Abstract
To identify novel biomarkers for HIV-1 resistance, including pathways that may be critical in anti-HIV-1 vaccine design, we carried out a gene expression analysis on blood samples obtained from HIV-1 highly exposed seronegatives (HESN) from a commercial sex worker cohort in Nairobi and compared their profiles to HIV-1 negative controls. Whole blood samples were collected from 43 HIV-1 resistant sex workers and a similar number of controls. Total RNA was extracted and hybridized to the Affymetrix HUG 133 Plus 2.0 micro arrays (Affymetrix, Santa Clara CA). Output data was analysed through ArrayAssist software (Agilent, San Jose CA). More than 2,274 probe sets were differentially expressed in the HESN as compared to the control group (fold change ≥1.3; p value ≤0.0001, FDR <0.05). Unsupervised hierarchical clustering of the differentially expressed genes readily distinguished HESNs from controls. Pathway analysis through the KEGG signaling database revealed a majority of the impacted pathways (13 of 15, 87%) had genes that were significantly down regulated. The most down expressed pathways were glycolysis/gluconeogenesis, pentose phosphate, phosphatidyl inositol, natural killer cell cytotoxicity and T-cell receptor signaling. Ribosomal protein synthesis and tight junction genes were up regulated. We infer that the hallmark of HIV-1 resistance is down regulation of genes in key signaling pathways that HIV-1 depends on for infection.
Introduction
The disease AIDS perhaps ranks as one of the most devastating scourges of mankind. Since it was identified in 1983 more than 30 million people have died and another 33 million currently live with the virus [1], [2]. The majority of HIV-1 infections occur in Sub-Saharan Africa, where in some countries prevalence rates of more than 40% have been documented among antenatal clinic attendees [3]. Despite advances made in antiretroviral therapy, only a third of those requiring treatment receive it and new infections far outstrip those on therapy [4]. Similar to other previous viral epidemics, prevention through vaccination shall be the best approach. However, the nature of the AIDS virus to integrate itself to the host genome and its ability to constantly mutate and shield immunogenic components that induce protective antibodies, continue to pose challenges for an effective vaccine. Though a recent prime boost vaccine strategy, RV 144, provided signals that a HIV-1 vaccine was possible, its efficacy was only a modest 30% [5].
The presence of individuals who are highly exposed to HIV-1 but do not get infected, provide hope for a better understanding of correlates for protection that may lead to a more effective vaccine strategy. Highly exposed seronegative (HESN) populations have been identified among intravenous drug users [6], children born to seropositive mothers [7], [8], discordant couples [9], [10], and commercial sex workers [11]–[13]. The findings in 1996 on the protective effect of the CCR5Δ32 allele in HIV-1 infection [14] shifted focus to host genetics as probable cause of HIV-1 resistance. Over the past 10 years, approximately 40 genes have been documented from HESN populations as possible candidates for differential host susceptibility to HIV-1 [15]. This include selected human leukocyte antigens, HLA [16], TRIM5α [17], specific KIR-HLA associations [18], and APOBEC3G [19]. However, only a fraction of HESN has the protective genotypes described in the literature. Previous gene discovery studies have focused on single gene approaches which may have missed other compelling gene candidates and overlook interactions between different genes or genetic pathways. Microarray expression profiling provides new opportunities for simultaneous analysis of thousands of messenger RNA expression patterns in tandem with their biological functions. Less data exist on the use of microarrays to identify biomarkers for HIV-1 susceptibility, including pathways that may be critical for successful anti-HIV-1 vaccine and therapeutic design. In a bid to determine biomarkers and genetic pathways that are unique to HESNs, we carried out a global whole blood gene expression profiling of HIV-1 highly exposed and uninfected individuals from a well-characterized female commercial sex worker cohort from Nairobi, Kenya.
Results
Gene expression outcome
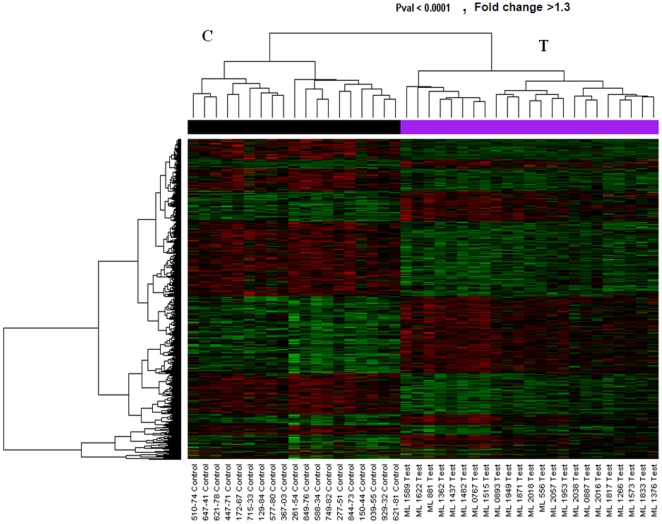
Our primary objective was to investigate if there is a predictive gene expression pattern that defines the HESN phenotype. Gene expression data from the 86 micro arrays were normalized and all were included in the analysis. An unsupervised hierarchical clustering algorithm was run on the data to ascertain if the two populations could be separated into two distinct groups based on their gene expression profiles. Figure 1 show a distinct separation between the HESN (Test) and HIV negative controls (Controls).
Figure 1. Heatmap of representative genes from a sample of HIV-1 highly exposed uninfected sex workers (Test) and susceptible HIV negative controls (control) evaluated through unsupervised hierarchical clustering algorithm.
Each row on the Y axis represents a single gene probe and the phylograms represent distinct signaling pathways. The red color denotes up regulated genes while the green are down regulated.
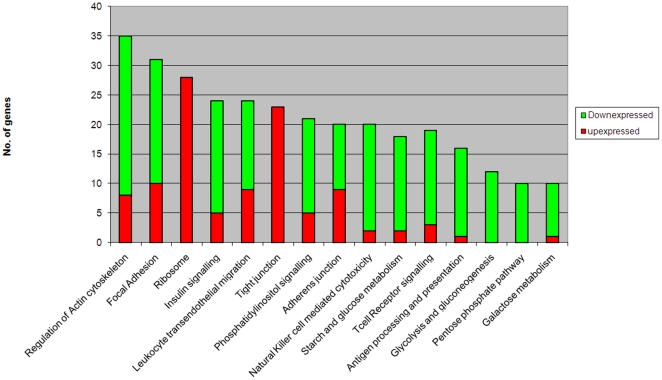
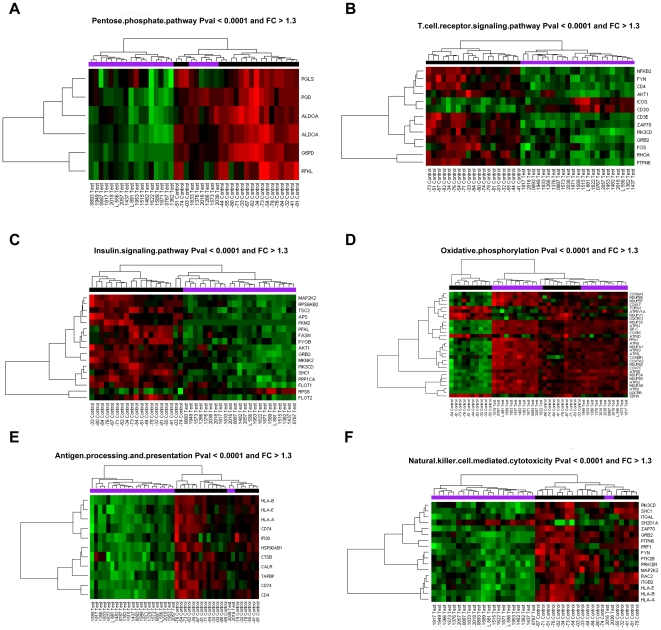
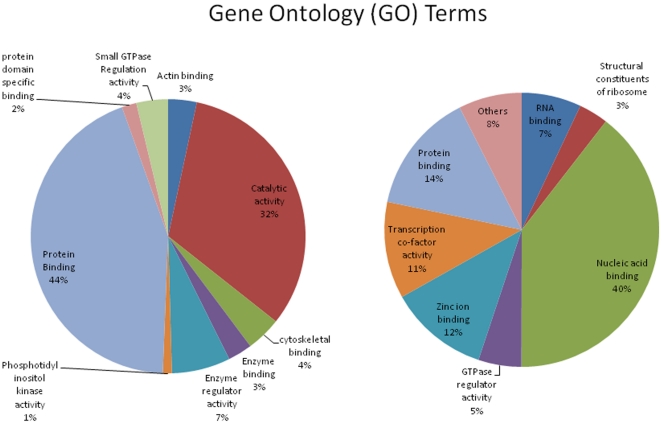
Of the 54675 gene probe sets represented on the Affymetrix U133 plus 2.0 gene chip, 2,274 probe sets were differentially expressed in the HESN group as compared with the control group (fold change ≥1.3; p value ≤0.05) after correction with multiple testing using the Benjamin-Hochberg false discovery rate (FDR<0.05). Of the total differentially expressed transcripts, 462 (20%) could be mapped onto the KEGG signaling database. Figure 2 shows the 15 signaling events with the highest number of genes identified through the KEGG analysis. Majority of these pathways (13 of 15, 87%) were significantly down-regulated in the HESN group. The top ranked down-regulated pathways belonged to energy metabolism, cell adhesion, signal transduction and immune signaling categories. Glucose/gluconeogenesis and pentose phosphate phosphorylation (p<0.0003) were the most down-regulated metabolic pathways, together with the immune signaling events of natural killer cell cytotoxicity (p<0.008), antigen processing and presentation (p<0.0013) and T-cell receptor signaling. The key up-regulated pathways were the ribosome protein synthesis and tight junction. Figures 3 (a to g) show the heatmaps of representative genes in the significantly impacted pathways. Gene ontology (GO) classification of the differentially expressed transcripts identified protein binding as the major functional group among the down-regulated genes (Fig. 4a). In contrast, up-regulated genes were significantly associated with nuclear binding (Fig. 4b).
Figure 2. Proportion of differentially expressed genes in significantly affected pathways.
Figure 3. Heatmaps of the most differentially regulated signaling pathways among the HIV -1 highly exposed (Test) and HIV negatives (Control).
Figure 3a -Pentose phosphate pathway; b) T cell receptor signaling; c) Insulin signaling, d) Oxidative phosphorylation, f) antigen processing and presentation, g) Natural Killer cell cytotoxicity. FC -fold change. Each row on the Y axis represents a single gene probe and the phylograms represent distinct signaling pathways. The red color denotes up regulated genes while the green are down regulated.
Figure 4. Gene ontology (GO) for downregulated genes (4a) and upregulated genes among the HIV-1 exposed uninfected sex workers.
Correlation with previous findings
More than 40% of genes that had earlier been reported to be associated with HIV-1 susceptibility in HESN populations were found to be differentially expressed in this dataset (Table 1). More significantly, genes that had been observed to be associated with the resistance phenotype in this cohort through single gene studies were confirmed to be differentially regulated. This included the Human leukocyte antigen (HLA) cluster (HLA-A, down-regulated, p value <0.001), Killer Immunoglobulin Receptors (KIR3DL1, down-regulated, p value <0.001) and Serpin B family (SerpinB13 up-regulated p<0.02). As noted in a previous study [20], expression of the CCR5 receptor cluster was not statistically different between the two populations (p value 0.6).
Table 1. Expression patterns of genes previously reported to be associated with HIV-1 Resistance.
| Gene | Regulation | Fold Change | p value | Reference |
| APOBEC3G | up | 1.1 | 0.7 | [19] |
| CCL3L1 | up | 1.2 | 0.4 | [65] |
| CCL4 | up | 1.1 | 0.6 | [19] |
| CCL5 | down | −1.1 | 0.6 | [19] |
| CCR5 | down | −1.1 | 0.6 | [14] |
| CD207(Langerin) | up | 1.2 | 0.4 | [66] |
| CD209(DC-SIGN) | down | −1.2 | 0.02 | [67] |
| CUL5 | up | 1.3 | 0.2 | [68] |
| CXCL12(SDF-1) | down | −1.2 | 0.6 | [69] |
| DEFA1 | up | 1.3 | 0.4 | [71] |
| DEFA3 | up | 1.3 | 0.4 | [71] |
| DEFB1 | up | 1.4 | 0.001 | [15] |
| DEFB4 | up | 1.2 | 0.5 | [15] |
| HLA-A | down | −1.1 | 0.01 | [16] |
| HLA-B | down | −1.1 | 0.04 | [16] |
| HLA-C | down | −1.1 | 0.03 | [16] |
| IL4 | down | −1.1 | 0.9 | [70] |
| KIR2DL1 | down | −1.9 | 0.02 | [18] |
| KIR2DL2 | down | −1.7 | 0.01 | [18] |
| KIR2DL3 | down | −1.8 | 0.07 | [18] |
| KIR2DL4 | down | −1.6 | 0.02 | [18] |
| KIR2DL5 | down | −1.6 | 0.02 | [18] |
| KIR2DS1 | down | −1.5 | 0.2 | [18] |
| KIR2DS2 | down | −1.8 | 0.1 | [18] |
| KIR2DS3 | down | −2.2 | 0.04 | [18] |
| KIR2DS4 | down | −1.6 | 0.1 | [18] |
| KIR2DS5 | down | −2.03 | 0.02 | [18] |
| KIR3DL1 | down | −1.9 | 0.01 | [72] |
| KIR3DL2 | down | −1.8 | 0.02 | [72] |
| KIR3DL3 | down | −1.8 | 0.08 | [72] |
| PPIA(CyclophilinA) | up | 1.5 | 0.002 | [24] |
| TRIM5alpha | down | −1.6 | 0.1 | [15], [17] |
| TSG101 | down | −1.1 | 0.3 | [15] |
| TLR2 | down | −1.04 | 0.7 | [73] |
| TLR4 | down | −1.2 | 0.07 | [73] |
| TLR8 | down | −1.4 | 0.001 | [73] |
Table 2 shows the list of the top 50 genes identified as differentially expressed with a fold change of 2 and above. This is the first time many of these genes have been shown to be associated with a HESN population.
Table 2. Top 50 novel genes identified by microarray analysis with a fold change expression of 2 and above between the HESN and control group.
| Affymetrix ID | Entrez | Gene Symbol | Chromosome | Regulation | pvalue |
| 237953_at | 1803 | Dpp4 | Chr2 | up | 1.99E-08 |
| 1569312_at | 7705 | ZNF146 | Chr19 | up | 1.17E-07 |
| 237051_at | 10463 | SLC30A9 | Chr4 | up | 1.27E-07 |
| 1559848_at | 387338 | NSUN4 | Chr1 | up | 2.23E-05 |
| 241798_at | 23244 | SCC-112 | Chr4 | up | 2.23E-05 |
| 244515_at | 5713 | PSMD7 | Chr16 | up | 2.37E-05 |
| 243683_at | 9643 | MORF4L2 | ChrX | up | 2.91E-05 |
| 1557575_at | 8987 | GENX-3414 | Chr4 | up | 3.09E-05 |
| 232279_at | 23338 | PHF15 | Chr5 | up | 4.94E-05 |
| 233127_at | 55422 | ZNF331 | Chr19 | up | 4.93E-05 |
| 225354_s_at | 83699 | SH3BGRL2 | Chr6 | down | 5.32E-05 |
| 223683_at | 84225 | ZMYND15 | Chr17 | down | 6.42E-05 |
| 212667_at | 6678 | SPARC | Chr5 | down | 6.57E-05 |
| 242191_at | 200030 | NBPF11 | Chr1 | up | 6.60E-05 |
| 233302_at | 64919 | BCLIIB | Chr14 | up | 7.06E-05 |
| 206494_s_at | 3674 | ITGA2B | Chr17 | down | 1.01E-04 |
| 213258_at | 7035 | TFP1 | Chr2 | down | 1.01E-04 |
| 221942_s_at | 2982 | GUCY1A3 | Chr4 | down | 1.01E-04 |
| 201108_s_at | 7057 | THBS1 | Chr15 | down | 1.41E-04 |
| 207114_at | 80740 | LY6G6C | Chr6 | down | 1.72E-04 |
| 227088_at | 8654 | PDE5A | Chr4 | down | 1.77E-04 |
| 1559126_at | 23223 | RRP12 | Chr10 | up | 1.83E-04 |
| 216580_at | 120872 | RPL7 | Chr12 | up | 2.08E-04 |
| 215859_at | 56926 | NCLN | Chr19 | up | 2.20E-04 |
| 240263_at | 51106 | TFB1M | Chr6 | up | 2.62E-04 |
| 227461_at | 85439 | STON2 | Chr14 | down | 2.63E-04 |
| 201059_at | 2017 | CTTN | Chr11 | down | 2.69E-04 |
| 1560043_at | 51706 | CYB5R1 | Chr1 | up | 2.79E-04 |
| 202729_s_at | 4052 | LTBP1 | Chr2 | down | 3.04E-04 |
| 230014_at | 51646 | YPEL5 | Chr2 | up | 3.18E-04 |
| 236841_at | 374666 | FAM39DP | Chr15 | up | 4.28E-04 |
| 239805_at | 9058 | SLC13A2 | Chr17 | down | 4.75E-04 |
| 232570_s_at | 80332 | ADAM33 | Chr20 | up | 6.13E-04 |
| 233087_at | 64839 | FBXL17 | Chr5 | up | 6.13E-04 |
| 202275_at | 2539 | G6PD | ChrX | down | 6.13E-04 |
| 229991_s_at | 94121 | SYTL4 | ChrX | down | 6.24E-04 |
| 230945_at | 10120 | ACTRIB | Chr2 | up | 6.80E-04 |
| 1558975_at | 124402 | FAM100A | Chr16 | up | 6.86E-04 |
| 1552750_at | 117286 | CIB3 | Chr19 | up | 6.94E-04 |
| 209676_at | 7035 | TFP1 | Chr2 | down | 7.28E-04 |
| 206254_at | 1950 | EGF1 | Chr4 | down | 1.10E-04 |
| 226303_at | 5239 | PGM5 | Chr9 | down | 1.33E-03 |
| 205524_s_at | 1404 | HAPLN1 | Chr5 | down | 1.36E-03 |
| 222319_at | 4676 | NAP1L4 | Chr11 | up | 1.41E-03 |
| 205409_at | 2355 | FOSL2 | Chr2 | down | 1.46E-03 |
| 200785_s_at | 4035 | LRP1 | Chr12 | down | 1.50E-03 |
| 212077_at | 800 | CALD1 | Chr7 | down | 1.62E-03 |
| 219232_s_at | 112399 | EGLN3 | Chr14 | down | 1.79E-03 |
| 1569542_at | 166647 | GP125 | Chr4 | up | 1.90E-03 |
| 200661_at | 5476 | CTSA | Chr20 | down | 2.00E-03 |
Quantitative RT-PCR validation
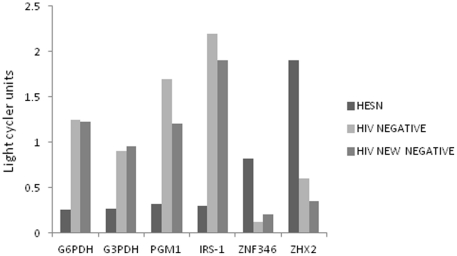
To validate the outcome of the microarray, representative down-regulated and up-regulated genes were picked for quantitative real time RT-PCR. Genes from one of the most down regulated signaling pathways- glucose\gluconeogenesis were randomly picked for qRTPCR analysis while a random sample of zinc finger proteins were selected to represent upregulated pathways. And to control for probable effect of sex work, samples from HIV seronegative sex workers (New Negatives) who had been involved in sex work for one year or less, were included in the qRTPCR analysis.
The expression differences of the selected genes in HESN women as compared to the controls were similar to the microarray data (Fig. 5). No significant differences were observed in expression of the validated genes between the HIV seronegative sex workers and the HIV negative maternal-child health clinic attendees.
Figure 5. Outcome of a comparative quantitative RTPCR validation on randomly selected genes from the microarray analysis.
Genes expression profiles from HESNs were compared with HIV uninfected non sex worker maternal child health clinic attendees (HIV negative) and HIV Negative new entrants into the sex trade (New Negatives). Each assay was a ratio gene quantity to its 18srRNA. G6PDH- glycose 6 phospate dehydrogenase; G3PDH-Glyceraldehyde 3 phosphate dehydrogenase; PGM-1-Phosphoglucomutase-1; IRS-1-Insulin receptor substrate-1; ZNF346-Zinc finger nuclease 346; ZHX2- Zinc finger and homeodomain protein 2.
Discussion
Our primary objective in this study was to investigate if there is a predictive gene expression signature pattern that defines the HESN phenotype in the Pumwani sex worker cohort. Apart from identifying candidate gene biomarkers, we used a systems biology approach to take into account the inherent complexity that may not be elucidated through single gene observations. We also used whole blood in an attempt to unravel the probable interplay between systemic intercellular networks that may be lost through individual cell type approaches. As controlling for HESN studies is often a challenge, we have attempted to include data from HIV negative new entrants into the sex worker trade as an additional control group to lessen confounding factors such as frequent exposures to other sexually transmitted infections. The overall outcome show a distinct difference in expression patterns between the HESN population and controls. Previous studies in this cohort have identified specific human leukocyte antigens (HLA) and interferon regulatory factors (IRF) as associated with resistance [16], [20], [21]. At the mucosal level, specific antibody responses as well as several antiproteases have been found to be elevated among the HIV-1 resistant women as compared to the controls [22], [23]. We noted a significantly differential expression pattern on the HLA types, confirming a probable role these markers may have on HIV-1 resistance. We also observed that some biomarkers, which had been identified in other HESN populations as possible contributors to the resistance phenotype, for example defensin β 1 [15] and certain polymorphs of cyclophilin A [24] were significantly up-regulated in our HESN group. These associations with previous findings reinforced our confidence in the platform we used.
The most up-regulated gene was the Dipeptidyl peptidase 4 (DPP4). The HESN group had a more than a 2 fold change over expression of this gene as compared to controls. DPP4, also known as CD26, is a 110 kDa protein that has been found to be one of the most ubiquitous soluble proteins. Its expression levels have been associated with cancer, diabetes and infectious disease [25]–[27]. The role of DPP4 in HIV-1 infection remains controversial. One of its known functions is to cleave dipeptides from the amino terminus of proteins containing a proline or alanine moiety at the ultimate position. A number of chemokines have this sequence at their termini. RANTES, a chemokine of the interleukin-8 superfamily, is a selective attractant for memory T lymphocytes and monocytes and has been shown to inhibit HIV infection by competitively binding CCR5 [28]. Proost et al [29] showed that truncated RANTES inhibited HIV-1 infection of mononuclear cells 5-fold more efficiently than intact RANTES. He confirmed that the truncation of RANTES was a result of the activity of DPP4 cleavage. Similarly, Morimoto and co-workers [30] have demonstrated that Jurkat T cell line expressing very high levels of DPP4 activity were resistant to HIV-1 infection. In contrast, several laboratories [31], [32] have reported a selective decrease in DPP4-expressing T cells in AIDS progression, suggesting that DPP4 rendered the cells more sensitive to HIV-1 infection. Our in vivo observations of clustering of high DPP4 expression among the HESN sex workers denote a possible HIV-1 protective effect.
In conformity with the high expression of DPP4, we observed a down-regulation of the insulin signaling pathway among the HESN women. Though we could not, in our studies, directly attribute the low expression of insulin genes to DPP4, the association of very high DPP4 with diabetes is a confirmed phenomenon [33], [34]. DPP4 inhibits the incretin hormones, glucagon-like peptide 1 and Gastric Inhibitory peptide, limiting release of insulin from the pancreatic cells and inducing a hyperglycemic state. Subsequently treatment protocols for type 2 diabetes have incorporated DPP4 inhibitors [35]. Though our follow up studies affirmed high DPP4 expression at the protein level [36], clinical evaluation of the HESN cohort did not confirm type 2 diabetes but an impaired fasting glucose state. This suggests that development of type 2 diabetes nay not be an automatic phenomenon in this cohort. We are carrying out further functional studies to determine the probable contribution and mechanism of DPP4 and insulin repression in the HESN phenotype.
The phospatidylinositol pathway was a key impacted signaling system among the HIV-1 resistant women (p<0.0001) This pathway consists of a cellular family of kinases that are activated by tyrosine phosphorylation and act as second messengers by regulating the phosphorylation of other kinases, including the ribosomal S6 kinases and the MAPK signaling network [37]. Because of its control of activation of many pathways, the inositol pathway is a critical mediator of various cellular processes. Down regulation of the phospatidylinositol system has been linked, to among others, low insulin levels. Insulin is a key regulator of the PI3K/AKT signaling system and insulin, through the insulin receptor substrate (IRS-1), provokes a rapid increase in levels of inositol phospholipids, PIP [38]. Additionally, PI3K was shown to be a key member of the pathway and was required for insulin induced glucose transport and glycogen synthesis. We postulate that the down expression of key immune pathways and cell adhesion molecule genes in our dataset may have been the result of the switch down of the PI3/AKT pathway, due in part by reduced insulin secretion. Furthermore evidence of direct effect of phosphotidylinositol signaling on HIV-1 infection has been noted. Francois et al [39], reported that inhibition of PI3K resulted in absence of HIV-1 infection of CD4+ T cells and macrophages with X4 and R5 pseudo typed viruses. Similarly Brown et al [40] has shown that blocking phosphotidylinositol phosphate (a substrate of P13K), with monoclonal antibodies inhibited HIV-1 infection by two HIV-1 primary isolates in human peripheral blood mononuclear cells. Chugh and colleagues [41] have demonstrated that blocking the PI3K/AKT pathway with Akt inhibitors dramatically reduced HIV-1 production from virus-infected macrophages. The implications of the observed down regulation of the phospatidylinositol pathway may hence be direct or through inhibition of HIV-1 dependency signaling events requiring PIP modulation.
In concordance with earlier findings from our group using specific cell subsets [42], we also observed lower basal T-cell gene expression, confirming a lowered immune activation state among the HESN group. Immune activation has been suggested to be the greater risk factor in HIV-1 infection and higher levels of activated T-cells in sub-Saharan African populations have been associated with higher HIV-1 prevalence [43]. This observation has been supported further by evidence of decrease in T-cell activation with successful antiretroviral use [44]. This low immune activation phenomenon, or immune quiescence, proposes a mechanistic profile of fewer activated cells presenting lesser targets to the AIDS virus and making it easier to clear infections that occur. Our findings of low basal gene expression in T-cells in our HESN population is also concordant with reports from similar populations that have shown lower levels of activated T-cells among seronegative partners of HIV-1 infected spouses [45]. Although these observations contrasts with reports from others [46]–[49], the gene expression patterns observed in our data bear a close metabolic signature pattern to that of quiescent T cells. Activated T cells have been noted to derive its ATP requirements from cytoplasmic glycolytic pathway [50], whereas quiescent T-cells derive metabolic needs through oxidative phosphorylation and other catabolic processes in the mitochondria [51], [52]. We noted that one of the few gene expression pathways that was up regulated by the HESNs was the mitochondrial oxidative phophorylation pathway (OXPHOS) with a concomitant profound down regulation of enzymes in the glycolysis pathway.
An up-regulation of DNA binding genes that have been associated with gene editing and silencing was a key phenomenon among the HESN. This included the Zinc finger (ZFN) and SMAD proteins encoding genes. Among the highly expressed ZFN genes were the Zinc Finger and Homeoboxes 2 (ZHX2) and Zinc Finger protein 20 (ZBTB20). ZHX2 has been identified as a transcription repressor gene [53] while ZBTB20 is a key repressor of alpha fetoprotein gene transcription [54]. SMAD3 is a strong enforcer of quiescence in T-lymphocytes through its antiproliferative and pro-survival signals [55]. In addition, the non-coding gene Xist, reported to trigger chromosome-wide gene repression [56], was highly up-regulated in the HESN women (p 2.80E−10).
In tandem with the overexpression of gene silencing factors at the transcriptional level, we observed dramatic down regulation of host factors reported to be important for successful HIV-1 infection. These factors included intergrins, a class of cell surface receptors that mediate linkages with the extracellular matrix and have been identified as HIV-1 receptors [57], catenins, components of the actin cytoskeleton shown to enhance HIV-1 infection by anchoring chemokine receptors CCR5 and CCR4 to the cell membrane [58], and NF- κB a known modulator of immune function that directly enhances HIV-1 transcription at the proviral level [59].
A previous in vitro genomewide screen identified more than 273 host genes that were essential for HIV-1 infection [60]. We noted that 66 of the genes identified in that study were among those differentially regulated by the HIV-1 resistant women at significant levels, most of which (61%) were down regulated (data not shown).
Our findings imply that a repertoire of genes acting in concert rather than a single determinant may be responsible for the observed HESN phenotypes in the Pumwani cohort. Our data also suggest that genes contributing to a lowered basal immune activation state, in tandem with a general repression of host HIV-1 dependency factors, may be a key contributor to the HIV-1 resistance phenomenon. Hence, the understanding of signaling and metabolic events that regulate immune activation may provide crucial information for the design of effective anti-HIV-1 preventive strategies.
Materials and Methods
Ethical Statement
This study was guided by the Helsinki Declaration on ethical principles for medical research involving human subjects. All studies were approved by University of Nairobi/Kenyatta National Hospital Ethical Review Board and the University of Manitoba Health Research Ethics Board. All patients provided written informed consent for collection of samples and subsequent analysis.
Subjects
The Nairobi (Pumwani) commercial sex worker cohort was established in 1985 and has provided vital data that there might be biological mediated resistance to HIV-1 infection [11], [16], [20], [21]. Despite repeated exposures to HIV-1, a number of women in this cohort have remained HIV-1 uninfected for long periods of time and have been epidemiologically defined as HIV resistant [20]. Resistance has not been linked to frequency and duration of condom use, sexually transmitted infections, or differing HIV-1 exposure rates [21] In this study, sex workers who had been followed up for more than 7 years and continue to be HIV-1 uninfected were included. As a control, samples were obtained from an age-matched low risk HIV-1 negative group of non-pregnant women in a monogamous relationship attending a maternal and child health (MCH) clinic To control for confounding effects due to sex work, the outcome was validated against samples from a group of HIV negative sex workers who were recently enrolled in the cohort (New Negatives) and had not met a definitive stage for HESN.
Sample collection and processing
Whole blood samples were collected in PaxGene blood tubes (Preanalytix, GMBH) and total RNA was extracted in Nairobi following the manufacturer's protocol (Qiagen). Briefly, pelleted nucleic acids were digested with proteinase K then shredded through a spin column to remove protein debris. The resulting supernatant was mixed with ethanol and RNA was selectively bound to a silica-based fibre matrix. Elution of RNA was done with RNASE-free water after washing with saline buffers and digestion with DNAse to remove DNA contamination. Assessment of RNA quality, integrity and purity were done through a Bionalyser 2100 (Agilent Technologies, Palo Alto CA). RNA Samples were considered for further analysis only if they had distinct 28 s and 18 s ribosomal peaks and had been processed or frozen at −70°C within 5 hours after collection.
Microarray analysis
RNA samples meeting the above criteria were shipped to The Centre For Applied Genomics, Hospitals for Sick Children, Toronto for analysis. 100 ng of RNA was amplified using the Affymetrix two cycle amplification kit (Affymetrix, Santa Clara, CA) incorporating the Ambion MEGAscript T7 amplification procedure. To minimize the challenges posed by abundant globin RNA transcripts, Nugen Whole Blood Solution (Nugen, San Carlos, CA) procedures were incorporated into the Affymetrix protocol. Amplification, labeling, hybridization onto the Affymetrix U133 Plus 2.0 Gene Chip, washing, and scanning were performed as per the manufacturer's protocol.
Expression analysis
Background subtraction and probe signal summarization was analyzed using the R Bioconductor software. The resulting log2 signal values were retransformed to linear scale and analyzed using the Affymetrix MAS5 package. The program's algorithm output files represented the differences in intensities between the perfect match and mismatch probe sets or a detection of present, marginal or absent calls. Raw image DAT data was processed to CEL files and analyzed using the ArrayAssist software (Stratagene, La Jolla CA). The ratio of the geometric means of the expression analysis of the relevant gene fragments was computed to yield a fold change analysis. Confidence intervals and corrected p values were calculated using a Rank Wilcoxson test. Significance was assessed using a 0.5 threshold of the false discovery rate (FDR) which accounts for multiple testing. Only genes with differences between the HESN and controls >1.3 fold and p value <0.05 and called as present in at least 50% of the samples tested were considered as differentially expressed. Heatmaps showing differential expression were generated using R and Bioconductor Software [61]. The Database for Annotation, Visualization and Integrated Discovery (DAVID) [62], [63] was used for pathway analysis of the significantly expressed genes.
Real Time PCR
Selected array results from HESN and controls were validated by quantitative PCR using LightCycler real time PCR (Roche Diagnostics). And to asses that the gene expression profiles may not have been confounded by sex work, blood samples from sex worker women who have been involved in sex work for one year and were HIV negative (New Negatives) were randomly selected and included as additional controls in qRT-PCR confirmation of microarray data.
Samples were analysed with Quantitect SYBR Green RT PCR assay kit as per the manufacturer's protocol (Qiagen). Briefly, 100 ng of each sample were ran in duplicate and normalized to 18 sRNA. Relative expression of each gene was determined from a standard curve of pooled cDNA from human peripheral blood mononuclear cells with total RNA extracted in the same way in a 10 series dilution to form the qRT-PCR standards [64]. p-values for significance were analyzed using a student t-test.
GeneBank
The microarray data have been deposited at the Genomic Spatial Event (GSE) database under numbers GSE 30155–30240 and GSE 33580.
Acknowledgments
We thank the volunteers of the Pumwani sex worker cohort for their invaluable time and contribution to this project. We sincerely acknowledge the support from the clinic staff of the University of Nairobi/University of Manitoba Collaborative Program for sample and data collection. Our gratitude to Mark Jordan and Brenda Oosterveen of Agriculture Canada for validating our microarray assays, Leslie Slaney and Ian Maclean of University of Manitoba for logistical support and Rose Odera and Salome Ngamau of Kenya Medical Research Institute for secretarial services.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was funded by Bill and Melinda Gates Foundation under Grand Challenges in Global Health (GCGH) initiative and the Canada Institutes of Health Research (CIHR). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Joint United Nations Programme on HIV/AIDS, UNAIDS. 2008. Report on the Global AIDS Epidemic. http://www.unaids.org. [PubMed]
- 2.Kallings LO. The first postmordern epidemic: 25 years of HIV/AIDS. J internal Med. 2008;263:218–243. doi: 10.1111/j.1365-2796.2007.01910.x. [DOI] [PubMed] [Google Scholar]
- 3.Stover J, Fidzani B, Molomo BC, Moeti T, Musuka G. Estimated HIV trends and program effects in Botswana. PLos ONE. 2008;3:e3729. doi: 10.1371/journal.pone.0003729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wainberg M, Kuan-Te Jeang. 25 years of HIV research-progress and perspectives. BMC Medicine. 2008;6:31. doi: 10.1186/1741-7015-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–20. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 6.Saez-Cirion A, Versmisse P, Truong LX, Chakrabarti L, Carpentier W, et al. Persistent resistance to HIV-1 infection in CD4 T cells from exposed uninfected Vietnamese individuals is mediated by entry and post entry blocks. Retrovirology. 2006;3:81. doi: 10.1186/1742-4690-3-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rowland- Jones SL, Nixon DF, Aldhous MC, Gotch F, Ariyoshi K, et al. HIV specific cytotoxic T-cell activity in a HIV-exposed but uninfected infant. Lancet. 1993;341:860–861. doi: 10.1016/0140-6736(93)93063-7. [DOI] [PubMed] [Google Scholar]
- 8.Clerici M, Sison AV, Berzofsky JA, Rakusan TA, Brandt CD, et al. Cellular immune factors associated with mother-to-infant tansmission of HIV. AIDS. 1993;7:1427–1433. doi: 10.1097/00002030-199311000-00004. [DOI] [PubMed] [Google Scholar]
- 9.Begaud E, Chartier L, Marechal V, Ipero J, Leal J, et al. Reduced CD4 T- cell activation and in vitro susceptibility to HIV-1 infection in exposed uninfected Central Africans. Retrovirology. 2006;3:35. doi: 10.1186/1742-4690-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelker HC, Seidlin M, Vogler M, Valentine FT. Lymphocytes from some long term seronegative heterosexual partners of HIV infected individuals proliferate in response to HIV antigens. AIDS Res Hum Retroviruses. 1992;8:1355–9. doi: 10.1089/aid.1992.8.1355. [DOI] [PubMed] [Google Scholar]
- 11.Fowke KR, Nagelkerke NJ, Kimani J, Simonsen N, Anzala AO, et al. Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet. 1996;348:1347–1351. doi: 10.1016/S0140-6736(95)12269-2. [DOI] [PubMed] [Google Scholar]
- 12.Rowland-Jones SJ, Sutton J, Ariyoshi K, Dong T, Gotch F, et al. HIV specific cytotoxic T-cells in HIV exposed but uninfected Gambian Women. Nat Med. 1995;1:59–64. doi: 10.1038/nm0195-59. [DOI] [PubMed] [Google Scholar]
- 13.Beyrer C, Artenstein AW, Rugpao S, Stephens H, VanCott TC, et al. Epidemiologic and biologic characterization of a cohort of human immunodeficiency virus type 1 highly exposed persistently seronegative female sex workers in Northen Thailand. J Infect Dis. 1999;179:59–67. doi: 10.1086/314556. [DOI] [PubMed] [Google Scholar]
- 14.Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply exposed individuals in HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 15.Lama J, Planelles V. Host factors influencing susceptibility to HIV infection and AIDS Progression. Retrovirology. 2007;4:52. doi: 10.1186/1742-4690-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacDonald KS, Fowke KR, Kimani J, Dunand VA, Nalgerke NJ, et al. Influence of HLA supertypes on susceptibility and resistance to human immunodeficiency virus type 1 infection. J Inf Dis. 2000;181:1581–1589. doi: 10.1086/315472. [DOI] [PubMed] [Google Scholar]
- 17.Carthagena L, Parise MC, Ringeard M, Chelbi-Alix MK, Hazan U, et al. Implications of TRIM alpha and TRIMCyp in interferon induced antiretroviral restriction activities. Retrovirology. 2008;5:59. doi: 10.1186/1742-4690-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyton RJ, Altmann DM. Natural Killer cells, killer immunoglobulin-like receptors and human leucocyte antigen Class I in disease. Clin Exp Immunol. 2007;149:1–8. doi: 10.1111/j.1365-2249.2007.03424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valcke HS, Bernard NF, Bruneau J, Alary M, Tsoukas CM, et al. APOBEC3G genetic variants and their association with risk of HIV infection in highly exposed Caucasians. AIDS. 2006;20:1984–1986. doi: 10.1097/01.aids.0000247124.35129.e1. [DOI] [PubMed] [Google Scholar]
- 20.Plummer FA, Ball TB, Kimani J, Fowke KR. Resistance to HIV-1 Infection among highly exposed sex workers in Nairobi: what mediates protection and why does it develop? Immunol Lett. 1999;66:27–34. doi: 10.1016/s0165-2478(98)00182-5. [DOI] [PubMed] [Google Scholar]
- 21.Ball TB, Hezhao J, Kimani J, Mclaren P, Marlin C, et al. Polymorphisms in IRF-1 associated with resistance to HIV-1 infection in highly exposed uninfected Kenyan sex workers. AIDS. 2007;21:1091–1101. doi: 10.1097/QAD.0b013e3280ef6ae1. [DOI] [PubMed] [Google Scholar]
- 22.Kaul R, Plummer F, Clerici M, Bomsel M, Lopalco L, et al. Mucosal IgA in exposed uninfected subjects: evidence for a role in protection against HIV infection. AIDS 2001. 2001;15:431–2. doi: 10.1097/00002030-200102160-00026. [DOI] [PubMed] [Google Scholar]
- 23.Burgener A, Boutilier J, Wachihi C, Kimani J, Carpenter M, et al. Identification of differentially expressed proteins in the cervical mucosa of HIV-1 resistant sex workers. Proteome Res. 2008;10:4446–54. doi: 10.1021/pr800406r. [DOI] [PubMed] [Google Scholar]
- 24.Rits AM, Van Dort KA, Kootsra NA. Polymorphisms in the regulatory region of the cyclophilin A gene influence the susceptibility for HIV-1 Infection. PLOS One. 2008;3:e3975. doi: 10.1371/journal.pone.0003975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Havre PA, Abe M, Urasaki Y, Ohnuma K, Morimoto C, et al. The role of CD26/dipeptidyl peptidase IV in cancer. Frontiers in Bioscience. 2008;13:1634–45. doi: 10.2741/2787. [DOI] [PubMed] [Google Scholar]
- 26.Boonacker E, Van Noordern CJ. The multifunctional or moonlighting protein CD26/DPPIV. European Journal of Cell Biology. 2003;82:53–73. doi: 10.1078/0171-9335-00302. [DOI] [PubMed] [Google Scholar]
- 27.Hildebrandt M, Rose M, Ruter J, Salama A, Monnikes A, et al. Dipetidylpeptidase IV activity in patients in patients with inflammatory bowel bowel disease. Scand J Gastroenterology. 2001;36:1067–1072. doi: 10.1080/003655201750422675. [DOI] [PubMed] [Google Scholar]
- 28.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–6. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 29.Proost P, Meester I, Schols D, Struyf S, Lambeir A, et al. Amino-terminal truncation of chemokines by CD26/dipeptidyl-peptidase IV- conversion of RANTES into a potent inhibitor of monocyte chemotaxis and HIV infection. J Biol Chem. 1998;273:7222–7. doi: 10.1074/jbc.273.13.7222. [DOI] [PubMed] [Google Scholar]
- 30.Morimoto C, Lord CI, Zhang C, Duke-Chohan J, Letvin N, et al. Role of CD26/dipeptidyl peptidase IV in human immunodeficiency virus type 1 infection and apotosis. Proc Natl Acad Sci USA. 1994;91:9960–4. doi: 10.1073/pnas.91.21.9960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blazquez M, Madueno J, Gonzalez R, Jurado R, Bachovchin W, et al. Selective decrease of CD26 expression in T-cells from HIV infected individuals. J Immunol. 1992;149:3073–7. [PubMed] [Google Scholar]
- 32.Vanham G, Kestens L, Van Hoof J, Peene C, Colebunders R, et al. Decreased expression of memory marker CD26 on both CD4+ and CD8+ T lymphocytes of HIV infected subjects. J Acquired Immune Defic Syndr. 1993;6:749–57. [PubMed] [Google Scholar]
- 33.Mentlein R, Gallwitz B, Schmidt W. Dipeptidyl peptidase IV hydrolyses gastric inhibitory polypeptide, glucagon like peptide 1 (7–36) amide peptide histidine methionine and is responsible for their degradation in human serum. Eur J Biochem. 1993;214:829–835. doi: 10.1111/j.1432-1033.1993.tb17986.x. [DOI] [PubMed] [Google Scholar]
- 34.Drucker D, Shi Q, Crivici A, Sumner-Smith M, Tavares W, et al. Regulation of the biological activity of glucagons-like peptide 2 in vivo by dipeptidyl peptidase IV. Nat Biotechnology. 1997;15:673–677. doi: 10.1038/nbt0797-673. [DOI] [PubMed] [Google Scholar]
- 35.Pratley RE, Salsali A. Inhibition of DPP-4: A new therapeutic approach for treatment of type 2 diabetes. Curr Med Res Opin. 2007;4:919–31. doi: 10.1185/030079906x162746. [DOI] [PubMed] [Google Scholar]
- 36.Songok EM, Osero B, Mackinnon L, Rono MK, Apidi W, et al. CD26/DPPIV is highly expressed in peripheral blood of HIV resistant female commercial sex workers. Virol J. 2010;7:343. doi: 10.1186/1743-422X-7-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isotani S, Hara K, Takunaga C, Inoue H, Avruch J, et al. Immunopurified mammalian target of rapamycin phophorylates and activates p70 S6 kinase alpha in vitro. J Biol Chem. 1999;274:34493–8. doi: 10.1074/jbc.274.48.34493. [DOI] [PubMed] [Google Scholar]
- 38.Uenio M, Carvalheira JB, Tambascia RC, Bezerra RM, Amaral ME, et al. Regulation of insulin signalling by hyperinsulinemia: role of IRS-1/2 serine phosphorylation and the Mtor/p70 S6K pathway. Diabetologia. 2005;48:506–18. doi: 10.1007/s00125-004-1662-6. [DOI] [PubMed] [Google Scholar]
- 39.Francois F, Klotman ME. Phosphatidylinositol 3-Kinase regulates human immunodeficiency virus type 1 replication following viral entry in primary CD4+ T-lymphocytes and macrophages. J Virol. 2003;77:2539–2549. doi: 10.1128/JVI.77.4.2539-2549.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown BK, Karasavvas N, Beck Z, Matyas GR, Birx DL, et al. Monoclonal antibodies to phoshatidylinositol phosphate neutralize human immunodeficiency virus type 1: Role of phosphate-binding subsites. Virol. 2007;81:2087–2091. doi: 10.1128/JVI.02011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chugh P, Bradel-Tretheway B, Monteiro-Filho CMR, Planelles V, Maggirwar SB, et al. AKt inhibitors as an HIV-1 infected macrophage-specific antiviral therapy. Retrovirology. 2008;5:11. doi: 10.1186/1742-4690-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mclaren PJ, Ball TB, Wachihi C, Jaoko W, Kelvin DJ, et al. HIV-exposed seronegative commercial sex workers show a quiescent phenotype in the CD4+ T cell compartment and reduced expression of HIV-dependent host factors. J Infect Dis. 2010;Suppl 3:S339–44. doi: 10.1086/655968. [DOI] [PubMed] [Google Scholar]
- 43.Bentwich Z, Kalinkovich A, Weisman Z. Immune activation is a dominant factor in the pathogenesis of African AIDS. Immunology Today. 1995;16:187–191. doi: 10.1016/0167-5699(95)80119-7. [DOI] [PubMed] [Google Scholar]
- 44.Koblavi-Deme S, Maran M, Kabran N, Borget MY, Kalou M, et al. Changes in levels of immune activation and reconstitution markers among HIV-1 infected Africans receiving antiretroviral therapy. AIDS. 2003;17:(suppl 3):S17–22. doi: 10.1097/00002030-200317003-00003. [DOI] [PubMed] [Google Scholar]
- 45.Begaud E, Chartier L, Marechal V, Ipero J, Leal J, et al. Reduced CD4 T- cell activation and in vitro susceptibility to HIV-1 infection in exposed uninfected Central Africans. Retrovirology. 2006;3:35. doi: 10.1186/1742-4690-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jennes W, Evertse D, Borget MY, Vuylsteke B, Maurice C, et al. Supressed cellular alloimune responses in HIV exposed seronegative female sex workers. Clin Exp Immunol. 2006;143:435–444. doi: 10.1111/j.1365-2249.2006.03017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biasin M, Caputo SL, Speciale L, Colombo F, Racioppi L, et al. Mucosal and systemic immune activation is present in human immunodeficiency virus exponed seronegative women. J infect Dis. 2000;182:1365–1374. doi: 10.1086/315873. [DOI] [PubMed] [Google Scholar]
- 48.Tran HK, Chartier L, Troung LX, Nguyen NN, Fontanet A, et al. Systemic immune activation in HIV-1 exposed uninfected Vietnamese intravascular drug users. AIDS Res Human Retroviruses. 2006;22:255–261. doi: 10.1089/aid.2006.22.255. [DOI] [PubMed] [Google Scholar]
- 49.Suy A, Castro P, Nomdedeu M, Garcia F, Lopez A, et al. Immunological profile of heterosexual highly HIV exposed uinfected individuals: predominant role of CD4 and CD8 T-cell activation. J Infect Dis. 2007;196:1191–1201. doi: 10.1086/521193. [DOI] [PubMed] [Google Scholar]
- 50.Roos D, Loos JA. Changes in carbohydrate metabolism of mitogenically stimulated human peripheral lymphocytes II. Relative importance of glycolysis and oxidative phosphorylation on phytohaemagglutinin stimulation. Exp Cell Res. 1973;77:127–135. doi: 10.1016/0014-4827(73)90561-2. [DOI] [PubMed] [Google Scholar]
- 51.Jones GR, Thomson BC. Revving the engine: Signal transduction fuels T-cell activation. Immunity. 2007;27:173–178. doi: 10.1016/j.immuni.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 52.Krauss S, Brand MD, Buttgereit F. Signalling takes a breath-new quantitative perspectives on bioenergetics and signal transduction. Immunity. 2001;15:497–502. doi: 10.1016/s1074-7613(01)00205-9. [DOI] [PubMed] [Google Scholar]
- 53.Kawata H, Yamada K, Shou Z, Mizutani T, Yazawa T, et al. : Zinc finger and homeoboxes (ZXH)2, a novel member of the ZHX family function as a transcriptional repressor. Biochem J. 2003;373:747–757. doi: 10.1042/BJ20030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie Z, Zhang H, Tsai W, Zhang Y, Du Y, et al. Zinc finger protein ZBTB20 is a key repressor of alfa-fetoprotein gene transcription in liver. Proc Natl Acad Sci USA. 2008;105:10859–10864. doi: 10.1073/pnas.0800647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Modiano JF, Johnson L, Bellgrau D. Negative regulators in homeostasis of naïve peripheral T-cells. Immunol Res. 2008;41:137–153. doi: 10.1007/s12026-008-8017-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wutz A, Gribnau J. X inactivation Xplained. Current Opin Genet Dev. 2007;5:387–93. doi: 10.1016/j.gde.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 57.Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, et al. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol. 2008;9:301–9. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- 58.Schweneker M, Bachmann AS, Moelling K. The HIV-1 co-receptor CCR5 binds to alpha-catenin, a component of the cellular cytoskeleton. Biochem Biophys Commun. 2004;325:751–7. doi: 10.1016/j.bbrc.2004.10.096. [DOI] [PubMed] [Google Scholar]
- 59.Alcami J, Lera DL, Folgueira L, Pedrazza MA, Jacque JM, et al. Absolute dependence on Kappa B responsive elements for initition and tat mediated amplification of HIV transcription in blood CD4 T-lymphocytes. EMBO. 1995;14:1552–1560. doi: 10.1002/j.1460-2075.1995.tb07141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–6. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 61.Dudoit S, Gentleman RC, Ouackenbush J. Open source software for analysis of microarray data. Biotechniques. 2003:Suppl 45–51. [PubMed] [Google Scholar]
- 62.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 63.Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, et al. DAVID: Database for Annotation Visualization, and Intergrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 64.Lester RT, Yao XD, Ball TB, Mckinnon LR, Kaul R, et al. Toll-like receptor expression and responsiveness are increased in viraemic HIV-1 infection. AIDS. 2008;22:685–94. doi: 10.1097/QAD.0b013e3282f4de35. [DOI] [PubMed] [Google Scholar]
- 65.Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, et al. The influence of CCL3L1 gene containing segmental duplications on HIV/AIDS suscpetibility. Science. 2005;174:5655–64. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 66.de witte L, Nabatov A, Pion M, Fluitsman D, de Jong MA, et al. Langerin is a natural barrier to HIV-1 transmission by Langerhan cells. Nat Med. 2007;13:367–371. doi: 10.1038/nm1541. [DOI] [PubMed] [Google Scholar]
- 67.Martin MP, Lederman MM, Hutchenson HB, Goedert JJ, Nelson GW, et al. Association of DC-SIGN promoter polymorphism with increased risk for parenteral, but not mucosal, acquisition of human immunodeficiency virus type 1 infection. J Virol. 2004;78:14053–14056. doi: 10.1128/JVI.78.24.14053-14056.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.An P, Duggal P, Wang LH, OBrien SJ, Donfield S, et al. Polymorphism of CUL5 are associated with CD4+ T cell loss in HIV-1 infected individuals. PLoS Genet. 2007;3:e19. doi: 10.1371/journal.pgen.0030019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winkler C, Modi W, Smith MW, Nelson GW, Wu X, et al. Genetic restriction of AIDS pathogenesis by a SDF-1 chemokine variant. Science. 1998;279:389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 70.Schuitmemaker H. IL4 and IL10 as potent inhibitors of HIV1 replication in macrophages in vitro: a role for cytokines in the in vivo virus host range? Res Immunol. 1994;145:588–92. doi: 10.1016/s0923-2494(05)80038-0. [DOI] [PubMed] [Google Scholar]
- 71.Chang TL, Vargas J, Jnr, DelPortillo A, Klotman ME. Dual Role of alpha-defensins-I in anti-HIV innate immunity. J Clin Invest. 2005;115:765–773. doi: 10.1172/JCI200521948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jennes W, Verhevden S, Demanet C, Adie-Toure CA, Vuylsteke B, et al. Cutting edge: Resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in absence of their HLA ligands. J Immunol. 2006;177:6588–92. doi: 10.4049/jimmunol.177.10.6588. [DOI] [PubMed] [Google Scholar]
- 73.Finberg RW, Wang JP, Kurt-Jones EA. Toll-like receptors and viruses. Rev Med Virol. 2007;17:35–43. doi: 10.1002/rmv.525. [DOI] [PubMed] [Google Scholar]