Abstract
Voltage-gated sodium channels initiate electrical signaling in excitable cells and are the molecular targets for drugs and disease mutations, but the structural basis for their voltage-dependent activation, ion selectivity, and drug block is unknown. Here, we report the crystal structure of a voltage-gated Na+-channel from Arcobacter butzleri (NavAb) captured in a closed-pore conformation with four activated voltage-sensors at 2.7 Å resolution. The arginine gating charges make multiple hydrophilic interactions within the voltage-sensor, including unanticipated hydrogen bonds to the protein backbone. Comparisons to previous open-pore potassium channel structures suggest that the voltage-sensor domains and the S4-S5 linkers dilate the central pore by pivoting together around a hinge at the base of the pore module. The NavAb selectivity filter is short, ~6.5 Å wide, and water-filled, with four acidic side-chains surrounding the narrowest part of the ion conduction pathway. This unique structure presents a high field-strength anionic coordination site, which confers Na+-selectivity through partial dehydration via direct interaction with glutamate side-chains. Fenestrations in the sides of the pore module are unexpectedly penetrated by fatty acyl chains that extend into the central cavity, and these portals are large enough for the entry of small, hydrophobic pore-blocking drugs.
Electrical signals (termed action potentials) encode and process information within the nervous system and regulate a wide range of physiological processes1,2. The voltage-gated ion channels (VGICs) that mediate electrical signaling have distinct functional roles1,2. Voltage-gated sodium (NaV) channels initiate action potentials. Voltage-gated calcium (CaV) channels initiate processes such as synaptic transmission, muscle contraction, and hormone secretion in response to membrane depolarization. Voltage-gated potassium (KV) channels terminate action potentials and return the membrane potential to its resting value. The NaV channels are mutated in inherited epilepsy, migraine, periodic paralysis, cardiac arrhythmia, and chronic pain syndromes3. These channels are molecular targets of drugs used in local anesthesia and in treatment of genetic and sporadic NaV channelopathies in brain, skeletal muscle, and heart4. The rapid activation, Na+-selectivity, and drug sensitivity of NaV channels are unique among VGICs2.
VGICs share a conserved architecture in which four subunits or homologous domains create a central ion-conducting pore surrounded by four voltage-sensors5. The voltage-sensing domain (VSD) is composed of the S1-S4 segments, while the pore module is formed by the S5 and S6 segments with a P-loop between them5. The S4 segments place charged amino acids within the membrane electric field that undergo outward displacement in response to depolarization and initiate opening of the central pore6,7. Although the architecture of KV channels has been established at high-resolution8,9, the structural basis for rapid, voltage-dependent activation of VGICs remains uncertain7,9, and the structures responsible for Na+-selective conductance and drug block in NaV channels are unknown. The primary pore-forming subunits of NaV and CaV proteins in vertebrates are composed of approximately 2,000 amino acid residues in four linked homologous domains5. The bacterial NaChBac channel family is an important model for structure-function studies of more complex vertebrate NaV and CaV channels10,11. NaChBac is a homotetramer, and its pharmacological profile is similar to NaV and CaV channels.10,12 Bacterial NaV channels are highly Na+-selective, but they can be converted into Ca2+-selective forms through simple mutagenesis13. The NaChBac family represents the likely ancestor of vertebrate NaV and CaV channels. Through analysis of the three-dimensional structure of NavAb from Arcobacter butzleri, we provide the first insights into the structural basis of voltage-dependent gating, ion selectivity, and drug block in NaV and CaV channels.
Structure of NavAb in a membrane environment
NavAb is a member of the NaChBac family and functions as a voltage-gated sodium-selective ion channel (Supplementary Fig. 1, 2). Vertebrate CaV channels require solubilization in digitonin and NaV channels require specific lipids to retain function when purified14,15. Accordingly, we solubilized NavAb in digitonin, crystallized it in a lipid-based bicelle system, and determined its structure at 2.70 Å resolution (Supplementary Fig. 3, 4, 5, 6; Table 1). NavAb crystallized as a dimer-of-dimers with 28 lipid molecules bound per tetramer (Supplementary Figure 3, 6b). Crystal-packing suggests a membrane-like environment (Supplementary Fig. 6a). NavAb VSDs interact noncovalently with the pore module of a neighboring subunit (Fig. 1a), and crystallographic temperature factors highlight their dynamic nature (Supplementary Fig. 6c).
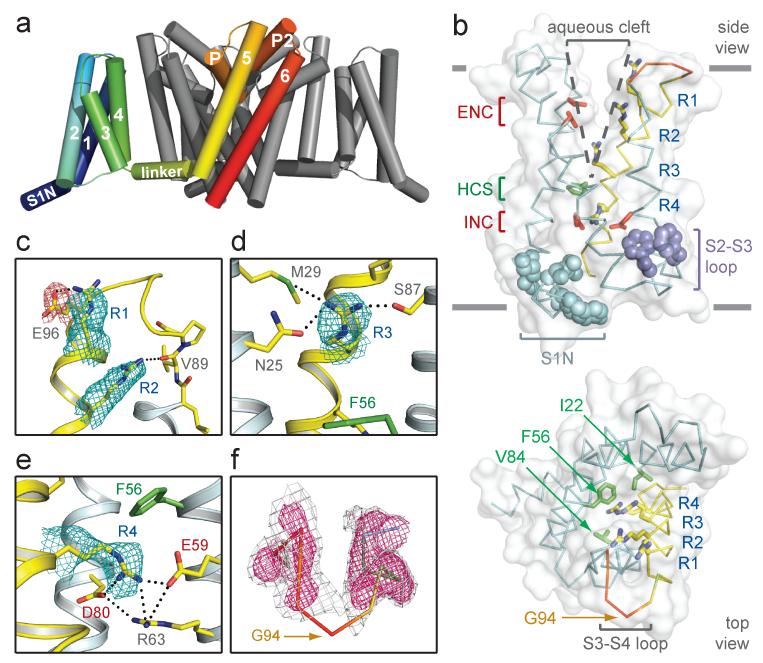
Figure 1. Structure of NavAb and the activated VSD.
a, Structural elements in NavAb. One subunit is highlighted (1-6, transmembrane segments S1-S6). The nearest VSD has been removed for clarity. b, Side and top-views of the VSD illustrating the extracellular negative charge-cluster (red, ENC), the intracellular negative charge-cluster (red, INC), hydrophobic constriction site (green, HCS), residues of the S1N helix (cyan) and phenylalanines of the S2-S3 loop (purple). S4 segment and gating charges (R1-R4) are in yellow. c-e, Hydrogen bonding of gating-charges, dotted lines (<3.5 Å). Fo-Fc omit maps are contoured over E96 and R1-R4 at 1, 1, 1.5, 2.5 and 1.75 σ, respectively. f, S3-S4 loop. Colored according to crystallographic temperature factors of the main-chain (blue < 50 Å2 to red > 150 Å2). An Fo-Fc omit map is contoured at 1.5 σ (grey) and 2.5 σ (pink).
Structure of the activated voltage-sensor
S4 segments in VSDs consist of repeated motifs of a positively charged residue, usually arginine, followed by two hydrophobic residues5-7. The R2 and R3 “gating charges” in NavAb are positioned to interact with a conserved extracellular negative-charge cluster (ENC), while the R4 gating charge interacts with a conserved intracellular negative-charge cluster (INC; Fig. 1b). These structural features, in conjunction with disulphide-locking experiments16,17, indicate that the VSDs are activated. These ion-pair interactions are expected to stabilize and catalyze S4 movement in the membrane electric field7,18,19. Highly conserved Arg63 in the S2 segment also interacts with R4 and the INC (Fig. 1e), which may stabilize the INC and modulate its electrostatics20. NavAb has a spectrum of additional gating charge interactions. R1 interacts with Glu96, R2 forms a hydrogen bond with the backbone carbonyl of Val89 in S3, and R3 forms hydrogen bonds with Asn25 and Met29 in S1, and Ser87 in S3 (Fig. 1c-e). This network of hydrogen bonds (Supplementary Fig. 7a) should complement exchange of ion-pair partners and provide a low energy pathway for S4 movement. The R2-backbone interaction would escape detection in mutagenesis experiments (Fig. 1c) and could have unrecognized significance in passage of gating charges through the gating pore (Fig. 1b).
The S4 segment in NavAb forms a 310-helix from R1 to R4. This conformation places all four gating charges in a straight line on one side of S4 (Fig. 1b), such that they could move linearly through the central portion of the gating pore, rather than in a spiral pattern7,17-19. The S3 segment is a straight α-helix, and the S3-S4 loop displays a dynamic connection to S4 (Fig. 1f). The lack of structural rigidity within the S3-S4 loop (Fig. 1f) suggests that it moves relatively freely in response to large S4 movements during activation.
Our structural analysis further reveals that the S1N helix and S2-S3 loop shield the intracellular surface of the VSD (Fig. 1b and Supplementary Fig. 8). The S2-S3 loop is conserved among VGICs, and two prominent Phe side-chains likely stabilize the VSD in the membrane during gating transitions (Fig. 1b, Supplementary Fig. 7, 8)9. The S1N-to-S3 region may behave as a modular unit during activation. In contrast to the sheltered intracellular surface of the VSD, a large aqueous cleft extends ~10 Å into the membrane above the hydrophobic constriction site (HCS, Fig. 1b). The HCS contains highly conserved residues (Ile22, Phe56 and Val84; Supplementary Fig. 7) that seal the VSD against ion leakage during S4 movement (Fig. 1b). The NavAb VSD therefore illustrates two important concepts from structure-function studies of NaV channels: a large external vestibule accessible to hydrophilic reagents and a focused membrane electric field over the intracellular half of the VSD6,7.
Despite their separation over one billion years of evolution, the VSDs of NavAb and KV1.2 display highly similar conformations (Supplementary Fig. 8a). R4 of NavAb is in an equivalent position to K5 in KV1.2 (Supplementary Fig. 8a), the most outward location of K5 during voltage-sensor activation20. This observation implies that the NavAb and KV1.2 VSDs are both activated.
The NavAb activation gate is closed
The pore of NavAb is closed, providing the first view of a closed pore in a VGIC (Fig. 2a and Supplementary Fig. 3). Met221 completely occludes the ion conduction pathway, as confirmed by unbiased experimentally-phased electron-density maps (Supplementary Fig. 4c). The S6 helices of NavAb superimpose well with other closed-pore structures and are distinct from the open-pore KV1.2 structure (Fig. 2a, b). A subtle iris-like dilation of the activation gate may be sufficient to open the pore, and the surrounding cuff of S4-S5 linkers may prevent larger pore opening (Fig. 2a-c).
Figure 2. NavAb pore module.
a, Pore-lining S6 helices of NavAb (yellow) and the closed MlotiK (3BEH), KcsA (1K4C) and NaK (2AHY) channels. Cα locations of Met221 defines a common radius for the closed activation gate (red circle). b, Comparison of S6 helices of NavAb and KV1.2/2.1 (2R9R). c, Interaction of S6 with S4-S5 linkers (NavAb, top; KV1.2/2.1, bottom). d, Architecture of the NavAb pore. Glu177 side-chains (purple sticks); pore volume, grey. e, Electrostatic potential colored from −10 to 10 kT (red to blue).
It is surprising to have a closed pore in a VGIC with activated voltage-sensors at 0 mV. Our NavAb structures were obtained by introducing a Cys at two locations near the intracellular end of S6 (Ile217Cys and Met221Cys). Evidently, these substitutions allowed us to trap the NavAb channel in the pre-open state previously invoked in kinetic models of VGIC gating (Supplementary Discussion)21-23.
Architecture of the pore and selectivity filter
VGICs are selective for specific cations yet conduct these ions at nearly the rate of free diffusion2. Our NavAb structure uncovers a basis for selectivity and high conductance of NaV channels. The NavAb pore module consists of an outer funnel-like vestibule, a selectivity filter, a central cavity, and an intracellular activation gate (Fig. 2d, Supplementary Fig. 4b). The large central cavity in NavAb accommodates a Na+ ion with one hydration shell and presents a hydrophobic surface over which ions should rapidly diffuse (Fig. 2e, Supplementary Figs. 1, 9). The pore (P)-helices are positioned to stabilize cations in the central cavity through helical-dipole interactions (Fig. 2d, Supplementary Fig. 4b), as for K+-channels24,25. Strikingly, a second pore-helix (P2-helix) forms an extracellular funnel in NavAb (Fig. 2d). This unique P2-helix is not seen in K+ channels and may represent a conserved structural element in the outer vestibule of NaV and CaV channels.
The ion conduction pathway in NavAb is strongly electronegative and the selectivity filter forms the narrowest constriction near the extracellular side of the membrane (Fig. 2d, e, 3; Supplementary Fig. 9). Classic permeation studies suggested a high field-strength anionic site with dimensions of ~3.1 × 5.1 Å for the selectivity filter in NaV channels26,27 and 5.5 × 5.5 Å in CaV channels28. Mutagenesis studies implicated Glu side-chains as key determinants of ion selectivity in these channels29-33. In NavAb, the four Glu177 side-chains form a ~6.5 × 6.5 Å rectangle with an orifice of ~4.6 × 4.6 Å defined by van der Waals surfaces (Fig. 3a, Supplementary Fig. 9d). Remarkably, Glu177 aligns with Glu residues that determine ion selectivity in NaV and CaV channels (Fig. 3e).
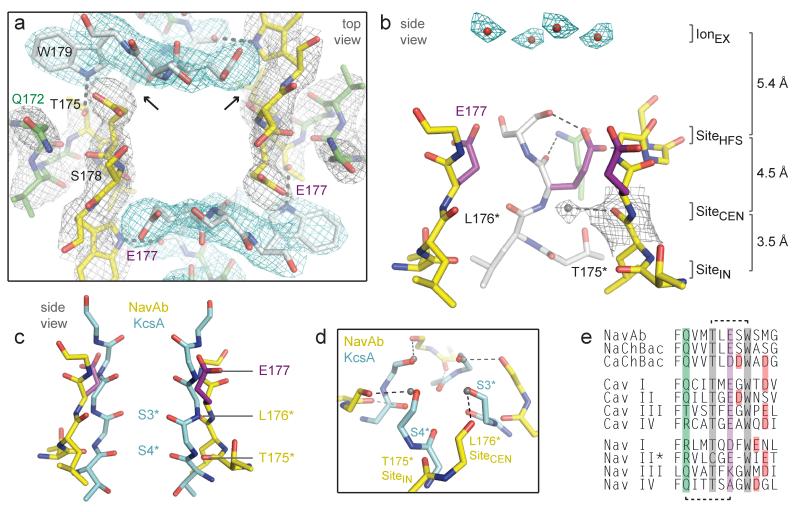
Figure 3. Structure of the NavAb selectivity filter.
a, Top view of the selectivity filter. Symmetry-related molecules are colored white and yellow; P-helix residues are colored green. Hydrogen bonds between Thr175 and Trp179 are indicated by grey dashes. Electron-densities from Fo-Fc omit maps are contoured at 4.0 σ (blue and grey) and subtle differences can be appreciated (small arrows). b, Side view of the selectivity filter. Glu177 (purple) interactions with Gln172, Ser178 and the backbone of Ser180 are shown in the far subunit. Fo-Fc omit map, 4.75 σ (blue); putative cations or water molecules (red spheres, IonEX). Electron-density around Leu176 (grey; Fo-Fc omit map at 1.75 σ) and a putative water molecule is shown (grey sphere). Na+-coordination sites: SiteHFS, SiteCEN and SiteIN. c, Superposition of NavAb and a K+-channel selectivity filter: NavAb Glu177 side-chains (purple) and backbone carbonyls (*); K+-channel (blue, 1K4C), site 3 and site 4 backbone carbonyls (ref. 35) (*). This structural alignment is based on P-helices. d, Enlarged view of SiteCEN and SiteIN: water molecules, grey spheres; dotted lines, ~2.5 Å. e, Selectivity filter sequence alignment. E177 homologs, purple; outer-ring of negatively charged residues (red).
The Glu177 side-chains of NavAb are supported by an elaborate architecture. The P-helix ends with the conserved Thr175, which accepts a hydrogen bond (3.0 Å) from the conserved Trp179 of a neighboring subunit (Fig. 3a). This landmark interaction staples together adjacent subunits at the selectivity filter. The residues between Thr175 and Trp179 form a tight turn and expose backbone carbonyls of Thr175 and Leu176 to conducted ions (Fig. 3b). The Glu177 side-chains form hydrogen bonds with the backbone amides of Ser180 (2.6 Å) and Met181 (3.1 Å) from the P2-helix (Fig. 3b, Supplementary Fig. 10). An extensive network of additional interactions (Supplementary Fig. 10), including hydrogen bonds between Gln172 from the P-helix and the carbonyl of Glu177 (Fig. 3a, b), further stabilizes the selectivity filter. Due to the dimer-of-dimers arrangement, the Glu177 and Ser178 side-chains of NavAb are in two slightly different environments (small arrows, Fig. 3a, Supplementary Fig. 11), consistent with functional nonequivalence of the corresponding glutamates in CaV channels31-33.
In agreement with the low affinity of NaV channels for permeant ions (Kd for Na+>350 mM34), no extra density was observed beside the Glu177 side-chains. Instead, strong electron densities were found above Glu177 at a distance of >4 Å. These densities likely represent cations or solvent molecules (IonEX, Fig. 3b) positioned above the selectivity filter by its intense electronegativity (Fig. 2e).
Ion permeation and selectivity
NavAb represents a prototype for understanding Na+ selectivity and permeation. Analysis of the pore radius indicates that a partially hydrated Na+ ion can be accommodated at the high field-strength site formed by the Glu177 side-chains (SiteHFS; Fig. 3a,b, Supplementary Fig. 9d). The much smaller K+-channel filter can fit inside the NavAb selectivity filter (Fig. 3c). Careful inspection of electron density suggests four well-bound water molecules 2.5 Å from the Leu176 carbonyls (SiteCEN, Fig. 3b). Remarkably, these four water molecules occupy the same positions as site 3 carbonyls from K+-channels (Fig. 3c, d)35. A distance of 2.5 Å is also found between the backbone carbonyls of Thr175 from NavAb and the site 4 carbonyls of K+-channels (Fig. 3c, d)35. Similar to other Na+ complexes (Supplementary Fig. 12)36-40, a Na+ ion surrounded by a square array of four water molecules could interact with backbone carbonyls of Leu176 (SiteCEN) or Thr175 (SiteIN) (Fig. 3d, Supplementary Fig. 12). Therefore, unlike K+-channels, the NavAb selectivity filter appears to select and conduct Na+ ions in a mostly hydrated form.
The NavAb structure fits closely with Hille’s single-ion pore model for NaV channels, in which a high-field-strength anion partially dehydrates the permeating ion2,34. According to Eisenman’s theory41, a Na+ ion would approach the SiteHFS more closely than the larger K+ ion, allowing more efficient removal of water and faster permeation (Fig. 3a, b)34. A Na+ ion could fit in-plane between the Glu177 side-chains, with one side-chain coordinating the Na+ ion directly and neighboring Glu177 side-chains acting as hydrogen bond acceptors for in-plane water molecule(s)26,27,34. With two waters remaining axial to the ion, this arrangement would approximate trigonal bipyramidal coordination38. Since only one Glu177 side-chain engages the permeating ion directly, this transient complex would be inherently asymmetric. When the permeating ion escapes SiteHFS, full rehydration occurs along the water-lined sites formed by the backbone carbonyls of Leu176 (SiteCEN) and Thr175 (SiteIN; Fig. 3b,d, Supplementary Fig. 12).
Free diffusion then allows the hydrated Na+ ion to enter the central cavity and move through the open activation gate into the cytoplasm34. The selectivity-filter structure of NavAb concentrates barriers to ion flow into ~5 Å (Fig. 3b, Supplementary Fig. 9d), which should promote high flux rates34. This permeation mechanism likely reflects the high free energy of Na+ hydration, where further removal of solvating waters would present too high an energy barrier. In sharp contrast, K+-selective channels conduct nearly fully-dehydrated K+ ions through direct interactions with backbone carbonyls in a long, narrow, multi-ion pore35,36. The selectivity filters of vertebrate NaV and CaV channels likely resemble NavAb, and amino acid substitutions within this framework must impart Na+ versus Ca2+selectivity (Supplementary Discussion)13,29-33.
Interaction sites of pore blockers
NavAb provides a foundation to interpret pharmacological mechanisms. From the extracellular side, the Glu177 side-chains of NavAb represent the blocking site of Nav channels by protons and guanidinium moieties of tetrodotoxin and saxitoxin2,42, as well as the site where divalent cations and protons bind and block CaV channels (Fig. 3)31-33. From the intracellular side, local anesthetics, antiarrhythmics, and antiepileptic drugs block NaV and CaV channels2,4 by entering through the open intracellular mouth of the pore and binding to an overlapping receptor site on the S6 segments43-45. Alignment of NavAb S6 segments with vertebrate NaV and CaV channels reveals a high degree of sequence similarity (Supplementary Fig. 7b), and drug molecules could easily fit into the large central cavity (Fig. 3, Supplementary Fig. 9). Use-dependent block is enhanced by repetitive opening of the pore to provide drug access2,46, and the local anesthetic etidocaine is an open-channel blocker of NaChBac12. The tight seal observed at the intracellular activation gate in NavAb illustrates why pore opening is required for access of large or hydrophilic drugs to the S6 receptor site (Fig. 2, Supplementary Fig. 4c).
Fenestrations provide hydrophobic access to the pore
Membrane lipids modulate the structure and function of VGICs8,9,47,48. However, NavAb presents a completely unexpected type of lipid interaction that has profound implications. The NavAb central cavity reveals four lateral openings leading from the membrane to the lumen of the closed pore (Fig. 4). These fenestrations measure ~8 × 10 Å, and could become larger depending upon nearby side-chain conformations (Phe203, Fig. 4). Lipids penetrate through these side portals and lie deep within the central cavity, occluding the ion conduction pathway in NavAb (Fig. 4, red). Because acyl-chain containing detergents were never used in preparation of NavAb crystals, these electron densities are assigned as acyl chains of membrane phospholipids. Similar fenestrations were not observed in the open-pore structure of KV1.2 (refs 8,9), raising the possibility that these lipid chains withdraw and the fenestrations close in the open state.
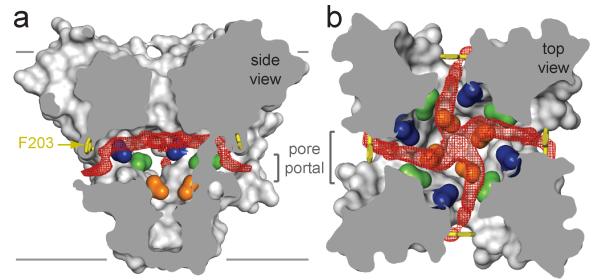
Figure 4. Membrane access to the central cavity in NavAb.
a, Side-view through the pore module illustrating fenestrations (portals) and hydrophobic access to central cavity. Phe203 side-chains, yellow sticks. Surface representations of NavAb residues aligning with those implicated in drug binding and block, Thr206, blue; Met209, green; Val213, orange. Membrane boundaries, grey lines. Electron-density from an Fo-Fc omit map is contoured at 2.0 σ. b, Top-view sectioned below the selectivity filter, colored as in a.
The lateral pore fenestrations in NavAb lead directly to the drug binding sites within the central cavity and abut residues that are important for drug binding in NaV and CaV channels (Fig. 4, Supplementary Fig. 7b)43,44. These NavAb portals appear compatible with passage of small neutral or hydrophobic drugs such as phenytoin49 and benzocaine46, which can gain access to their receptor site in closed channels2,46. We propose that pore fenestrations may be directly involved in voltage-dependent drug block according to the “modulated receptor model”46. Our findings highlight the potential for lipids and other hydrophobic molecules to influence the function of ion channels from the lipid phase of the membrane.
Structural basis for central pore gating
The domain-swapped arrangement of the VSD around the pore allows the S4-S5 linker to couple S4 movements to activation of VGICs (Fig. 1a)9. Kinetic models indicate that all four voltage-sensors activate and then the central pore opens in a concerted transition21-23. An essential element of this gating model is a state in which all four VSDs have activated but the pore remains closed21-23. It is likely that we have captured this pre-open state in our crystals (Supplementary Discussion). NavAb therefore provides a unique opportunity to consider the structural basis for coupling of VSD activation to pore-opening.
When activated VSDs of NavAb and KV1.2 are overlaid (Supplementary Fig. 8a), the S4-S5 linkers superimpose precisely, but the pore domains diverge at the foot of S5 (Fig. 5a). Superposition of the pore domains demonstrates an equivalent displacement of the VSDs (Supplementary Fig. 13). These comparisons lead to a working model for pore opening. First, during activation, the S4-S5 linker and VSD move together as a modular unit (Fig. 5a). Second, a single molecular hinge at the base of S5 mediates the closed-to-open pore transition (Fig. 5a, b). Third, tight structural-coupling is maintained between the S5 and S6 segments (Supplementary Fig. 13a). This model suggests that rotation of the VSD and S4-S5 linker as a structural unit pulls the S5-S6 helices outward to open the pore (Fig. 5b, Supplementary Fig. 13b). Because of their tight structural-coupling, displacement of the S5-S6 segments from one subunit forces the neighboring subunits to move similarly, leading to concerted pore opening. During this transition, the amphipathic S4-S5 linker pivots along the plane of the membrane interface (Fig. 5b, Supplementary Fig. 7, 13b). In contrast to KV1.2, the S6 helices in NavAb have not fully engaged their interaction site on the S4-S5 linker (Fig. 2c), in agreement with NavAb’s pre-open state. The rolling motion of the VSDs around the pore produces displacements up to ~10 Å at the intracellular side (Fig. 5b, Supplementary Fig. 13b), which may influence movements of the S1N helix and the conserved S2-S3 loop.
Figure 5. Model for activation gate opening.
a, Superposition of the NavAb and KV1.2/2.1 based on their VSDs (cylinders). b, Superposition of NavAb and KV1.2/2.1 tetrameric pore modules (PM) viewed from the membrane: S5 gating hinge, *. c-d, S1 interaction with P-helix. .S1 Thr is P-helix is 2.9 Å neighboring subunit in KV1.2/2.1 (), but >4.5 Å in NavAb.
In NavAb, a 310-helix extends from R1 to R4 (Fig. 1b). In KV1.2, a 310-helix encompasses R3 to K5 (equivalent to NavAb R2 to R4), but the remaining S4 segment is α-helical9. Conceivably, energy derived from voltage-driven translocation of S4 may be stored in the higher-energy 310-helix, and then released to help drive pore opening. The VSDs in KV1.2 are displaced outward (~2 Å) compared to the pre-open NavAb structure (Fig. 5b), which could account for the small gating current associated with concerted pore-opening6. At the extracellular side of the VSD, an S1 threonine residue hydrogen bonds (2.9 Å) with the P-helix of a neighboring subunit in KV1.2 (Fig. 5d), providing a conserved contact point that allows the VSD to perform mechanical work on the pore50. The equivalent S1 threonine in NavAb has not yet engaged the P-helix (Fig. 5c). This interaction may therefore represent an essential step in activation gating that has not yet occurred in the pre-open state of NavAb.
Conclusion
The structure of NavAb provides key insights into the molecular basis of voltage-sensing, ion conductance, and voltage-dependent gating in a historic class of ion channels1,2. A new network of interactions within the VSD appears well-positioned to catalyze gating charge movements during activation. Our model for electromechanical coupling reveals a rolling motion of the VSD and its connecting S4-S5 linker around the pore. The NavAb selectivity filter illustrates the basis for selective Na+ conductance through a water-lined pore featuring a high-field-strength anionic site. Finally, hydrophobic access from the membrane phase has been uncovered as a potentially important pathway for drug binding and modulation of VGICs.
Supplementary Material
SUMMARY.
The structure of a NaV channel reveals insights into gating, ionic selectivity and pharmacological action.
Methods Summary.
NavAb was expressed in insect cells and purified using anti-Flag resin and size exclusion chromatography, reconstituted into DMPC:CHAPSO bicelles, and crystallized over an ammonium sulphate solution containing 0.1 M Na-citrate, pH4.75. Cysteine mutants were complexed with mercury to obtain initial experimental phases. A SAD data set from a mercury-free SeMet-substituted protein crystal expedited model building. Standard crystallographic refinement procedures and structural analyses were performed. Electrophysiological experiments on NavAb were performed in tsA-201 cells using standard protocols.
Acknowledgements
We thank Dr. Bertil Hille for comments on a draft of the manuscript and members of the Zheng and Catterall groups for their support throughout this project. We are grateful to investigators who provided genomic DNA and the beamline staff at the Advanced Light Source (BL8.2.1 and BL8.2.2) for their assistance during data collection. J.P. acknowledges support from a Canadian Institutes of Health Research fellowship and the encouragement of Emily Payandeh. This work was supported by grants from the National Institutes of Health (R01 NS15751 and U01 NS058039 to W.A.C.) and by the Howard Hughes Medical Institute (N. Z.).
Appendix
Methods
Protein expression and purification
After exploring traditional expression approaches in Escherichia coli51, the NavAb channel from Arcobacter butzleri was cloned into the pFASTBac-Dual vector behind the polyhedron promoter using the BamHI and NotI restriction sites preceded by an N-terminal Flag-tag. Recombinant baculovirus were generated using the Bac-to-Bac system (Invitrogen) and insect cells were infected for large-scale protein production. Cells were harvested 72 h post-infection and resuspended in 50 mMTris pH 8.0, 200 mM NaCl (Buffer A) supplemented with protease inhibitors and DNase. Following sonication, digitonin (EMD Biosciences) was added to 1% and solubilization was carried out for 1-2 h at 4° C. Following centrifugation, clarified supernatant was gently agitated with anti-Flag M2-agarose resin (Sigma) pre-equilibrated with Buffer B (Buffer A supplemented with 0.12% digitonin) for 1-2 h at 4° C. Flag-resin was collected in a column by gravity flow, washed with ten column volumes of Buffer B, and eluted with two column volumes of Buffer B supplemented with 0.1 mg/mL Flag peptide. The eluate was passed over a Superdex 200 column (GE Healthcare) in 10 mM Tris pH 8, 100 mM NaCl and 0.12% digitonin and peak fractions containing NavAb were concentrated using a Vivaspin (30K MWKO) centrifugal device. Site-directed mutagenesis was performed using the standard QuikChange protocol (Stratagene) and all constructs were confirmed by DNA sequencing. Selenomethionine-labeled proteins were expressed using established protocols52, except cells were washed and starved for methionine at 8 h following infection, followed by SeMet (Anatrace) supplementation at 12 h after infection. SeMet-labeled proteins were purified as described above.
Heavy atom screening and labeling
During our efforts to identify useful derivatives for crystallographic phasing, we ultimately turned to the fluorescence detection of heavy atom labeling (FD-HAL) method53. Over thirty NavAb single-site cysteine mutations were rapidly screened using the FD-HAL method, and many of these mutant proteins were subsequently crystallized, presumably as covalent mercury-channel complexes. The NavAb-Ile217Cys and NavAb-Met221Cys mutants that yielded useful SAD-data sets were prepared as follows: proteins were purified as described above and concentrated to ~1 mg/mL; HgCl2 was added to a final concentration of 10 mM and the mixture was incubated at room temperature for 2 h. The protein buffer was subsequently exchanged (into mercury-free buffer) through five rounds of concentration and dilution using Vivaspin (30K MWKO) centrifugal devices. Following structure determination, it became apparent that Met221 lines the narrowest portion of the closed NavAb pore (e.g. Supplementary Fig. 4c).
NavAb crystallization and data collection
Prior to crystallization, NavAb was concentrated to ~20 mg/mL and reconstituted into DMPC:CHAPSO (Anatrace) bicelles according to standard protocols54,55. The NavAb-bicelle preparation was mixed in a 1:1 ratio and setup in a hanging-drop vapor-diffusion format over a well solution containing 1.8-2.1 M ammonium sulfate, 100 mM Na-citrate pH 4.75. The mercury-free proteins, the mercury-complexes, and the SeMet-labeled proteins all crystallized under essentially identical conditions. Crystals were typically passed through solutions containing 2 M ammonium sulfate, 100 mM Na-citrate pH 4.75 and 28% glucose (wt/v) in increments of ~6% glucose during harvesting. Crystals were plunged into liquid nitrogen and maintained at 100 K during all data collection procedures.
Over 1,000 crystals were screened and nearly 100 diffraction data sets were collected at a synchrotron radiation source (Advanced Light Source, BL8.2.1 and BL8.2.2). A single anomalous dispersion (SAD) data set collected near the mercury absorption edge (λ = 1.005 Å) from a mercury-containing complex of the NavAb-Ile217Cys mutant was ultimately used to determine initial experimental phases. Our highest resolution SeMet-SAD data set was collected near the selenium absorption edge (λ = 0.9795 Å) from a mercury-free NavAb-Met221Cys SeMet-labled crystal. Subsequent native (i.e. mercury-free) data sets were collected at standard wavelengths. Because the NavAb crystals were small (typically < 0.15 mm × 0.15 mm × 0.15 mm), contained a high solvent content (~80%), were weakly diffracting, and radiation sensitive, special care was taken to minimize exposure times and to orient the crystals in order to maximize data completeness and quality.
Structure determination and refinement
X-ray diffraction data were integrated and scaled with the HKL2000 suite or DENZO/SCALEPACK56 and, when required, further processed with the CCP4 package57. Experimental phases were determined using a 3.4 Å SAD-data set from a Hg-containing NavAb-Ile217Cys crystal. The SOLVE/RESOLVE58 software were run in a standard setting and the first map, calculated at 3.7 Å, is shown in Supplementary Fig. 3. Ideal poly-alanine α-helices were manually fitted into this map and the model was subsequently used in combined SAD-molecular replacement (MR) protocols within the Phenix software59 using a 3.3 Å SAD-data set obtained from a SeMet-labeled NavAb-Met221Cys crystal. SAD-MR and MR-SAD-based maps were calculated and compared, allowing for complete register and amino-acid assignment of the NavAb model. Higher resolution native data sets were ultimately obtained and phased by MR-methods using the CNS suite60 (although our best native NavAb-Met221Cys data set is actually from a SeMet-containing crystal). Reiterative rounds of model building in O 61 were guided by inspection of omit maps and refinement with CNS60 was performed with strict NCS-restraints, which were later relaxed during final rounds of refinement. Two strong densities (one per protein chain) assigned as solvent molecules (near the pore turret loop; not discussed in the main text) and all lipid molecules were added to the models at very late stages of refinement. Although trace amounts of digitonin are present in the crystallization condition, digitonin molecules were not readily observed in any electron density map. Refinement statistics, scaling statistics, and overall map quality were ultimately used to assign the NavAb space group as I222, although the data were found to closely mimic I422 (Rwork/Rfree stall at ~32% in I422).
Structure analysis
The geometry of NavAb structural models was assessed using PROCHECK62. The pore radius of NavAb was calculated using standard settings in the MOLE software63. Electrostatic surface calculations were performed with the APBS software64, calculated with 150 mM NaCl in the solvent. Structural alignments were performed using LSQMAN65 and O61, where all channels were independently aligned onto NavAb based on the amino acid positions at the very beginning (i.e. N-terminal portion) of their P-helices. The superposition of the atomic resolution Na+-complex structure40 shown in Supplementary Fig. 12 was positioned manually, but the K+-channel and NaK-channel superpositions (Fig. 2, Fig. 3, Fig. 5b, Supplementary Fig. 12, 13b) were obtained by simply aligning P-helices, as described above. All Fo-Fc omit maps shown throughout the main text and supplement have been calculated using standard settings and appropriate buffers in the CNS program60. The Fo-Fc omit map shown in Fig. 3b specifically derives from the 2.7 Å NaAb-Ile217Cys data set and amino acids 170-183 were omitted from the calculation box. All structural figures were prepared with the PyMol software66.
Electrophysiology
NavAb was cloned into the CDM8 vector and transfected into tsA-201 cells (along with a CD8 marker construct) using standard protocols. Whole-cell currents were recorded with continuous perfusion of extracellular solution using an Axopatch 200 amplifier (Molecular Devices, Foster City, CA) with glass pipettes polished to 2-4 Ω resistance. The intracellular pipette solution contained (in mM): 10 NaCl, 105 CsF, 20 TEA, 10 EGTA, 10 HEPES pH 7.4 (adjusted with CsOH). The extracellular Na+ solution contained (in mM): 100 NaCl, 1 CaCl2, 1 MgCl2, 1 KCl, 50 TEA, 10 HEPES pH 7.4 (CsOH). For K-containing and Cs-containing extracellular solutions, NaCl was replaced with KCl or CsCl, respectively. The extracellular NMDG solution contained (in mM): 100 NMDG, 1 CaCl2, 1 MgCl2, 1 KCl, 50 TEA, 10 HEPES pH 7.4 (HCl) and the extracellular Ca2+ solution contained (in mM): 75 CaCl2, 1 MgCl2, 1 KCl, 50 TEA, 10 HEPES pH 7.4 (CsOH).Voltage clamp pulses were generated and currents were recorded using Pulse software controlling an Instrutech ITC18 interface (HEKA, Great Neck, NY). Data were analyzed using Igor Pro 6.2 (WaveMetrics, Lake Oswego, OR).
- 51.Koth CM, Payandeh J. Strategies for the cloning and expression of membrane proteins. Adv Protein Chem Struct Biol. 2009;76:43–86. doi: 10.1016/S1876-1623(08)76002-4. [DOI] [PubMed] [Google Scholar]
- 52.Cronin CN, Lim KB, Rogers J. Production of selenomethionyl-derivatized proteins in baculovirus-infected insect cells. Protein Sci. 2007;16:2023–2029. doi: 10.1110/ps.072931407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaptal V, et al. Fluorescence Detection of Heavy Atom Labeling (FD-HAL): A rapid method for identifying covalently modified cysteine residues by phasing atoms. J Struct Biol. 2010;171:82–87. doi: 10.1016/j.jsb.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 54.Faham S, Bowie JU. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J Mol Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 55.Faham S, et al. Crystallization of bacteriorhodopsin from bicelle formulations at room temperature. Protein Sci. 2005;14:836–840. doi: 10.1110/ps.041167605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Vol. 276. Academic Press; 1997. [DOI] [PubMed] [Google Scholar]
- 57.CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 58.Terwilliger T. SOLVE and RESOLVE: Automated Structure Solution and Density Modification. Vol. 374. Elsevier; 2003. pp. 22–37. [DOI] [PubMed] [Google Scholar]
- 59.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 61.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 62.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 63.Petrek M, Kosinova P, Koca J, Otyepka M. MOLE: a Voronoi diagram-based explorer of molecular channels, pores, and tunnels. Structure. 2007;15:1357–1363. doi: 10.1016/j.str.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 64.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kleywegt GJ. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr D Biol Crystallogr. 1996;52:842–857. doi: 10.1107/S0907444995016477. [DOI] [PubMed] [Google Scholar]
- 66.PyMOL . Version 1.2r3pre. [Google Scholar]
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Author contributions
J.P., N.Z. and W.A.C. conceived and J.P. conducted the protein purification and crystallization experiments. J.P. and N.Z. determined and analyzed the structures of NavAb. J.P. and T.S. performed functional studies of NavAb. J.P., N.Z. and W.A.C. wrote the manuscript.
Author information
Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 3RVY, 3RVZ and 3RWO. Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hille B. Ion channels of excitable membranes. 3rd edn Sinauer Associates, Inc.; 2001. [Google Scholar]
- 3.Ryan DP, Ptacek LJ. Episodic neurological channelopathies. Neuron. 2010;68:282–292. doi: 10.1016/j.neuron.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Catterall WA. Common modes of drug action on Na+ channels: local anesthetics, antiarrhythmics and anticonvulsants. Trends Pharmacol Sci. 1987;8:57–65. [Google Scholar]
- 5.Yu FH, Catterall WA. The VGL-chanome: a protein superfamily specialized for electrical signaling and ionic homeostasis. Sci STKE. 2004;2004:re15. doi: 10.1126/stke.2532004re15. [DOI] [PubMed] [Google Scholar]
- 6.Bezanilla F. The action potential: from voltage-gated conductances to molecular structures. Biol Res. 2006;39:425–435. doi: 10.4067/s0716-97602006000300005. [DOI] [PubMed] [Google Scholar]
- 7.Catterall WA. Ion channel voltage sensors: structure, function, and pathophysiology. Neuron. 2010;67:915–928. doi: 10.1016/j.neuron.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 9.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 10.Ren D, et al. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- 11.Koishi R, et al. A superfamily of voltage-gated sodium channels in bacteria. J Biol Chem. 2004;279:9532–9538. doi: 10.1074/jbc.M313100200. [DOI] [PubMed] [Google Scholar]
- 12.Zhao Y, Scheuer T, Catterall WA. Reversed voltage-dependent gating of a bacterial sodium channel with proline substitutions in the S6 transmembrane segment. Proc Natl Acad Sci U S A. 2004;101:17873–17878. doi: 10.1073/pnas.0408270101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yue L, Navarro B, Ren D, Ramos A, Clapham DE. The cation selectivity filter of the bacterial sodium channel, NaChBac. J Gen Physiol. 2002;120:845–853. doi: 10.1085/jgp.20028699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curtis BM, Catterall WA. Reconstitution of the voltage-sensitive calcium channel purified from skeletal muscle transverse tubules. Biochemistry. 1986;25:3077–3083. doi: 10.1021/bi00359a002. [DOI] [PubMed] [Google Scholar]
- 15.Feller DJ, Talvenheimo JA, Catterall WA. The sodium channel from rat brain. Reconstitution of voltage-dependent scorpion toxin binding in vesicles of defined lipid composition. J Biol Chem. 1985;260:11542–11547. [PubMed] [Google Scholar]
- 16.DeCaen PG, Yarov-Yarovoy V, Zhao Y, Scheuer T, Catterall WA. Disulfide locking a sodium channel voltage sensor reveals ion pair formation during activation. Proc Natl Acad Sci U S A. 2008;105:15142–15147. doi: 10.1073/pnas.0806486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeCaen PG, Yarov-Yarovoy V, Sharp EM, Scheuer T, Catterall WA. Sequential formation of ion pairs during activation of a sodium channel voltage sensor. Proc Natl Acad Sci U S A. 2009;106:22498–22503. doi: 10.1073/pnas.0912307106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catterall WA. Molecular properties of voltage-sensitive sodium channels. Annu Rev Biochem. 1986;55:953–985. doi: 10.1146/annurev.bi.55.070186.004513. [DOI] [PubMed] [Google Scholar]
- 19.Yarov-Yarovoy V, Baker D, Catterall WA. Voltage sensor conformations in the open and closed states in ROSETTA structural models of K+ channels. Proc Natl Acad Sci U S A. 2006;103:7292–7297. doi: 10.1073/pnas.0602350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tao X, Lee A, Limapichat W, Dougherty DA, MacKinnon R. A gating charge transfer center in voltage sensors. Science. 2010;328:67–73. doi: 10.1126/science.1185954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zagotta WN, Hoshi T, Aldrich RW. Shaker potassium channel gating. III: Evaluation of kinetic models for activation. J Gen Physiol. 1994;103:321–362. doi: 10.1085/jgp.103.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuzmenkin A, Bezanilla F, Correa AM. Gating of the bacterial sodium channel, NaChBac: voltage-dependent charge movement and gating currents. J Gen Physiol. 2004;124:349–356. doi: 10.1085/jgp.200409139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao Y, Yarov-Yarovoy V, Scheuer T, Catterall WA. A gating hinge in Na+ channels; a molecular switch for electrical signaling. Neuron. 2004;41:859–865. doi: 10.1016/s0896-6273(04)00116-3. [DOI] [PubMed] [Google Scholar]
- 24.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 25.Jogini V, Roux B. Electrostatics of the intracellular vestibule of K+ channels. J Mol Biol. 2005;354:272–288. doi: 10.1016/j.jmb.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 26.Hille B. The permeability of the sodium channel to organic cations in myelinated nerve. J Gen Physiol. 1971;58:599–619. doi: 10.1085/jgp.58.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hille B. The permeability of the sodium channel to metal cations in myelinated nerve. J Gen Physiol. 1972;59:637–658. doi: 10.1085/jgp.59.6.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCleskey EW, Almers W. The Ca channel in skeletal muscle is a large pore. Proc Natl Acad Sci U S A. 1985;82:7149–7153. doi: 10.1073/pnas.82.20.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heinemann SH, Terlau H, Stuhmer W, Imoto K, Numa S. Calcium channel characteristics conferred on the sodium channel by single mutations. Nature. 1992;356:441–443. doi: 10.1038/356441a0. [DOI] [PubMed] [Google Scholar]
- 30.Favre I, Moczydlowski E, Schild L. On the structural basis for ionic selectivity among Na+, K+, and Ca2+ in the voltage-gated sodium channel. Biophys J. 1996;71:3110–3125. doi: 10.1016/S0006-3495(96)79505-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Ellinor PT, Sather WA, Zhang JF, Tsien RW. Molecular determinants of Ca2+ selectivity and ion permeation in L-type Ca2+ channels. Nature. 1993;366:158–161. doi: 10.1038/366158a0. [DOI] [PubMed] [Google Scholar]
- 32.Ellinor PT, Yang J, Sather WA, Zhang JF, Tsien RW. Ca2+ channel selectivity at a single locus for high-affinity Ca2+ interactions. Neuron. 1995;15:1121–1132. doi: 10.1016/0896-6273(95)90100-0. [DOI] [PubMed] [Google Scholar]
- 33.Chen XH, Bezprozvanny I, Tsien RW. Molecular basis of proton block of L-type Ca2+ channels. J Gen Physiol. 1996;108:363–374. doi: 10.1085/jgp.108.5.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hille B. Ionic selectivity, saturation, and block in sodium channels. A four-barrier model. J Gen Physiol. 1975;66:535–560. doi: 10.1085/jgp.66.5.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morais-Cabral JH, Zhou Y, MacKinnon R. Energetic optimization of ion conduction rate by the K+ selectivity filter. Nature. 2001;414:37–42. doi: 10.1038/35102000. [DOI] [PubMed] [Google Scholar]
- 36.Ye S, Li Y, Jiang Y. Novel insights into K+ selectivity from high-resolution structures of an open K+ channel pore. Nat Struct Mol Biol. 2010;17:1019–1023. doi: 10.1038/nsmb.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alam A, Jiang Y. Structural analysis of ion selectivity in the NaK channel. Nat Struct Mol Biol. 2009;16:35–41. doi: 10.1038/nsmb.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doi M, et al. Caged and clustered structures of endothelin inhibitor BQ123, cyclo(-D-Trp-D-Asp--Pro-D-Val-Leu-)-Na+, forming five and six coordination bonds between sodium ions and peptides. Acta Crystallogr D Biol Crystallogr. 2001;57:628–634. doi: 10.1107/s0907444901002797. [DOI] [PubMed] [Google Scholar]
- 39.Harding MM. Metal-ligand geometry relevant to proteins and in proteins: sodium and potassium. Acta Crystallogr D Biol Crystallogr. 2002;58:872–874. doi: 10.1107/s0907444902003712. [DOI] [PubMed] [Google Scholar]
- 40.Phillips K, Dauter Z, Murchie AI, Lilley DM, Luisi B. The crystal structure of a parallel-stranded guanine tetraplex at 0.95 Å resolution. J Mol Biol. 1997;273:171–182. doi: 10.1006/jmbi.1997.1292. [DOI] [PubMed] [Google Scholar]
- 41.Eisenman G, Horn R. Ionic selectivity revisited: the role of kinetic and equilibrium processes in ion permeation through channels. J Membr Biol. 1983;76:197–225. doi: 10.1007/BF01870364. [DOI] [PubMed] [Google Scholar]
- 42.Noda M, Suzuki H, Numa S, Stuhmer W. A single point mutation confers tetrodotoxin and saxitoxin insensitivity on the sodium channel II. FEBS Lett. 1989;259:213–216. doi: 10.1016/0014-5793(89)81531-5. [DOI] [PubMed] [Google Scholar]
- 43.Hockerman GH, Peterson BZ, Johnson BD, Catterall WA. Molecular determinants of drug binding and action on L-type calcium channels. Annu Rev Pharmacol Toxicol. 1997;37:361–396. doi: 10.1146/annurev.pharmtox.37.1.361. [DOI] [PubMed] [Google Scholar]
- 44.Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. Molecular determinants of state-dependent block of Na+ channels by local anesthetics. Science. 1994;265:1724–1728. doi: 10.1126/science.8085162. [DOI] [PubMed] [Google Scholar]
- 45.Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. Common molecular determinants of local anesthetic, antiarrhythmic, and anticonvulsant block of voltage-gated Na+ channels. Proc Natl Acad Sci U S A. 1996;93:9270–9275. doi: 10.1073/pnas.93.17.9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hille B. Local anesthetics: hydrophilic and hydrophobic pathways for the drug-receptor reaction. J Gen Physiol. 1977;69:497–515. doi: 10.1085/jgp.69.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oliver D, et al. Functional conversion between A-type and delayed rectifier K+ channels by membrane lipids. Science. 2004;304:265–270. doi: 10.1126/science.1094113. [DOI] [PubMed] [Google Scholar]
- 48.Delmas P, Coste B, Gamper N, Shapiro MS. Phosphoinositide lipid second messengers: new paradigms for calcium channel modulation. Neuron. 2005;47:179–182. doi: 10.1016/j.neuron.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 49.Morello RS, Begenisich T, Yeh JZ. Determination of the active form of phenytoin. J Pharmacol Exp Ther. 1984;230:156–161. [PubMed] [Google Scholar]
- 50.Lee SY, Banerjee A, MacKinnon R. Two separate interfaces between the voltage sensor and pore are required for the function of voltage-dependent K+ channels. PLoS Biol. 2009;7:e47. doi: 10.1371/journal.pbio.1000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.