines with constitutively low or undetectable levels of endogenous MITF (HT144 and C32, respectively26, Fig. 2d). At the phenotypic level, induction of wild-type or E318K MITF led to increased proliferation compared to uninduced controls for each of the cell lines, although there was no significant difference in growth rate between the cells expressing the different isoforms (data not shown). We examined whole-genome expression profiles in these cells following induction of either wild-type or E318K MITF for 48 h. Of the 37 genes commonly regulated by wild-type and E318K MITF in both cell lines (Supplementary Fig. 1a, b; see Methods for analysis details), 28 (76%) had previously been identified as MITF target genes (Supplementary Table 5)27,28, and 17 showed ≥1.25-fold differences in expression between the wild-type and E318K isoforms (Supplementary Fig. 1b). We also identified two gene products that were uniquely differentially regulated compared to uninduced cells by the induction of wild-type MITF but not MITF E318K in both parental cell lines, and 16 gene products after induction of MITF E318K but not wild-type MITF (Supplementary Table 6). Of these, 61% (11/18) have previously been reported as MITF targets (Supplementary Table 6)27,28. Collectively, these data indicate that the MITF E318K mutant exhibits differential transcriptional activity against some, although not all, target genes. In agreement with the reporter assays (Fig. 2c), we identified transcriptional differences in gene products known to be involved in pigmentation (DCT, MLANA), in which the differences were more marked with expression of the E318K variant in comparison to wild-type MITF. These were validated by quantitative polymerase chain reaction with reverse transcription (qRT–PCR) in the cell lines used for microarray analysis (Fig. 2e), as well as in an additional melanoma cell line constitutively expressing wild-type or E318K mutant MITF (Fig. 2f and Supplementary Fig. 2). In keeping with the increase in expression of these pigmentation genes, we detected a 22% increase in melanin content in HT-144 melanoma cells 72 h after induction of MITF E318K compared to wild-type MITF (data not shown). This is also consistent with our observation that carriers are more likely to have darker (that is, non-blue) eye colour (Supplementary Table 3) but, notably, these data contrast with other previously reported ‘fair-skin-associated’ melanoma risk variants, such as those in MC1R or TYR. It is uncertain whether the enhanced expression of pigment genes may contribute to melanomagenesis, perhaps by increasing oxidative stress and an increase in oxidative DNA damage29, or alternatively may simply reflect increased MITF activity, which (separately) promotes tumorigenesis, as MITF is a previously recognized amplified melanoma oncogene20.
We adopted the approach of whole-genome sequencing of patients from melanoma families and identified a novel germline mutation of MITF. This mutation was found to be present in numerous melanoma families, as well as the general population, in which its association with melanoma has an effect size similar to red-hair-causing variants of MC1R23. The melanoma susceptibility genes discovered through GWAS so far account for only a minority of inherited disease risk. A proportion of this ‘missing heritability’ may be due to rare sequence variants, which are poorly detected by GWAS using SNP arrays. The new MITF variant reported here shows reasonably strong linkage to melanoma (lod score 2.7) but crucially not a high enough signal to be clearly visible in previous genome-wide linkage scans. We also provide in vitro data supporting a functional mechanism by which this mutation may mediate melanoma risk, specifically abrogation of MITF sumoylation and differential transcription of select MITF target genes. Although the individual changes in transcription induced by the mutant E318K MITF in comparison to wild-type MITF are modest, the orchestrated change in the levels of multiple MITF target genes is likely to be biologically important, especially over the lifetime of a person. This study offers a rare glimpse of a complex functionality whereby a risk-conferring SNP affects the post-translational processing of a crucial lineage-specific survival and differentiation gene. This study demonstrates the utility of performing whole-genome and exome resequencing in appropriate affected individuals to identify such novel rare disease-specific variants and functionally characterize variants associated with complex disease not otherwise detectable via GWAS or linkage approaches.
Methods Summary
The collection of the Australian melanoma families used for the study, as well as the Queensland and AMFS case–control sets are described elsewhere and in Methods. Likewise the UK studies from Leeds and Cambridge as well as the panel of melanoma cell lines. Whole-genome sequencing, assembly and variant calling were performed by Complete Genomics, as described previously14. Genotyping of MITF E318K was performed using the Sequenom MassArray system (Australian studies) or a custom TaqMan assay (UK studies), with DNA from the affected family member in which E318K was identified included multiple times as a positive control. Statistical analyses are described in detail in Methods. Co-segregation analyses were performed in melanoma families via Sanger sequencing using the primers: forward, 5′-CAGGCTCGAGCTCATGGA-3′; reverse, 5′-TGGGGACACTATAGGCTTGG-3′. MITF sumoylation and TRPM1 reporter assays were performed as previously described25.
Online Methods
Melanoma families
Ascertainment and clinical evaluation of Australian and UK pedigrees with familial melanoma is described elsewhere31,32,33,34,35. In an effort to maximize the chance of identifying a novel melanoma susceptibility gene, we restricted inclusion in our study to pedigrees that were mutation negative for both CDKN2A and CDK4, by single-strand conformation polymorphism (SSCP) and/or direct sequencing.
Melanoma case–control studies
Australia. Cases were drawn from (1) a population-based sample from Queensland, unselected for age at onset (Queensland study of Melanoma: Environment and Genetic Associations; Q-MEGA36) and (2) a population-based case–control family study of melanoma diagnosed before age 40 years, ascertained in Brisbane, Melbourne and Sydney (AMFS)37. Two sets of Australian controls were used. First, a sample of unrelated individuals was ascertained; they were mainly parents of adolescent twins (80% of the sample), together with a smaller number of twins and their siblings, recruited through schools to participate in the Brisbane Adolescent Twin Study38. Additionally, a set of controls from AMFS were available for genotyping37. The AMFS and twin controls did not have a personal history of melanoma. Approval for these as well as the melanoma family studies was obtained from the Human Research Ethics Committees of Queensland Institute of Medical Research (QIMR), University of Sydney, University of Melbourne and cancer registries of New South Wales, Victoria and Queensland. Informed consent was obtained from all participants. Most samples had: questionnaire-based naevus count based on a four point scale: ‘none’, ‘a few’, ‘moderate’ and ‘very many’ (Queensland); or ‘none’, ‘few’, ‘some’ and ‘many’ for AMFS; data on pigmentation variables, including: hair colour (fair, light brown, red, dark brown, black), eye colour (blue/grey, green hazel, brown/black), skin colour (light, medium, dark) and number of freckles (none, few, some/moderate, many).
United Kingdom. The Leeds-based case-control study recruited population-based incident melanoma cases diagnosed between September 2000 and December 2006 from a geographically defined area of Yorkshire and the Northern region of the UK (63% response rate)9,12,39. Cases were identified by clinicians and pathology registers and via the Northern and Yorkshire Cancer Registry and Information Service to ensure overall ascertainment. For all but 18 months of the study period, recruitment was restricted to patients with Breslow thickness of at least 0.75 mm. Controls were ascertained by contacting general practitioners to identify eligible individuals. These controls were frequency-matched with cases for age and sex from general practitioners who had also had cases as a part of their patient register. Overall there was a 55% response rate for controls. The first 960 of the cases recruited and all controls were examined by trained interviewers who performed a standardized examination of the skin, recording naevi by anatomical site and size. For subsequent cases, self-reported information on naevi was obtained. In total we utilized 1,549 cases and 495 controls from this study. We also used 380 cases and 373 controls recruited by the University of Cambridge who were genotyped in the replication series. The cases and controls were recruited as a part of the SEARCH study40,41, an ongoing population-based study in Eastern England. Cases were ascertained through the Eastern Cancer Registry and Information Centre, and were aged between 18 and 70 years at diagnosis. Controls were drawn from SEARCH and EPIC-Norfolk. The control set was supplemented with 1,245 additional controls from the Wellcome Trust Blood Services control set. Details of these studies have been previously published40,41,42. Approvals for the Leeds-based family studies, the Leeds case–control study and the Leeds Melanoma Cohort study were obtained from the National Research Ethics Service, Northern and Yorkshire Research Ethics Committee. Approvals for Cambridge case–control studies were obtained from the Cambridgeshire 4 Research Ethics Committee and the Norwich Local Research Ethics Committee. Informed consent was obtained from all participants.
Melanoma cell lines
The melanoma tumour cell lines used in this study were derived from primary cutaneous melanomas or melanoma metastases, as described previously43. DNA was extracted using QIAGEN QIamp Blood Maxi kits (Qiagen) according to the manufacturer’s instructions.
DNA Sequencing (whole genome)
One affected member from Family 1 was sequenced by Complete Genomics using a nanoarray-based short-read sequencing-by-ligation strategy. Sequencing, genome assembly (to genome build hg18/NCBI36.1), and variant calling were performed as described previously14.
Sequenom MassARRAY genotyping of E318K
MITF E318K was genotyped in Australian samples as part of a multiplex assay designed using the Sequenom MassARRAY Assay Design software (version 3.0). The SNP was typed using Sequenom iPLEX chemistry on a MALDI-TOF Compact Mass Spectrometer (Sequenom). The 2.5 μl PCR reactions were performed in standard 384-well plates using 12.5 ng genomic DNA, 0.8 units of Taq polymerase (HotStarTaq, Qiagen), 500 μmol of each dNTP, 1.625 mM of MgCl2 and 100 nmol of each PCR primer (Bioneer). Standard PCR thermal cycling conditions and post-PCR extension reactions were carried out as described previously10. The iPLEX reaction products were desalted by diluting samples with 15 µl of water and adding 3 µl of resin. The products were spotted on a SpectroChip (Sequenom), and data were processed and analysed by MassARRAY TYPER 3.4 software (Sequenom). DNA from the Family 1 sample in which E318K was identified was included multiple times as a positive control. As an additional quality control measure, we also confirmed the genotype of multiple individuals from the Australian case–control study, particularly those with a family history of melanoma, via Sanger sequencing, identifying no genotyping errors.
TaqMan genotyping of E318K
UK samples were genotyped for E318K using a custom TaqMan assay. DNA from the Australian family member in which E318K was identified was included as a positive control. Leeds melanoma pedigrees also were re-screened as a quality control measure using high-resolution melting (HRM) primers designed to assay E318K, with the results from HRM and TaqMan matching exactly.
Statistical analyses
Parametric linkage analysis on the multiple case melanoma families was conducted assuming a dominant model of inheritance, with penetrances specified as 5%, 95%, 95% for the three genotype classes. The allele frequency at the disease and marker locus was specified to be 0.004. Computations were done using the standard (lod score) parametric linkage routine implemented in GENEHUNTER-MODSCORE44. For melanoma case–control status, number of cases carrying the variant was compared to number of controls carrying the variant using Fisher’s exact test. P values given are two-sided except where noted otherwise. Sub-phenotypes were tested similarly, with family history, age at onset and multiple primary melanoma carrier rates compared with that in controls. Number of blue-eyed individuals carrying the variant was compared to number of non-blue eyed individuals carrying the variant using Fisher’s exact test. Meta-analysis for disease/sub-phenotype status and eye colour was done by combining the raw data. Association with mole count (high versus low where high was Australian Q-MEGA ‘moderate/many’, Australian AMFS ‘some/many’, UK ‘>25 moles’ and low was Australian Q-MEGA ‘none/few’, Australian AMFS ‘none/few’, UK ‘<25 moles’) was tested using Fisher’s exact test for each sample individually, due to differences in the exact definition of low/high. Meta-analysis results for mole count were generated by combining the individual sample ORs (where the natural log of the odds ratio was weighted for each sample by the inverse variance of the natural log of the odds ratio). The case–control analysis corrected for naevus count and eye colour was computed by fitting a logistic regression with naevus count and eye colour categories as covariates.
Sanger sequencing for E318K
Australian pedigrees for which the proband was identified as carrying the E318K variant via Sequenom, as well as the panel of melanoma cell lines, were screened for E318K via Sanger sequencing using the following primers: forward, 5′-CAGGCTCGAGCTCATGGA-3′; reverse, 5′- TGGGGACACTATAGGCTTGG-3′.
MITF sumoylation assay
His–MITF and/or HA–SUMO1 expression vectors, previously described25, were transfected into COS-7 or NAE cells. The cells were harvested after three days and 10 μg total protein was applied to each lane. Western blots were performed with anti-MITF monoclonal antibody (C5).
TRPM1 reporter assay
TRPM1 reporter plasmid (100 ng), Renilla luciferase vector (1 ng) (as internal control), and MITF expression vector (0, 175 or 350 ng) were transfected into UACC62 human melanoma cells as previously described25. After 2 days, luciferase activity was determined. Transfection efficiency was normalized to Renilla luciferase activity. Results are reported as the average of at least four independent experiments.
Construction of inducible MITF-expressing cell lines
Stable inducible expression of MITF was achieved using the ViraPower T-REx Lentiviral Expression System (Invitrogen). Briefly, cell pools were transduced with lentiviral particles containing pLENTI6/TR, and selected with 3 µg ml−1 blasticidin. Wild-type MITF was cloned into pLENTI4/TO/V5-DEST using Gateway recombination. Site-directed mutagenesis of the putative sumolyation site was carried out using a QuickChange II XL site-directed mutagenesis kit (Stratagene/Agilent Technologies), and the following primer pair: E318K change: E318K forward, 5′- GGTGAATCGGATCATCAAGCAAAAACCCGTTCTTG -3′ and E318K reverse, 5′- CAAGAACGGGTTTTTGCTTGATGATCCGATTCACC -3′. The negative control construct was identical except that it encodes the β-galactosidase gene (Invitrogen). Lentivirus particles were packaged in 293FT cells, before being titred using MM96L cells. Target melanoma cell lines were transduced with MITF and mutant particles using a multiplicity of infection of less than 1, and selected with 500 µg ml−1 zeocin and 1 µg ml−1 blasticidin for 3 weeks. Cells were maintained on 100 µg ml−1 zeocin and 0.5 µg ml−1 blasticidin for all experiments.
Induction of MITF
Cells were treated the day after seeding with 1 µg ml−1 tetracycline to induce MITF expression, and harvested 24 or 48 h afterwards. RNA was extracted using Qiagen RNeasy mini kit as per manufacturer’s instructions. RNA concentration was determined using the NanoDrop 2000 (Thermo Scientific). For mRNA analyses 0.5 µg of total RNA was reverse transcribed using Superscript III according to the manufacturer’s instructions (Invitrogen) and MITF, DCT, MLANA, TBSP1 and GAPDH expression analysed using SYBR Green PCR Master Mix (Applied Biosystems). Primer sequences were MITF forward, 5′-CCAAGTACCACATACAGCAAGC-3′ and MITF reverse, 5′-TCATCCATCTGCATACAGGACG-3′, DCT forward 5′-CGACTCTGATTAGTCGGAACTCA-3′ and DCT reverse, 5′-GGTGGTTGTAGTCATCCAAGC-3′, MLANA forward, 5′- GAGAAAAACTGTGAACCTGTGGT-3′ and MLANA reverse, 5′- GACTGTTCTGCAGAGAGTTTCTCAT-3′, THBS1 forward, 5′-CGGTCCAGACACGGACCTGC-3′ and THBS1 reverse 5′-GGCTTTGGTCTCCCGCGCTT-3′, GAPDH forward, 5′-GGCTCTCCAGAACATCATCCCTGC-3′ and GAPDH reverse, 5′-GGGTGTCGCTGTTGAAGTCAGAGG -3′. qRT–PCR was performed using a 7900HT Fast Real Time PCR System (Applied Biosystems).
Generation of constitutive MITF-expressing melanoma cell lines
pcDNA4.1-HisMax-hMITF (wild type, E318K or K182R/K316R) and pEGFP-C1 (for selection with G418) were transfected into the UACC62 melanoma cell line. After 5 days of selection with G418, total RNA was collected and MITF target gene transcript levels assessed by quantitative RT–PCR using the Pfaffl method45.
Expression profiling and analysis
Biotinylated cRNA was prepared with the Illumina TotalPrep RNA Amplification Kit (Ambion). Labelled cRNA was hybridized to HumanHT-12 v4 BeadChip Arrays (Illumina), and then washed and scanned according to standard Illumina protocols. Data were extracted in GenomeStudio (Illumina) using default analysis settings and no normalization method. Resulting data were imported into GeneSpring GX v11.5 (Agilent Technologies). Expression values were normalized using quantile normalization with default settings.
Entities at least 1.5 fold up- or downregulated following induction of either wild-type or E318K MITF were determined (C32 wild type, 491; C32 E318K, 705; HT144 wild type, 273; HT144 E318K, 450). Targets similarly up-or downregulated only by E318K and not wild-type MITF in both of the melanoma cell lines were identified using Venn diagram (n = 16 entities). Also targets similarly up- or downregulated only by wild-type and not E318K MITF in both of the melanoma cell lines were identified using Venn diagram (n = 2 entities). Entities similarly up- or downregulated by induction of wild-type and E318K MITF were identified (C32 wild type and E318K, 244; HT144 wild type and E318K, 150), and those targets consistently differentially expressed in both of the cell lines identified by Venn diagram (n = 43 entities representing 37 gene products).
NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/) accession numbers are as follows: GEO Series reference, GSE31269; GSM774914 HT144 wild-type uninduced; GSM774915 HT144 wild-type 48 h induced; GSM774916 HT144 E318K uninduced; GSM774917 HT144 E318K 48 h induced; GSM774918 C32 wild-type uninduced; GSM774919 C32 wild-type 48 h induced; GSM774920 C32 E318K uninduced; GSM774921 C32 E318K 48 h induced.
Melanin quantification assay
Melanin levels were determined46 after 72 h induction of wild-type or E318K MITF, and normalized to amounts in uninduced control cells. Values are from duplicate readings from triplicate independent experiments.
Supplementary information
This file contains Supplementary Results, Supplementary Tables 1-6, Supplementary Figures 1-3 with legends and additional references. (PDF 860 kb)
Acknowledgements
This work was supported by team science awards by the Melanoma Research Alliance (J.M.T., N.K.H., H.T. and D.E.F.), the American Cancer Society (K.M.B., RSG-08-200-01), the National Institutes of Health (NIH; D.E.F., AR043369-14; N.K.H., CA88363; H.T., K24CA149202 and P50CA93683), Doris Duke Medical Foundation (D.E.F.), Dr Miriam and Sheldon G. Adelson Medical Research Foundation (D.E.F.), US-Israel Binational Science Foundation (D.E.F.), and the Division of Cancer Epidemiology and Genetics of the National Cancer Institute (K.M.B.). N.K.H., D.L.D., S.M. and G.W.M. are supported by National Health and Medical Research Council of Australia (NHMRC) research fellowships. M.H.L. is supported by Cancer Australia grant 1011143. The collection of samples in the Leeds-based case–control study (the Melanoma Cohort Study) was funded by Cancer Research UK (Project Grant C8216/A6129 and Programme awards C588/A4994 and C588/A10589) and by the NIH (R01 CA83115). Recruitment was facilitated by the UK National Cancer Research Network. We thank S. Leake, S. Haynes, S. Waseem for Leeds case–control data collection; and H. Snowdon and C. Taylor from the Leeds Cancer Research UK Cancer Centre Genomics Facility for the genotyping of the UK samples. AMFS was supported by the NHMRC (project grants 566946, 107359, 211172 and program grant number 402761 to G.J.M. and R.F.K.); the Cancer Council New South Wales (project grant 77/00, 06/10), the Cancer Council Victoria and the Cancer Council Queensland (project grant 371); and the NIH (via RO1 grant CA-83115-01A2 to the international Melanoma Genetics Consortium—GenoMEL). The University of Cambridge SEARCH study was supported by Cancer Research UK Programme awards (C490/A11021 and C8197/A10123). A.E.C. is the recipient of an NHMRC public health postdoctoral fellowship (520018) and a Cancer Institute NSW Early Career Development Fellowship (10/ECF/2-06). B.K.A. is supported by a University of Sydney Medical Foundation Program Grant and J.L.H. is an Australia Fellow of the NHMRC. We gratefully acknowledge all of the participants, the work and dedication of the research coordinators, interviewers, examiners and data management staff, including J. Arbuckle, S. Columbus, M. Lang, H. Rodais, C. Ellis (Centre for MEGA Epidemiology); E. A. Holland, C. Agha-Hamilton, C. El Hayek, L. Morgan, J. Roland, E. Tyler, J. Barton, C. Watts and L. Porter (Westmead Institute of Cancer Research); M. Hillcoat, K. Holland, P. Saunders, J. Roberts and S. Tait (Viertel Centre for Research in Cancer Control); A. Kurien, C. Patterson, C. Thoo, S. de Zwaan, A. Sklavos, S. Manoharan, J. Cahill and S. Brennand (skin examiners).
PowerPoint slides
Author Contributions
K.M.B., N.K.H. and J.M.T. designed, analysed and managed sequencing and genotyping aspects of the study. K.M.B., S.M., S.L.W. and N.K.H. wrote the paper. D.E.F. designed functional studies and contributed to manuscript preparation. J.M.T. designed genetic studies. H.T. designed functional studies, sequenced MITF in melanoma cell lines and contributed to manuscript preparation. Analysis of whole-genome sequence data was performed by K.H., M.S.S., N.K.H. and K.M.B. Genotyping of MITF E318K in all Australian samples was performed by V.Z., with statistical analyses performed by S.M. and M.H.L. D.C.W., D.L.D., G.W.M., N.K.H. and N.G.M oversaw collection of the Queensland samples and contributed to statistical analyses, data interpretation and manuscript preparation. G.J.M., E.A.H., H.S., J.A.M., J.J., M.F., M.A.J., R.F.K., G.G.G., B.K.A, J.F.A. and J.L.H. oversaw collection and contributed to phenotypic analyses of the AMFS study, with statistical analysis performed by A.E.C. D.T.B., J.A.N.-B., P.D.P., D.F.E. and A.M.D. designed and managed aspects of genotyping in UK case–control studies, with genotyping performed by M.H. and statistical analyses performed by J.C.T. J.M.P. assisted in analysis of Australian melanoma pedigrees. Sequencing of MITF in and analysis of Australian melanoma pedigrees and cell lines were performed by L.G.A., S.L.W., M.G., K.D.-R., V.B., M.S.S., J.S., C.S., C.L. and L.O’C. S.Y. and R.H. performed the sumoylation and MITF-transactivation studies and contributed to manuscript preparation. G.M.B. generated inducible-MITF melanoma cell lines, performed phenotypic and global transcript analyses and contributed to manuscript preparation.
Accession codes
Primary accessions
Gene Expression Omnibus
Data deposits
Expression microarray data are available through the NCBI GEO website under accession GSE31269. Data for the full genome sequenced from FAM1 has been deposited in NCBI dbGAP under accession phs000419.v1.p1.
Competing interests
The authors declare no competing financial interests.
Footnotes
Satoru Yokoyama, Susan L. Woods, Glen M. Boyle, Lauren G. Aoude, Stuart MacGregor, Victoria Zismann, Hensin Tsao, Jeffrey M. Trent, David E. Fisher, Nicholas K. Hayward and Kevin M. Brown: These authors contributed equally to this work.
References
- 1.Goldstein AM, et al. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J. Med. Genet. 2007;44:99–106. doi: 10.1136/jmg.2006.043802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zuo L, et al. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nature Genet. 1996;12:97–99. doi: 10.1038/ng0196-97. [DOI] [PubMed] [Google Scholar]
- 3.Naldi L, et al. Cutaneous malignant melanoma in women. Phenotypic characteristics, sun exposure, and hormonal factors: a case–control study from Italy. Ann. Epidemiol. 2005;15:545–550. doi: 10.1016/j.annepidem.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Titus-Ernstoff L, et al. Pigmentary characteristics and moles in relation to melanoma risk. Int. J. Cancer. 2005;116:144–149. doi: 10.1002/ijc.21001. [DOI] [PubMed] [Google Scholar]
- 5.Holly EA, Aston DA, Cress RD, Ahn DK, Kristiansen JJ. Cutaneous melanoma in women. I. Exposure to sunlight, ability to tan, and other risk factors related to ultraviolet light. Am. J. Epidemiol. 1995;141:923–933. doi: 10.1093/oxfordjournals.aje.a117359. [DOI] [PubMed] [Google Scholar]
- 6.Holly EA, Aston DA, Cress RD, Ahn DK, Kristiansen JJ. Cutaneous melanoma in women. II. Phenotypic characteristics and other host-related factors. Am. J. Epidemiol. 1995;141:934–942. doi: 10.1093/oxfordjournals.aje.a117360. [DOI] [PubMed] [Google Scholar]
- 7.Bataille V, et al. Risk of cutaneous melanoma in relation to the numbers, types and sites of naevi: a case–control study. Br. J. Cancer. 1996;73:1605–1611. doi: 10.1038/bjc.1996.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang YM, et al. A pooled analysis of melanocytic nevus phenotype and the risk of cutaneous melanoma at different latitudes. Int. J. Cancer. 2009;124:420–428. doi: 10.1002/ijc.23869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bishop DT, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nature Genet. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown KM, et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nature Genet. 2008;40:838–840. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duffy DL, et al. IRF4 variants have age-specific effects on nevus count and predispose to melanoma. Am. J. Hum. Genet. 2010;87:6–16. doi: 10.1016/j.ajhg.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falchi M, et al. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nature Genet. 2009;41:915–919. doi: 10.1038/ng.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gudbjartsson DF, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nature Genet. 2008;40:886–891. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- 14.Drmanac R, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 15.Gillanders E, et al. Localization of a novel melanoma susceptibility locus to 1p22. Am. J. Hum. Genet. 2003;73:301–313. doi: 10.1086/377140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han J, et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008;4:e1000074. doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nan H, et al. Genome-wide association study of tanning phenotype in a population of European ancestry. J. Invest. Dermatol. 2009;129:2250–2257. doi: 10.1038/jid.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sulem P, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nature Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 19.Zhu G, et al. A genome-wide scan for naevus count: linkage to CDKN2A and to other chromosome regions. Eur. J. Hum. Genet. 2007;15:94–102. doi: 10.1038/sj.ejhg.5201729. [DOI] [PubMed] [Google Scholar]
- 20.Garraway LA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 21.Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2007;67:2632–2642. doi: 10.1158/0008-5472.CAN-06-4152. [DOI] [PubMed] [Google Scholar]
- 22.Cronin JC, et al. Frequent mutations in the MITF pathway in melanoma. Pigment Cell Melanoma Res. 2009;22:435–444. doi: 10.1111/j.1755-148X.2009.00578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams PF, Olsen CM, Hayward NK, Whiteman DC. Melanocortin 1 receptor and risk of cutaneous melanoma: a meta-analysis and estimates of population burden. Int. J. Cancer. 2010;129:1730–1740. doi: 10.1002/ijc.25804. [DOI] [PubMed] [Google Scholar]
- 24.Murakami H, Arnheiter H. Sumoylation modulates transcriptional activity of MITF in a promoter-specific manner. Pigment Cell Res. 2005;18:265–277. doi: 10.1111/j.1600-0749.2005.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller AJ, Levy C, Davis IJ, Razin E, Fisher DE. Sumoylation of MITF and its related family members TFE3 and TFEB. J. Biol. Chem. 2005;280:146–155. doi: 10.1074/jbc.M411757200. [DOI] [PubMed] [Google Scholar]
- 26.Boyle GM, et al. Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Res. 2011;24:525–537. doi: 10.1111/j.1755-148X.2011.00849.x. [DOI] [PubMed] [Google Scholar]
- 27.Strub T, et al. Essential role of microphthalmia transcription factor for DNA replication, mitosis and genomic stability in melanoma. Oncogene. 2011;30:2319–2332. doi: 10.1038/onc.2010.612. [DOI] [PubMed] [Google Scholar]
- 28.Hoek KS, et al. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment Cell Melanoma Res. 2008;21:665–676. doi: 10.1111/j.1755-148X.2008.00505.x. [DOI] [PubMed] [Google Scholar]
- 29.Smit NP, et al. Increased melanogenesis is a risk factor for oxidative DNA damage—study on cultured melanocytes and atypical nevus cells. Photochem. Photobiol. 2008;84:550–555. doi: 10.1111/j.1751-1097.2007.00242.x. [DOI] [PubMed] [Google Scholar]
- 30.Hemesath TJ, Price ER, Takemoto C, Badalian T, Fisher DE. MAP kinase links the transcription factor Microphthalmia to c-Kit signalling in melanocytes. Nature. 1998;391:298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- 31.Kefford RF, Salmon J, Shaw HM, Donald JA, McCarthy WH. Hereditary melanoma in Australia. Variable association with dysplastic nevi and absence of genetic linkage to chromosome 1p. Cancer Genet. Cytogenet. 1991;51:45–55. doi: 10.1016/0165-4608(91)90007-h. [DOI] [PubMed] [Google Scholar]
- 32.Nancarrow DJ, et al. Linkage mapping of melanoma (MLM) using 172 microsatellite markers. Genomics. 1992;14:939–947. doi: 10.1016/s0888-7543(05)80115-6. [DOI] [PubMed] [Google Scholar]
- 33.Harland M, et al. Germline mutations of the CDKN2 gene in UK melanoma families. Hum. Mol. Genet. 1997;6:2061–2067. doi: 10.1093/hmg/6.12.2061. [DOI] [PubMed] [Google Scholar]
- 34.Harland M, et al. Mutation screening of the CDKN2A promoter in melanoma families. Genes Chromosom. Cancer. 2000;28:45–57. [PubMed] [Google Scholar]
- 35.Harland M, Mistry S, Bishop DT, Bishop JA. A deep intronic mutation in CDKN2A is associated with disease in a subset of melanoma pedigrees. Hum. Mol. Genet. 2001;10:2679–2686. doi: 10.1093/hmg/10.23.2679. [DOI] [PubMed] [Google Scholar]
- 36.Baxter AJ, et al. The Queensland Study of Melanoma: environmental and genetic associations (Q-MEGA); study design, baseline characteristics, and repeatability of phenotype and sun exposure measures. Twin Res. Hum. Genet. 2008;11:183–196. doi: 10.1375/twin.11.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cust AE, et al. Population-based, case-control-family design to investigate genetic and environmental influences on melanoma risk: Australian Melanoma Family Study. Am. J. Epidemiol. 2009;170:1541–1554. doi: 10.1093/aje/kwp307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGregor B, et al. Genetic and environmental contributions to size, color, shape, and other characteristics of melanocytic naevi in a sample of adolescent twins. Genet. Epidemiol. 1999;16:40–53. doi: 10.1002/(SICI)1098-2272(1999)16:1<40::AID-GEPI4>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 39.Newton-Bishop JA, et al. Melanocytic nevi, nevus genes, and melanoma risk in a large case–control study in the United Kingdom. Cancer Epidemiol. Biomarkers Prev. 2010;19:2043–2054. doi: 10.1158/1055-9965.EPI-10-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pooley KA, et al. Common single-nucleotide polymorphisms in DNA double-strand break repair genes and breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 2008;17:3482–3489. doi: 10.1158/1055-9965.EPI-08-0594. [DOI] [PubMed] [Google Scholar]
- 41.Pooley KA, et al. No association between TERT-CLPTM1L single nucleotide polymorphism rs401681 and mean telomere length or cancer risk. Cancer Epidemiol. Biomarkers Prev. 2010;19:1862–1865. doi: 10.1158/1055-9965.EPI-10-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature447, 661–678 (2007) [DOI] [PMC free article] [PubMed]
- 43.Pavey S, et al. Microarray expression profiling in melanoma reveals a BRAF mutation signature. Oncogene. 2004;23:4060–4067. doi: 10.1038/sj.onc.1207563. [DOI] [PubMed] [Google Scholar]
- 44.Strauch K. Parametric linkage analysis with automatic optimization of the disease model parameters. Am. J. Hum. Genet. 2003;73:A2624. [Google Scholar]
- 45.Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oancea E, et al. TRPM1 forms ion channels associated with melanin content in melanocytes. Sci. Signal. 2009;2:ra21. doi: 10.1126/scisignal.2000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This file contains Supplementary Results, Supplementary Tables 1-6, Supplementary Figures 1-3 with legends and additional references. (PDF 860 kb)
Data Availability Statement
Primary accessions
Gene Expression Omnibus
Data deposits
Expression microarray data are available through the NCBI GEO website under accession GSE31269. Data for the full genome sequenced from FAM1 has been deposited in NCBI dbGAP under accession phs000419.v1.p1.


