SUMMARY
A key step in bacterial endospore formation is engulfment, during which one bacterial cell engulfs another in a phagocytosis-like process that normally requires SpoIID, SpoIIM, and SpoIIP (DMP). We here describe a second mechanism involving the zipper-like interaction between the forespore protein SpoIIQ and its mother cell ligand SpoIIIAH, which are essential for engulfment when DMP activity is reduced or SpoIIB is absent. They are also required for the rapid engulfment observed during the enzymatic removal of peptidoglycan, a process that does not require DMP. These results suggest the existence of two separate engulfment machineries that compensate for one another in intact cells, thereby rendering engulfment robust. Photobleaching analysis demonstrates that SpoIIQ assembles a stationary structure, suggesting that SpoIIQ and SpoIIIAH function as a ratchet that renders forward membrane movement irreversible. We suggest that ratchet-mediated engulfment minimizes the utilization of chemical energy during this dramatic cellular reorganization, which occurs during starvation.
INTRODUCTION
In contrast to the eukaryotic cytoplasmic membrane, which is routinely internalized during receptor-mediated endocytosis and phagocytosis, the internalization of bacterial cytoplasmic membranes is limited to a handful of lineages that synthesize membrane-bound organelles, such as magnetosomes (Komeili et al., 2006; Scheffel et al., 2006). One dramatic example of endocytosis in bacteria occurs during the starvation-induced sporulation pathway of Bacillus subtilis and its relatives. In these endospore-forming bacteria, sporulation commences with an asymmetrically positioned cell division event that generates two adjacent daughter cells of unequal size: the smaller forespore, which will become the spore, and the larger mother cell, which ultimately lyses to release the spore into the environment (Figure 1A, and reviewed by Errington, 2003; Hilbert and Piggot, 2004). The future spore is then internalized by a phagocytosis-like process known as engulfment, during which the mother cell membrane migrates around the forespore, until the leading edges of the engulfing membrane meet and fuse to release the forespore into the mother cell cytoplasm. Following engulfment, the forespore is bounded by two membranes, its own cytoplasmic membrane and an external membrane derived from the engulfing mother cell membrane, while the mother cell (unlike most bacterial cells) now has two separate membrane systems: the mother cell cytoplasmic membrane, which faces the environment, and the outer forespore membrane, which encloses the forespore.
Figure 1. The Process of Engulfment during B. subtilis Sporulation.
(A) The smaller forespore (FS) and larger mother cell (MC) initially lie side by side. Engulfment commences with septal thinning (i), during which septal peptidoglycan (light gray) is degraded. The mother cell membrane then migrates around the forespore (steps ii–iii), until it meets and fuses to release the forespore into the mother cell cytoplasm (step iv).
(B) Engulfment in intact cells requires three mother cell membrane proteins, SpoIID (Pac-Man), SpoIIM (dotted box), and SpoIIP (shaded lollipop), that localize to the septum and leading edge of the engulfing membrane. SpoIID is a peptidoglycan hydrolase, suggesting that engulfment might be mediated by the processive hydrolysis of the peptidoglycan adjacent to the forespore membrane, which could move the mother cell membrane around the forespore (in direction indicated by the arrow). Figure based on Abanes-De Mello et al. (2002).
(C) The zipper-like interaction between the forespore membrane protein SpoIIQ (Q; gradient) and the mother cell membrane protein SpoIIIAH (AH; gray) localizes SpoIIIAH (Blaylock et al., 2004; Doan et al., 2005), which recruits additional mother cell proteins (Doan et al., 2005; Jiang et al., 2005).
(D) A mechanical ratchet: the stationary pawl engages the ratchet teeth (or cogs), thereby preventing backward rotation of the wheel, allowing movement only in the direction indicated by the arrow. In a Brownian ratchet, random thermal energy is rectified, resulting in unidirectional rotation of the ratchet.
Engulfment is critical for sporulation, as it mediates the cellular rearrangements necessary to allow spore assembly to proceed in the mother cell cytoplasm, providing a more controlled environment than the external milieu and perhaps contributing to the extreme durability of the resulting spores. Engulfment also serves as a morphological checkpoint for activation of the late forespore- and mother cell-specific transcription factors σG and σK, respectively (reviewed by Errington, 2003; Hilbert and Piggot, 2004; Kroos and Yu, 2000; Rudner and Losick, 2001). The forespore-expressed SpoIIQ protein plays a central role in this checkpoint, as it mediates localization of proteins required for activation of late transcription in both cells. The extracellular domain of SpoIIQ directly interacts with that of SpoIIIAH (Figure 1C), a mother cell protein required for σG activity (Blaylock et al., 2004; Doan et al., 2005). SpoIIIAH recruits, perhaps indirectly, two mother cell proteins required for σK activity, SpoIVFA and SpoIVFB (Doan et al., 2005; Jiang et al., 2005). This is likely the main essential role of SpoIIQ, which is dispensable for engulfment under some conditions but essential for late transcription under all conditions (Jiang et al., 2005; Sun et al., 2000). SpoIIQ shows dynamic localization during engulfment, assembling a focus at the septal midpoint, tracking the engulfing mother cell membrane around the forespore, and assembling foci that appear as helical arcs in three dimensions (Rubio and Pogliano, 2004). This localization pattern is similar to that of two mother cell proteins SpoIIQ recruits to the septum, SpoIIIAH and SpoIVFB (Blaylock et al., 2004; Jiang et al., 2005), suggesting that the foci might serve as sites for intracellular signal transduction during the activation of late transcription.
Engulfment shares a superficial similarity to eukaryotic phagocytosis in that the engulfed cell plays a relatively passive role in its fate, since mother cell-specific, but not forespore-specific, gene expression is essential for engulfment (Sun et al., 2000). The critical nature of mother cell gene expression is also evidenced by the fact that of the three proteins known to be essential for engulfment, two are expressed only in the mother cell (SpoIID and SpoIIM; Driks and Losick, 1991; Lopez-Diaz et al., 1986; Smith and Youngman, 1993), while one (SpoIIP) is expressed in both cells, but is only required in the mother cell (Abanes-De Mello et al., 2002; Dworkin and Losick, 2005). SpoIID, SpoIIM, and SpoIIP (DMP) play a crucial role in the first step of engulfment, septal thinning, during which septal peptidoglycan is degraded (Figure 1). The main peptidoglycan hydrolase required for septal thinning is SpoIID (Abanes-De Mello et al., 2002), although peptidoglycan hydrolases involved in cell separation during growth can also contribute (Perez et al., 2006).
Septal thinning is necessary for movement of the engulfing membrane around the forespore. Thus far, no proteins specifically required for membrane migration have been identified, although some mutations in spoIID and spoIIP slow both septal thinning and membrane migration, suggesting that these proteins are required throughout engulfment (Abanes-De Mello et al., 2002). This conclusion is supported by the localization of SpoIID, SpoIIM, and SpoIIP to the leading edge of the engulfing membrane (Abanes-De Mello et al., 2002). The failure to identify additional proteins involved in membrane migration (Abanes-De Mello et al., 2002; Eichenberger et al., 2003) suggests that either DMP proteins are the only dispensable proteins required for membrane migration, or that the requirement for additional proteins is masked either by their requirement at an earlier step in sporulation (such as polar septation) or by the presence of additional proteins that can compensate for their absence. In the absence of evidence for additional engulfment proteins, we previously proposed that the hydrolytic activity of the DMP proteins mediates membrane migration by processively degrading the peptidoglycan adjacent to the forespore, thereby pulling the mother cell membrane around the forespore (Figure 1B; Abanes-De Mello et al., 2002). In this model, the bacterial cell wall provides a stationary pathway along which the hydrolase complex moves. This proposed role for the DMP proteins in membrane movement is similar to the recently described “burnt-bridge” Brownian ratchet, in which the random diffusion of a protein along a linear track (such as peptidoglycan) is converted into biased diffusion if the protein destroys its preferred binding site, so that backward diffusion is inhibited (Antal and Krapivsky, 2005; Mai et al., 2001). When adapted to engulfment, this model suggests that directional movement of the DMP proteins around the forespore is generated with thermal, rather than chemical energy, and that the hydrolytic activity of SpoIID renders this movement unidirectional.
We here provide evidence for a DMP-independent mechanism for membrane migration. Specifically, we have made the unexpected observation that when peptidoglycan is enzymatically removed from sporulating B. subtilis cells, about half rapidly complete membrane migration. This process, which we call nascent protoplast engulfment, requires two proteins that are dispensable for engulfment in intact cells, SpoIIQ and SpoIIIAH, but does not require the engulfment proteins DMP, which are essential in intact cells. Our data suggest that SpoIIQ and SpoIIIAH might mediate engulfment by a ratchet-like mechanism. Ratchets are mechanical devices that ensure the unidirectional movement of a toothed wheel by virtue of a stationary pawl that inserts between the teeth to prevent backward rotation (Figure 1D). Several cellular proteins have recently been proposed to have ratchet-like activities that contribute directional movement to processes including translation, transcription, and the translocation of proteins across membranes (Bar-Nahum et al., 2005; Krantz et al., 2006; Okamoto et al., 2002; Saffarian et al., 2004; Schuwirth et al., 2005). While in some of these cases the movement that is rendered unidirectional (i.e., rectified) might be generated by the expenditure of chemical energy, Brownian ratchets instead convert random movement generated by thermal energy into directional movement. They thereby provide an attractive mechanism for engulfment, which occurs during a starvation-induced differentiation pathway and in an extracellular milieu likely devoid of the chemical energy available in the cytoplasm.
We also provide evidence that the SpoIIQ/SpoIIIAH (Q-AH) ratchet acts in intact cells, because it is essential for engulfment in mutants with either reduced DMP activity or a lack of the SpoIIB protein necessary for rapid engulfment. We propose that there are multiple protein systems or modules that contribute to engulfment, including the DMP septal thinning module and the Q-AH module. The development of separate modules that contribute to membrane migration during B. subtilis engulfment likely contributes robustness to the system, allowing engulfment to proceed with high fidelity in a changing environment.
RESULTS
Lysozyme-Treated Sporangia Complete Engulfment
To test the model that the DMP proteins act as a peptidoglycan-directed motor that drives membrane migration during engulfment (Abanes-De Mello et al., 2002), we used lysozyme to degrade the peptidoglycan of sporulating cells in an osmotically protected medium. Membranes were stained with FM 4–64 and time lapse fluorescence microscopy was used to determine if protoplasts completed engulfment. When growing bacterial cells are treated with lysozyme, they lose their rod shape and become spherical. When sporulating cells are treated with lysozyme, they also become spherical and the smaller forespore remains attached to the larger mother cell, resulting in a snowman-like structure (Figure 2A). In intact cells, the fluorescent membrane stain FM 4–64 provides a quantitative indicator of septation and engulfment (Pogliano et al., 1999), with engulfed forespores staining three times as brightly as the cytoplasmic membrane due to the presence of three membrane layers around the forespore (see Figure 1A). After the addition of lysozyme, ~60% of protoplasts apparently completed engulfment, since the forespore stained about 3-fold more brightly than the mother cell cytoplasmic membrane (Figures 2A, 2F, and 2L).
Figure 2. Time Lapse Fluorescence Microscopy Showing Protoplast Engulfment.
Protoplasts were prepared from cells harvested 2.5 hr after the initiation of sporulation (t2.5) and stained with FM 4–64 (red). Images were collected at 45 s intervals. Time after first image is shown in bottom right corner of each frame (min:s). Arrows indicate sporangia that complete engulfment; arrowheads indicate sporangia with retracting membranes; yellow arrowheads indicate forespore of sporangia that separate; double yellow arrowheads indicate the mother cell of such sporangia. (A and C) FM 4–64-stained membranes (red) and mother cell produced SpoIIIJ-GFP (green; strain KP10068). (B and D) Surface plots showing the fluorescence intensity of FM 4–64-stained membranes (B) and mother cell GFP (D) from panels in (A) and (C) at the first (0 min) and last time points. Scale bar, 2 μm. (E) Alternate fates of mother cell-expressed SpoIIIJ-GFP. After protoplast engulfment, SpoIIIJ-GFP (green line) surrounds the forespore (upper pathway), while if engulfment does not occur, SpoIIIJ-GFP is restricted to the larger mother cell protoplast. (F–K) Time lapse microscopy showing protoplast engulfment in various mutants. Scale bar is 2 μm. (F) Wild-type (PY79), (G) spoIID298, spoIIM∷Tn917, ΔspoIIP∷tet (KP4188), (H) ΔspoIIQ∷spc (KP575), (I) spoIIIAG-HΩkan (KP896), (J) spoIIQ rbm13 (KP4124), (K) spoIIQ rbm9 (KP4138). (L) Quantification of protoplast engulfment, showing percent protoplasts that engulf (green), retract but remain attached (blue), or retract but separate (red). More than 50 protoplasts were scored for each strain.
A strain expressing a GFP-tagged membrane protein in the mother cell (PspoIID-SpoIIIJ-GFP) (Rubio et al., 2005) was used to determine if the increased fluorescence intensity around the forespore was an artifact of FM 4–64 staining or a consequence of engulfment. If the increased FM 4–64 staining of some forespores reflects engulfment, then SpoIIIJ-GFP should move around the forespore after lysozyme treatment (Figure 2E). In protoplasts with brightly staining forespore membranes, GFP fluorescence moved around the forespore (Figures 2A and 2C, arrow). In contrast, in protoplasts with forespores that were not brightly stained, GFP fluorescence remained in the mother cell membrane (Figures 2A and 2C, arrowhead). Therefore, the enhanced FM 4–64 staining intensity following lysozyme treatment is a consequence of the advancement of the mother cell membrane around the forespore in an engulfment-like manner.
Interestingly, the rate of engulfment during protoplast formation was extremely rapid and appeared limited only by cell wall degradation. While engulfment in intact cells requires ~45 min (Pogliano et al., 1999; Sharp and Pogliano, 1999), engulfment was complete 5 to 20 min after lysozyme addition and occurred as soon as the cells began to lose the rod-like shape that depends on the cell wall (Figure 2F, arrow). Indeed, the release of the lectin wheat germ agglutinin (which binds to the cell wall component N-acetylglucosamine; Pogliano et al., 1997) from the cell wall during membrane advancement (data not shown) suggests that protoplast engulfment occurs during, rather than after, cell wall removal. This process of nascent protoplast engulfment will hereafter be referred to as protoplast engulfment for brevity.
Protoplast Engulfment Does Not Require SpoIID, SpoIIM, or SpoIIP
To determine if proteins required for engulfment in intact cells are also required for protoplast engulfment, we examined the ability of a spoIID, spoIIM, and spoIIP triple mutant to complete engulfment after lysozyme treatment. Protoplast engulfment occurred at almost wild-type levels in the triple mutant (Figures 2F, 2G, and 2L; 43% versus 63% in wild-type). Other proteins not required for protoplast engulfment are SpoIIB (not shown), the absence of which causes uneven septal thinning but allows engulfment in intact cells (Margolis et al., 1993; Perez et al., 2000), and SpoIVFA and SpoIVFB (Figure 2L), required for σK activation (Cutting et al., 1991). Thus, protoplast engulfment is mediated by a DMP-independent mechanism.
Protoplast Engulfment Requires the Interacting Proteins SpoIIQ and SpoIIIAH
In eukaryotes, it has been proposed that zipper-like interactions between the phagocytic cell and the engulfed cell contribute to migration of the phagocytic membrane (Griffin et al., 1975; Griffin and Silverstein, 1974; Isberg and Van Nhieu, 1995; McAbee and Grinnell, 1985; Swanson and Hoppe, 2004). We therefore tested if two proteins that establish a zipper-like connection between the forespore and the mother cell, SpoIIQ and SpoIIIAH (Blaylock et al., 2004; Doan et al., 2005), were required for protoplast engulfment. Neither protein is essential for engulfment in intact cells, although strains lacking SpoIIQ engulf more slowly than wild-type and are engulfment-defective under some conditions (Londono-Vallejo et al., 1997; Sun et al., 2000). SpoIIQ was essential for protoplast engulfment, since spoIIQ protoplasts failed to complete engulfment. In addition, the spoIIQ strain showed a high frequency of forespores that had dissociated from their respective mother cells (hereafter called daughter cell separation), which occurred in 82% of spoIIQ protoplasts versus 12% of wild-type (Figures 2H and 2L). The spoIIIAG-AH double mutant, which lacks SpoIIIAH and another protein encoded by the operon (SpoIIIAG), also showed no protoplast engulfment and high levels of daughter cell separation (Figure 2I and 2L). This phenotype was due to the absence of the SpoIIQ binding protein SpoIIIAH, since a spoIIIAH single mutant was identical to the spoIIIAG-AH double mutant, while a spoIIIAG single mutant was similar to wild-type (see Figure S2 in the Supplemental Data).
These results indicate that protoplast engulfment depends on SpoIIQ and SpoIIIAH, two interacting proteins produced in adjacent daughter cells, but not on DMP. The strong interaction between SpoIIQ and SpoIIIAH suggests that they might comprise a ratchet that prevents the backward movement of the engulfing mother cell membrane. In this model, protoplast engulfment could be mediated by a Brownian ratchet, in which the random thermal motion of the engulfing membrane is captured by the successive securing of the forespore to the mother cell via the Q-AH zipper. This model predicts that there should be a threshold level of SpoIIQ protein required for protoplast engulfment (the level necessary to extend around the forespore), below which the mother cell and forespore would remain attached, but no protoplast engulfment would be observed. In contrast, if SpoIIQ were acting as an enzyme during protoplast engulfment, reducing its levels should progressively slow engulfment. The ratchet model also predicts that the forespore protein SpoIIQ assembles a stable scaffold capable of resisting backward movement of the engulfing mother cell membrane. These predictions are tested in the following sections.
Protoplast Engulfment and Stability Require a Threshold Amount of SpoIIQ
To test if reduced SpoIIQ levels inhibit protoplast engulfment and stability, mutations were introduced into the spoIIQ ribosome binding site to produce varying amounts of the wild-type protein (see Table S1 in the Supplemental Data). SpoIIQ levels were assessed by immunoblot using SpoIIQ- and FtsZ-specific antibodies (the latter as a loading control; Figure 3). Several SpoIIQ activities showed a requirement for a threshold amount of SpoIIQ protein (Figure 3B). For example, protoplast engulfment remained roughly constant at 50%–60% as SpoIIQ levels dropped, but in mutants that produced less than half the usual amount of SpoIIQ, protoplast engulfment dropped to 5%–12% (Figure 3B). Similarly, the association of the forespore and mother cell was supported in all mutants with detectable SpoIIQ protein (5%–15% daughter cell separation) and abruptly dropped in mutants without detectable SpoIIQ (55%–75% daughter cell separation). Spore titer was constant until levels dropped to <35% of wild-type (as observed for rbm14), after which a steady decline was observed. Thus, as levels of SpoIIQ protein dropped, protoplast engulfment was blocked first, then spore production, and finally, adherence of the forespore and mother cell was abolished only by the absence of detectable protein. Thus, as predicted by the ratchet model, a threshold level of SpoIIQ was required to support protoplast engulfment.
Figure 3. Protoplast Engulfment Exhibits a Dosage-Dependent Requirement for SpoIIQ.

(A) Immunoblot analysis of SpoIIQ in ribosome binding site mutants expressed in strains defective in SpoIIQ proteolysis (spoIVB)at t1, t2, t3, and t4. ΔspoIIQ∷spec (KP4195), rbm2 (KP4203), rbm1 (KP4202), rbm12 (KP4201), rbm9 (KP4198), rbm14 (KP4199), rbm10 (KP4200), rbm11 (KP4196), rbm13 (KP4197), and wild-type (KP4194).
(B) Quantification of protoplast engulfment (t2.5), spore titers (t24), SpoIIQ levels (t4), and the ability to support engulfment in the spoIIB mutant (t3.5) as described in Experimental Procedures.
Two of the results described above also demonstrate that holding the mother cell and forespore in close proximity is not the only role of SpoIIQ and SpoIIIAH in protoplast engulfment. First, in wild-type cells, half the sporangia fail to complete protoplast engulfment, although the two cells remain held together by the Q-AH complex (Figure 2). We suspect that these cells do not have sufficient levels of SpoIIQ (or SpoIIIAH) to support protoplast engulfment, as is supported by the observation that early sporangia, which will have lower levels of SpoIIQ and SpoIIIAH, complete protoplast engulfment less efficiently (Figure S1). Second, several ribosome binding site mutations produce levels of SpoIIQ that support daughter cell attachment but not protoplast engulfment (Figure 3). Thus, the Q-AH mediated attachment of the forespore and mother cell is not sufficient for protoplast engulfment, which requires higher levels of SpoIIQ.
GFP-SpoIIQ Forms a Stationary Structure in Intact Cells
The ratchet model predicts that SpoIIQ assembles a stable structure capable of resisting backward movement of the engulfing mother cell membrane. We used fluorescence recovery after photobleaching (FRAP) analysis to compare the mobility of GFP-SpoIIQ to that of a forespore-expressed control protein, MalF-GFP. A region of the forespore was bleached, and movement of protein from the unbleached region into the bleached region was assessed by comparing the image intensity in the two regions for 5 min (see Experimental Procedures). The MalF-GFP control protein showed complete equilibration between the bleached and unbleached regions within 4 s (Figures 4I and 4J). In contrast, GFP-SpoIIQ recovered much more slowly or not at all, depending on the stage of engulfment (Figures 4A–4D; Figure 4I). During engulfment, most bleached areas (15/21) showed no recovery (defined as failing to reach half the equilibration point within 5 min; Figure 4C), while some (6/21) showed partial recovery (reaching half the equilibration point between 50 and 300 s; Figures 4B and 4I). In contrast, before engulfment, most bleached areas (6/8) partially or completely recovered within 5 min (Figures 4A and 4I), and after engulfment, half of the bleached regions (3/6) showed partial or complete recovery within 5 min (Figure 4D), while half (3/6) showed no recovery (Figure 4I). Thus, movement of GFP-SpoIIQ is highly restricted during engulfment, as required for the forespore component of a ratchet, which must resist backward force.
Figure 4. Photobleaching Experiments Demonstrate that GFP-SpoIIQ Assembles a Static Structure.
After photobleaching, images were collected, quantified, and plotted (see Experimental Procedures) to show the adjusted mean pixel intensity of the bleached (black squares) and unbleached (unfilled squares) regions and the theoretical pixel intensity value following equilibration between these regions (dashed line). Images of GFP-SpoIIQ (green) and FM 4–64-stained membranes (red) of photobleached cells are shown to the right of each graph. Images below each plot show GFP-SpoIIQ during the experiment. Bleached regions are indicated by a yellow circle. (A–D) FRAP of GFP-SpoIIQ (ΔspoIIQ∷spc, gfp-spoIIQ) (KP845). (E–H) FRAP of GFP-SpoIIQ in the presence of SpoIIQ (gfp-spoIIQ) (KP866). (I) Frequency with which the various phenotypes were observed in individual cells subjected to FRAP. Recovery includes cells that reached ≥75% of the theoretical equilibration point; partial recovery reached 50%–75%; no recovery was less than 50%. (J) FRAP of MalF-GFP (malF-gfp) (KP834).
When GFP-SpoIIQ is expressed in the presence of native SpoIIQ, it no longer assembles foci and is slightly mislocalized, with a small amount of fluorescence ahead of the engulfing membranes (Jiang et al., 2005; Sun et al., 2000). This suggests that either wild-type SpoIIQ prevents GFP-SpoIIQ from assembling normally, or that the increased total SpoIIQ protein produced by the merodiploid strain cannot be fully immobilized by the unidentified mother cell binding partner that restricts SpoIIQ to the septum (Rubio and Pogliano, 2004). FRAP analysis demonstrated that the presence of wild-type SpoIIQ also increased the mobility of GFP-SpoIIQ during engulfment, as sporangia showed either partial (13/15) or complete (2/15) recovery of fluorescence intensity within 5 min (Figures 4E–4H; Figure 4I). Thus, when GFP-SpoIIQ is expressed together with wild-type SpoIIQ, it fails to be immobilized and it also fails to assemble foci, suggesting that the two features might be linked.
The above results suggest that the forespore protein SpoIIQ and the mother cell protein SpoIIIAH comprise a ratchet that mediates protoplast engulfment. In intact cells, the Q-AH ratchet would engage behind the engulfing membranes, whose movement around the forespore requires the DMP proteins. Protoplast engulfment might be mediated by a Brownian ratchet-like mechanism in which random thermal movement of the mother cell membrane (rotation of the toothed wheel) is rectified by the Q-AH ratchet to result in movement around the forespore. Alternatively, Q-AH might rectify forward movement generated by some unidentified force-generating mechanism.
Evidence for Ratchet Function in Intact Cells
Although neither SpoIIQ nor SpoIIIAH are essential for engulfment in intact cells, one might expect the Q-AH ratchet to improve the processivity of engulfment; therefore, it might be essential for engulfment in mutants that reduce the activity of DMP. To test this hypothesis, we investigated the effects of a spoIIQ null mutation on engulfment in a temperature-sensitive spoIID mutation (spoIID31)at the semipermissive temperature (Abanes-De Mello et al., 2002), using an in vivo membrane fusion assay to monitor the completion of engulfment (Sharp and Pogliano, 1999). During engulfment, the membrane-impermeable stain FM 4–64 stains the forespore membranes (Figure 5A, arrowhead; Figure 5M), while after engulfment it is excluded from the forespore, which stains only with the membrane-permeable stain Mitotracker Green (Figure 5A, arrow; Figure 5M). The absence of SpoIIQ blocked engulfment in spoIID31 at the semipermissive temperature, with no sporangia completing engulfment in the double mutant (Figure 5F) versus 16% in spoIID31 (Figure 5E), 32% in spoIIQ (Figure 5B), and 61% in wild-type (Figure 5A). SpoIIIAH was also required for engulfment in the spoIID31 strain (Figure 5G). Furthermore, SpoIIQ and SpoIIIAH were necessary for engulfment in cells expressing GFP fusions to SpoIIM or SpoIIP (Figure S3). These GFP fusion proteins support wild-type levels of spore formation (Abanes-De Mello et al., 2002) and engulfment, although not at completely wild-type rates. Together these results indicate that SpoIIQ and SpoIIIAH are essential for engulfment when the activity of the DMP engulfment proteins is reduced.
Figure 5. SpoIIQ and SpoIIIAH Contribute to Engulfment in Intact Cells.
(A–L) Samples were harvested at t3 and a fusion assay was performed to assess the completion of engulfment. FM 4–64 is membrane impermeable (red); Mitotracker Green is permeable (green; MTG shown in overlay). Fused sporangia, indicated by arrows, have completed engulfment; the unfused sporangia, indicated by arrowheads, have not. Percent of sporangia fused at t3 (A–G) or percent fused relative to ΔspoIIQ∷spc, amyE∷spoIIQΩcat, spoIIB∷erm (KP4369) at t3.5 (H–L) is shown (>200 sporangia scored for each strain). (A) Wild-type (PY79), (B) ΔspoIIQ∷spc (KP575), (C) ΔspoIIIAH (MO1429), (D) ΔspoIIIAG-HΩkan (KP10359), (E) spoIID31 (KP38), (F) spoIID31, ΔspoIIQ∷spc (KP4270), (G) spoIID31, ΔspoIIIAH (KP4296), (H) ΔspoIIQ∷spc, amyE∷spoIIQ, spoIIB∷erm (KP4369), (I) ΔspoIIQ∷spc, spoIIB∷erm (KP4276), (J) ΔspoIIQ∷spc, amyE∷spoIIQ(rbm9), spoIIB∷erm (KP4373), (K) ΔspoIIQ∷spc, amyE∷spoIIQ(rbm10), spoIIB∷erm (KP4377), (L) ΔspoIIQ∷spc, amyE∷spoIIQ(rbm13), spoIIB∷erm (KP4370). (M) Cartoon of membrane fusion assay showing FM 4–64 (red) and Mitotracker Green (green).
If SpoIIQ acts like a ratchet in intact cells, one would expect that its ability to support engulfment in strains with slow engulfment would require a threshold level of SpoIIQ. In contrast, if SpoIIQ acted as an enzyme, decreasing levels of SpoIIQ would cause a gradual reduction in engulfment efficiency. We tested the effect of the spoIIQ ribosome binding site mutations on engulfment in intact cells lacking SpoIIB, which is required for efficient septal thinning and membrane migration (Margolis et al., 1993; Perez et al., 2000). The spoIIB mutant slows engulfment (16% fused at t3.5), while the additional inactivation of spoIIQ abolishes engulfment, indicating that SpoIIQ is essential for engulfment in the absence of SpoIIB (Figures 5H and 5I). Those spoIIQ ribosome binding site mutations that supported protoplast engulfment also supported engulfment in the spoIIB strain (Figure 5L; Figure 3B, striped bars), while those that failed to support protoplast engulfment also failed to support engulfment in the spoIIB strain (Figures 5J and 5K; Figure 3B). The finding that protoplast engulfment and engulfment in the absence of SpoIIB require identical levels of SpoIIQ suggests that SpoIIQ contributes to these processes by a similar mechanism.
We previously noted that while the absence of SpoIIQ only modestly slowed engulfment in resuspension media, it almost completely blocked engulfment when sporulation was induced by nutrient exhaustion in Difco Sporulation Medium (DSM; Sun et al., 2000). If SpoIIQ and SpoIIIAH played a ratchet-like role in intact cells, then SpoIIIAH should also be required for engulfment when sporulation was induced by nutrient exhaustion. This was indeed the case, as the absence of either SpoIIQ or SpoIIIAH similarly compromised the completion of engulfment membrane fusion in DSM (61% complete engulfment in wild-type by t3, versus 9% in spoIIQ and 17% in spoIIIAH; Figure S3). These data suggest that SpoIIQ and SpoIIIAH act together to mediate engulfment under these conditions.
DISCUSSION
Two Distinct Mechanisms for Membrane Migration during B. subtilis Engulfment
We here demonstrate the existence of a second mechanism for mediating membrane migration during engulfment that is independent of the DMP proteins required in intact cells. Surprisingly, when peptidoglycan is enzymatically removed from sporulating cells, more than half the sporangia rapidly complete engulfment. Protoplast engulfment depends on two proteins, the forespore protein SpoIIQ and the mother cell protein SpoIIIAH, that establish a zipper-like connection between the two cells (Blaylock et al., 2004; Doan et al., 2005). These proteins also play an important role in engulfment in intact cells, as both SpoIIQ and SpoIIIAH are necessary for efficient engulfment in some media (Londono-Vallejo et al., 1997; Sun et al., 2000), and our data demonstrates that both proteins are required when the activity of the DMP engulfment proteins is compromised. The observation that protoplast engulfment does not depend on the DMP proteins required for engulfment in intact cells indicates that the two engulfment processes occur by distinct mechanisms. We therefore propose that DMP and Q-AH comprise two distinct modules for engulfment, with the DMP module mediating septal thinning and membrane migration and the Q-AH module contributing to membrane migration via a separate mechanism (discussed below). The existence of two independently acting modules for engulfment likely contributes robustness to the process and suggests that engulfment might have evolved from two rudimentary engulfment mechanisms.
Evidence for a Ratchet Comprised of the SpoIIQ and SpoIIIAH Membrane Proteins
SpoIIQ and SpoIIIAH are interacting membrane proteins synthesized in adjacent cells of the sporangium (Blaylock et al., 2004; Doan et al., 2005). The robust interaction between the extracellular domains of these two proteins across the septum throughout engulfment suggests that they might facilitate engulfment by assembling a ratchet that resists the backward movement of the engulfing membrane. Three lines of evidence support this proposal. First, photobleaching analysis demonstrates that GFP-SpoIIQ is stationary during engulfment, as required of the forespore ratchet component, which must resist backward movement of the engulfing mother cell membrane. Second, a threshold level of SpoIIQ is required for protoplast engulfment and for engulfment in intact cells lacking SpoIIB, as would be expected if the process depended on sufficient SpoIIQ to wrap around the forespore. Finally, the interaction between SpoIIQ and SpoIIIAH can bear the mechanical load required to hold the forespore and mother cell together after removal of the wall, suggesting that it can also bear the load required to resist backward motion of the mother cell membrane during protoplast engulfment. We therefore suggest that SpoIIQ provides the stationary component(s) of a ratchet that rectifies forward membrane movement via its interaction with the mother cell protein SpoIIIAH (Figure 6A). Protoplast engulfment might occur via a Brownian ratchet mechanism, with random thermal motion of the mother cell membrane trapped by the Q-AH ratchet to result in directional membrane migration.
Figure 6. Models for Engulfment in Protoplasts and Intact Cells.
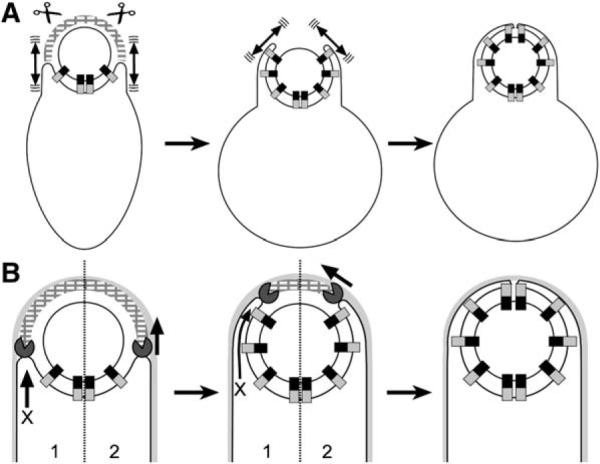
(A) Brownian ratchet model for protoplast engulfment. During protoplast formation, peptidoglycan hydrolases (scissors) remove the peptidoglycan (hatched arcs) that blocks the engulfing membrane, allowing the Q-AH ratchet to engage and prevent backward membrane movement. The mother cell membrane could move by Brownian motion (double-headed arrows).
(B) Models for engulfment in intact cells. (1) Missing motor model. The DMP proteins (Pac-Man) might act like lysozyme, degrading the peptidoglycan that blocks engulfment. Since SpoIIQ and SpoIIIAH are not essential for engulfment in whole cells, this model requires additional unidentified force-generating proteins (X arrow). (2) Dual ratchet model. The membrane-anchored DMP proteins might act as a burnt-bridge Brownian ratchet, in which the enzymatic activity of SpoIID renders complex diffusion along the peptidoglycan unidirectional, thereby moving the engulfing membrane around the forespore. The Q-AH ratchet could contribute to the directionality and processivity of the DMP ratchet.
Potential Roles of the DMP Proteins: a Burnt-Bridge Brownian Ratchet?
Protoplast engulfment appears to commence as soon as the cell wall is removed, suggesting that peptidoglycan provides a barrier to the engulfing membrane. These results raise the possibility that the DMP proteins that degrade septal peptidoglycan might contribute to membrane migration by removing this barrier to the engulfing membranes, rather than by contributing to membrane movement itself. We cannot rule out this possibility, although it requires the existence of additional, unidentified proteins that generate the force necessary to move the mother cell membranes (Figure 6B, model 1) because the Q-AH ratchet is not always essential for engulfment in intact cells. It also remains possible that peptidoglycan is both a barrier to the engulfing membrane and a linear track along which the DMP proteins move via a burnt-bridge Brownian ratchet (Figure 6B, model 2). Indeed, this combined barrier and track is seen with collagen fibers, along which proteases move via a burnt-bridge Brownian ratchet mechanism as they degrade collagen to remove barriers to the migration of cells through tissues (Saffarian et al., 2004). Interestingly, some of these proteases interact with membranes, suggesting that they work with the cytoskeleton to generate and stabilize cellular protrusions. The burnt-bridge Brownian ratchet provides an attractive mechanism for the translocation of the DMP hydrolase around the forespore, as it does not require the utilization of chemical energy supplies that are limited in the extracellular milieu and during starvation. Thus, our data suggest that engulfment might be mediated by two ratchets, the DMP burnt-bridge ratchet and the Q-AH ratchet, which together create forward membrane movement and render this movement irreversible.
DMP and Q-AH Independently Contribute to the Robustness of Engulfment in Cells
Although engulfment can occur in the absence of the Q-AH module, membrane migration is slowed (Figure 5, Figure S3; Londono-Vallejo et al., 1997; Sun et al., 2000). This suggests that the Q-AH module facilitates engulfment in intact cells, where in addition to serving as a ratchet, it might contribute directionality to engulfment by favoring migration of the mother cell membrane around the forespore, where additional Q-AH interactions can form. Indeed, the Q-AH module appears to compensate for reduced activity of the DMP module, as a synergistic engulfment defect is noted when spoIIQ or spoIIIAH mutations are introduced into strains with reduced DMP activity. Importantly, an identical level of SpoIIQ is required for this compensatory engulfment activity and for protoplast engulfment, and both SpoIIQ and SpoIIIAH are required for efficient engulfment in nutrient exhaustion, suggesting that the Q-AH ratchet also operates in intact cells. The endospore-forming bacteria inhabit a wide variety of environments, some at extremes of temperature or salinity. We speculate that the environmental robustness of engulfment is achieved by the combinatorial activities of the independently acting DMP and Q-AH modules.
EXPERIMENTAL PROCEDURES
Strains and Culture Conditions
Strains (Table S2) are derivatives of B. subtilis PY79, except MO1422 and MO1429, which are derivatives of JH642. Mutations were introduced by transformation (Dubnau and Davidoff-Abelson, 1971). Oligonucleotides used in cloning are in Table S1. Sporulation was induced by resuspension (Sterlini and Mandelstam, 1969)at 37°C (unless otherwise noted). CH medium was prepared using casein hydrolysate from EMD, due to variability in the ability of oxoid casein hydrolysate to support sporulation. Spore titers were determined as described (Perez et al., 2000).
Microscopy and Deconvolution
Membrane fusion assays (Sharp and Pogliano, 1999) and FM 4–64 staining (Rubio and Pogliano, 2004) were performed as described, using an Applied Precision Spectris microscope equipped with a Quantified Laser Module (Liu et al., 2006). Image files were deconvolved using SoftWoRx (15 iterations on conservative setting); medial focal planes are shown. Membrane fusion assays for quantification used PY79 as an internal control, and data were normalized to 61% wild-type sporangia fused at t3. Photobleaching experiments to assess GFP-SpoIIQ or MalF-GFP mobility used cells from t2 or t3 of sporulation; these were concentrated, stained with FM 4–64, and applied to poly-L-lysine treated coverslips. FM 4–64 and GFP images were collected after photobleaching. Photobleaching was achieved using a 0.05 s pulse of a 488 nm argon laser at 50% power, and subsequent GFP images were collected at 15 s intervals for 5 min for GFP-SpoIIQ or as quickly as possible for MalF-GFP. Exposure times were limited to 2 to 3 s.
Protoplast Engulfment Assay
Sporulating cells were collected by microcentrifugation at 9000 rpm for 10 s and resuspended in 1/20 volume SMM buffer (0.5 M sucrose, 20 mM maleic acid, 20 mM MgCl2 [pH 6.5]). Samples were placed on a slide, mixed with FM 4–64 and lysozyme (final concentrations 5 μg/mL and 1 mg/mL, respectively), and immobilized on a poly-L-lysine treated coverslip. Images were collected at 45 s intervals for 25 min at room temperature. To score engulfment, pixel intensity data was quantified using the Data Inspector function of SoftWoRx. Protoplasts were scored as completing engulfment if the pixel intensity at the forespore pole was 2- to 4-fold higher than that of the mother cell's cytoplasmic membrane. Pilot experiments indicated that the efficiency of protoplast engulfment depended on the stage of sporulation, with more engulfment later in sporulation (Figure S1). Experiments in Figure 2 and Figure 3 used samples collected 2.5 hr after the initiation of sporulation.
3D Graphs of Pixel Intensity Values
To quantitatively visualize the pixel intensity data (as in Figure 2), pixel intensity data from the medial focal plane (of deconvolved but unadjusted images) was exported to Microsoft Excel and graphed using the surface plot function.
Site-Directed Mutagenesis and Characterization of spoIIQ RBS Mutations
Mutations were constructed (Sawano and Miyawaki, 2000) in pCH505 (pDG1662 with a 2 kb spoIIQ region) using primers (Table S1) to modify the ribosome binding site. Mutations were verified by sequencing and integrated into the chromosome at amyE (Guerout-Fleury et al., 1996) and transformed into the ΔspoIIQ strain KP575. To analyze SpoIIQ levels, mutations were transformed into a strain lacking the SpoIVB protease responsible for SpoIIQ proteolysis (Jiang et al., 2005). Western blot samples were prepared (Pogliano et al., 1997) 1, 2, 3, and 4 hr after the start of sporulation; blots were then probed with rabbit anti-SpoIIQ (Jiang et al., 2005) and sheep anti-FtsZ (kindly provided by Liz Harry), scanned, and analyzed using Adobe Photoshop. Total pixel intensity from a region of defined size from the SpoIIQ immunoblot was compared to a corresponding region on the FtsZ immunoblot, creating a SpoIIQ/FtsZ ratio. The ratio from PY79 at t4 was designated as 100%; all samples were normalized to this value.
Quantification of Fluorescence Recovery in Small Cells
FRAP quantification methods normally assume that photobleaching has not notably affected the total fluorescence in the cell, so full recovery can be defined as the time at which the bleached area returns to its prebleach fluorescence intensity. While some eukaryotic cells might be large enough for total cellular fluorescence to remain essentially unchanged after bleaching, this is not the case for small bacterial cells or eukaryotic organelles. In these cases, a limited pool of fluorescent protein is confined to a small area, and photobleaching significantly decreases total fluorescence (we observe a 50%–80% reduction in total fluorescence). In these cases, a freely diffusing protein will never recover to its prebleach fluorescence intensity, and atypical, nonhyperbolic recovery curves result. To account for this loss in total fluorescence, we assessed recovery as the ability of adjusted mean fluorescence of the bleached and unbleached regions to approach the theoretical fluorescence intensity that would occur when the remaining fluorescent protein equilibrated between the two regions.
Using the “edit polygon” function of the Delta Vision version 2.10 software, we defined individual polygons to represent (1) the background fluorescence (a cell-free region), (2) the entire membrane region of the forespore, and (3) completely unbleached or (4) bleached regions of the forespore. Average mean fluorescence from the background region was subtracted from all time points prior to data processing. Mean fluorescence for each data region was adjusted for losses incurred by photobleaching during image acquisition by using a technique in which postbleach values are corrected by the application of a ratio created by dividing the first whole-cell fluorescence post-bleach value by the nth whole-cell fluorescence postbleach value. Data were plotted as time versus mean fluorescence for the unbleached region, the bleached region, and the whole cell fluorescence. The pixel intensity expected for the theoretical equilibration point was calculated by application of each whole-cell fluorescence ratio to its corresponding mean whole-cell fluorescence value, and it is represented on the graphs as a dotted line. The equilibration value was designated as F∞ for estimation of t1/2 recovery.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shinobu Chiba for providing plasmid pCH505, which was used to generate the spoIIQ RBS mutations; Patrick Stragier, for generously providing strains MO1422 and MO1429; Liz Harry, for providing FtsZ-specific antibodies; and Jonathan Dworkin, for suggesting the use of EMD casein hydrolysate. We also thank Rachel Larsen, Joe Pogliano, Shinobu Chiba, Aileen Rubio, and Amber Dance for providing helpful comments on this manuscript. This work was supported by the National Institute of Health (GM 57045).
Footnotes
Supplemental Data The Supplemental Data for this article can be found online at http://www.cell.com/cgi/content/full/126/5/917/DC1/, and includes the following: Table S1 (oligonucleotides) and Table S2 (strain list); movies showing protoplast engulfment (Movies S1–S4); and three figures showing the efficiency of protoplast engulfment at different times of sporulation (Figure S1), protoplast engulfment of ΔspoIIIAH and ΔspoIIIAG strains (Figure S2), and engulfment efficiency of spoIIQ and spoIIIAH strains during resuspension or nutrient depletion (Figure S3).
REFERENCES
- Abanes-De Mello A, Sun YL, Aung S, Pogliano K. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 2002;16:3253–3264. doi: 10.1101/gad.1039902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antal T, Krapivsky PL. “Burnt-bridge” mechanism of molecular motor motion. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2005 doi: 10.1103/PhysRevE.72.046104. in press. Published online October 6, 2005. 10.1103/PhysRevE.72.046104. [DOI] [PubMed] [Google Scholar]
- Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- Blaylock B, Jiang X, Rubio A, Moran CP, Jr., Pogliano K. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev. 2004;18:2916–2928. doi: 10.1101/gad.1252704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutting S, Roels S, Losick R. Sporulation operon spoIVF and the characterization of mutations that uncouple mother-cell from forespore gene expression in Bacillus subtilis. J. Mol. Biol. 1991;221:1237–1256. doi: 10.1016/0022-2836(91)90931-u. [DOI] [PubMed] [Google Scholar]
- Doan T, Marquis KA, Rudner DZ. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol. Microbiol. 2005;55:1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- Driks A, Losick R. Compartmentalized expression of a gene under the control of sporulation transcription factor sigma E in Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 1991;88:9934–9938. doi: 10.1073/pnas.88.22.9934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D, Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J. Mol. Biol. 1971;56:209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- Dworkin J, Losick R. Developmental commitment in a bacterium. Cell. 2005;121:401–409. doi: 10.1016/j.cell.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Eichenberger P, Jensen ST, Conlon EM, van Ooij C, Silvaggi J, Gonzalez-Pastor JE, Fujita M, Ben-Yehuda S, Stragier P, Liu JS, Losick R. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 2003;327:945–972. doi: 10.1016/s0022-2836(03)00205-5. [DOI] [PubMed] [Google Scholar]
- Errington J. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 2003;1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- Griffin FM, Jr., Silverstein SC. Segmental response of the macrophage plasma membrane to a phagocytic stimulus. J. Exp. Med. 1974;139:323–336. doi: 10.1084/jem.139.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin FM, Jr., Griffin JA, Leider JE, Silverstein SC. Studies on the mechanism of phagocytosis. I. Requirements for circumferential attachment of particle-bound ligands to specific receptors on the macrophage plasma membrane. J. Exp. Med. 1975;142:1263–1282. doi: 10.1084/jem.142.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Hilbert DW, Piggot PJ. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 2004;68:234–262. doi: 10.1128/MMBR.68.2.234-262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg RR, Van Nhieu GT. The mechanism of phagocytic uptake promoted by invasin-integrin interaction. Trends Cell Biol. 1995;5:120–124. doi: 10.1016/s0962-8924(00)88962-x. [DOI] [PubMed] [Google Scholar]
- Jiang X, Rubio A, Chiba S, Pogliano K. Engulfment-regulated proteolysis of SpoIIQ: evidence that dual checkpoints control σK activity. Mol. Microbiol. 2005;58:102–115. doi: 10.1111/j.1365-2958.2005.04811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. [DOI] [PubMed] [Google Scholar]
- Krantz BA, Finkelstein A, Collier RJ. Protein translocation through the anthrax toxin transmembrane pore is driven by a proton gradient. J. Mol. Biol. 2006;355:968–979. doi: 10.1016/j.jmb.2005.11.030. [DOI] [PubMed] [Google Scholar]
- Kroos L, Yu YT. Regulation of sigma factor activity during Bacillus subtilis development. Curr. Opin. Microbiol. 2000;3:553–560. doi: 10.1016/s1369-5274(00)00140-5. [DOI] [PubMed] [Google Scholar]
- Liu NJ, Dutton RJ, Pogliano K. Evidence that the SpoIIIE DNA translocase participates in membrane fusion during cytokinesis and engulfment. Mol. Microbiol. 2006;59:1097–1113. doi: 10.1111/j.1365-2958.2005.05004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londono-Vallejo JA, Frehel C, Stragier P. SpoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol. Microbiol. 1997;24:29–39. doi: 10.1046/j.1365-2958.1997.3181680.x. [DOI] [PubMed] [Google Scholar]
- Lopez-Diaz I, Clarke S, Mandelstam J. spoIID operon of Bacillus subtilis: cloning and sequence. J. Gen. Microbiol. 1986;132:341–354. doi: 10.1099/00221287-132-2-341. [DOI] [PubMed] [Google Scholar]
- Mai J, Sokolov IM, Blumen A. Directed particle diffusion under “burnt bridges” conditions. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2001 doi: 10.1103/PhysRevE.64.011102. in press. Published online June 11, 2001. 10. 1103/PhysRevE.64.011102. [DOI] [PubMed] [Google Scholar]
- Margolis PS, Driks A, Losick R. Sporulation gene spoIIB from Bacillus subtilis. J. Bacteriol. 1993;175:528–540. doi: 10.1128/jb.175.2.528-540.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAbee DD, Grinnell F. Binding and phagocytosis of fibronectin-coated beads by BHK cells: receptor specificity and dynamics. J. Cell. Physiol. 1985;124:240–246. doi: 10.1002/jcp.1041240211. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Brinker A, Paschen SA, Moarefi I, Hayer-Hartl M, Neupert W, Brunner M. The protein import motor of mitochondria: a targeted molecular ratchet driving unfolding and translocation. EMBO J. 2002;21:3659–3671. doi: 10.1093/emboj/cdf358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez AR, Abanes-De Mello A, Pogliano K. SpoIIB localizes to active sites of septal biogenesis and spatially regulates septal thinning during engulfment in Bacillus subtilis. J. Bacteriol. 2000;182:1096–1108. doi: 10.1128/jb.182.4.1096-1108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez AR, Abanes-De Mello A, Pogliano K. Suppression of engulfment defects in bacillus subtilis by elevated expression of the motility regulon. J. Bacteriol. 2006;188:1159–1164. doi: 10.1128/JB.188.3.1159-1164.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J, Osborne N, Sharp MD, Abanes-De Mello A, Perez A, Sun YL, Pogliano K. A vital stain for studying membrane dynamics in bacteria: a novel mechanism controlling septation during Bacillus subtilis sporulation. Mol. Microbiol. 1999;31:1149–1159. doi: 10.1046/j.1365-2958.1999.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano K, Hofmeister AE, Losick R. Disappearance of the σE transcription factor from the forespore and the SpoIIE phosphatase from the mother cell contributes to establishment of cell-specific gene expression during sporulation in Bacillus subtilis. J. Bacteriol. 1997;179:3331–3341. doi: 10.1128/jb.179.10.3331-3341.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio A, Pogliano K. Septal localization of forespore membrane proteins during engulfment in Bacillus subtilis. EMBO J. 2004;23:1636–1646. doi: 10.1038/sj.emboj.7600171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio A, Jiang X, Pogliano K. Localization of translocation complex components in Bacillus subtilis: enrichment of the signal recognition particle receptor at early sporulation septa. J. Bacteriol. 2005;187:5000–5002. doi: 10.1128/JB.187.14.5000-5002.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Losick R. Morphological coupling in development: lessons from prokaryotes. Dev. Cell. 2001;1:733–742. doi: 10.1016/s1534-5807(01)00094-6. [DOI] [PubMed] [Google Scholar]
- Saffarian S, Collier IE, Marmer BL, Elson EL, Goldberg G. Interstitial collagenase is a Brownian ratchet driven by proteolysis of collagen. Science. 2004;306:108–111. doi: 10.1126/science.1099179. [DOI] [PubMed] [Google Scholar]
- Sawano A, Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 2000;28:E78. doi: 10.1093/nar/28.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schuler D. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature. 2006;440:110–114. doi: 10.1038/nature04382. [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Sharp MD, Pogliano K. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. USA. 1999;96:14553–14558. doi: 10.1073/pnas.96.25.14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Youngman P. Evidence that the spoIIM gene of Bacillus subtilis is transcribed by RNA polymerase associated with σE. J. Bacteriol. 1993;175:3618–3627. doi: 10.1128/jb.175.11.3618-3627.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlini JM, Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YL, Sharp MD, Pogliano K. A dispensable role for forespore-specific gene expression in engulfment of the forespore during sporulation of Bacillus subtilis. J. Bacteriol. 2000;182:2919–2927. doi: 10.1128/jb.182.10.2919-2927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson JA, Hoppe AD. The coordination of signaling during Fc receptor-mediated phagocytosis. J. Leukoc. Biol. 2004;76:1093–1103. doi: 10.1189/jlb.0804439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






