Abstract
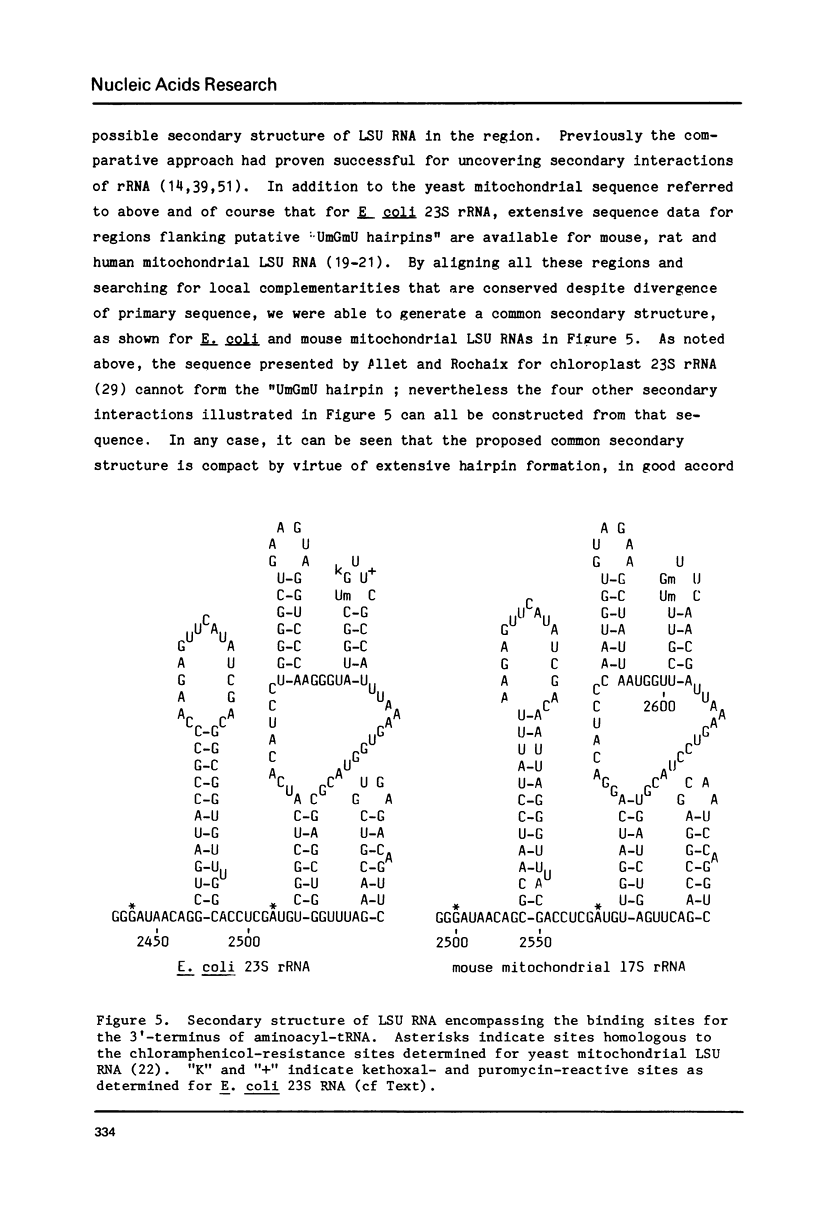
The positions of post-transcriptionally methylated residues within hamster mitochondrial ribosomal RNA have been established. Comparisons with other mitochondrial rRNA, and with bacterial, eucaryotic and chloroplast rRNA show that the methylated regions i) are comprised of conserved primary sequences and/or secondary structures and ii) are situated at the subunit interface of the ribosome. The comparative analyses also reveal that the ribose-methylated sequence UmGmU of hamster mitochondrial large ribosomal subunit (LSU1) RNA lies in a universally conserved hairpin loop which contains a putative puromycin-reactive nucleotide. The "UmGmU hairpin" is within 100 nucleotides of two chloramphenicol-resistance residues of LSU RNA. We present a secondary structure for this region which is conserved in LSU RNAs. This structure allows physical juxtaposition of the three antibiotic-interacting loci and thus defines RNA components of the ribosomal-binding site for the 3'-terminus of aminoacyl-tRNA.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allet B., Rochaix J. D. Structure analysis at the ends of the intervening DNA sequences in the chloroplast 23S ribosomal genes of C. reinhardii. Cell. 1979 Sep;18(1):55–60. doi: 10.1016/0092-8674(79)90353-2. [DOI] [PubMed] [Google Scholar]
- Bonen L., Doolittle W. F., Fox G. E. Cyanobacterial evolution: results of 16S ribosomal ribonucleic acid sequence analyses. Can J Biochem. 1979 Jun;57(6):879–888. doi: 10.1139/o79-108. [DOI] [PubMed] [Google Scholar]
- Brand R. C., Gerbi S. A. Fine structure of ribosomal RNA. II. Distribution of methylated sequences within Xenopus laevis rRNA. Nucleic Acids Res. 1979 Nov 24;7(6):1497–1511. doi: 10.1093/nar/7.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Noller H. F. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jan;77(1):201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Palmer M. L., Kennedy P. J., Noller H. F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon P., Ehresmann C., Ehresmann B., Ebel J. P. The complete nucleotide sequence of the ribosomal 16-S RNA from Excherichia coli. Experimental details and cistron heterogeneities. Eur J Biochem. 1979 Oct 15;100(2):399–410. doi: 10.1111/j.1432-1033.1979.tb04183.x. [DOI] [PubMed] [Google Scholar]
- Chapman N. M., Noller H. F. Protection of specific sites in 16 S RNA from chemical modification by association of 30 S and 50 S ribosomes. J Mol Biol. 1977 Jan 5;109(1):131–149. doi: 10.1016/s0022-2836(77)80049-1. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Timkovich R., Almassy R. J. The cytochrome fold and the evolution of bacterial energy metabolism. J Mol Biol. 1976 Feb 5;100(4):473–491. doi: 10.1016/s0022-2836(76)80041-1. [DOI] [PubMed] [Google Scholar]
- Dubin D. T., Taylor R. H., Davenport L. W. Methylation status of 13S ribosomal RNA from hamster mitochondria: the presence of a novel riboside, N4-methylcytidine. Nucleic Acids Res. 1978 Nov;5(11):4385–4397. doi: 10.1093/nar/5.11.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin D. T., Taylor R. H. Modification of mitochondrial ribosomal RNA from hamster cells: the presence of GmG and late-methylated UmGmU in the large subunit (17S) RNA. J Mol Biol. 1978 Jun 5;121(4):523–540. doi: 10.1016/0022-2836(78)90398-4. [DOI] [PubMed] [Google Scholar]
- Dujon B. Sequence of the intron and flanking exons of the mitochondrial 21S rRNA gene of yeast strains having different alleles at the omega and rib-1 loci. Cell. 1980 May;20(1):185–197. doi: 10.1016/0092-8674(80)90246-9. [DOI] [PubMed] [Google Scholar]
- Eckerman D. J., Symons R. H. Sequence at the site of attachment of an affinity-label derivative of puromycin on 23-S ribosomal RNA of Escherichia coli ribosomes. Eur J Biochem. 1978 Jan 2;82(1):225–234. doi: 10.1111/j.1432-1033.1978.tb12015.x. [DOI] [PubMed] [Google Scholar]
- Eladari M. E., Hampe A., Galibert F. Nucleotide sequence neighbouring a late modified guanylic residue within the 28S ribosomal RNA of several eukaryotic cells. Nucleic Acids Res. 1977 Jun;4(6):1759–1767. doi: 10.1093/nar/4.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon I. C., Anderson S., Nierlich D. P. Distinctive sequence of human mitochondrial ribosomal RNA genes. Nature. 1980 Jul 31;286(5772):460–467. doi: 10.1038/286460a0. [DOI] [PubMed] [Google Scholar]
- Fellner P., Sanger F. Sequence analysis of specific areas of the 16S and 23S ribosomal RNAs. Nature. 1968 Jul 20;219(5151):236–238. doi: 10.1038/219236a0. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Magrum L. J., Balch W. E., Wolfe R. S., Woese C. R. Classification of methanogenic bacteria by 16S ribosomal RNA characterization. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4537–4541. doi: 10.1073/pnas.74.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G. E., Stackebrandt E., Hespell R. B., Gibson J., Maniloff J., Dyer T. A., Wolfe R. S., Balch W. E., Tanner R. S., Magrum L. J. The phylogeny of prokaryotes. Science. 1980 Jul 25;209(4455):457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Woese C. R. 5S RNA secondary structure. Nature. 1975 Aug 7;256(5517):505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- Frisby D. Oligonucleotide mapping of non-radioactive virus and messenger RNAs. Nucleic Acids Res. 1977 Sep;4(9):2975–2996. doi: 10.1093/nar/4.9.2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwell P., Harris R. J., Symons R. H. Affinity labelling of 23-S ribosomal RNA in the active centre of Escherichia coli peptidyl transferase. Eur J Biochem. 1974 Dec 2;49(3):539–544. doi: 10.1111/j.1432-1033.1974.tb03858.x. [DOI] [PubMed] [Google Scholar]
- Hashimoto S., Sakai M., Muramatsu M. 2'-O-methylated oligonucleotides in ribosomal 18S and 28S RNA of a mouse hepatoma, MH 134. Biochemistry. 1975 May 6;14(9):1956–1964. doi: 10.1021/bi00680a024. [DOI] [PubMed] [Google Scholar]
- Herr W., Chapman N. M., Noller H. F. Mechanism of ribosomal subunit association: discrimination of specific sites in 16 S RNA essential for association activity. J Mol Biol. 1979 Jun 5;130(4):433–449. doi: 10.1016/0022-2836(79)90433-9. [DOI] [PubMed] [Google Scholar]
- Herr W., Noller H. F. Protection of specific sites in 23 S and 5 S RNA from chemical modification by association of 30 S and 50 S ribosomes. J Mol Biol. 1979 Jun 5;130(4):421–432. doi: 10.1016/0022-2836(79)90432-7. [DOI] [PubMed] [Google Scholar]
- Khan M. S., Salim M., Maden B. E. Extensive homologies between the methylated nucleotide sequences in several vertebrate ribosomal ribonucleic acids. Biochem J. 1978 Mar 1;169(3):531–542. doi: 10.1042/bj1690531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klootwijk J., Klein I., Grivell L. A. Minimal post-transcriptional modification of yeast mitochondrial ribosomal RNA. J Mol Biol. 1975 Sep 25;97(3):337–350. doi: 10.1016/s0022-2836(75)80044-1. [DOI] [PubMed] [Google Scholar]
- Klootwijk J., Planta R. J. Analysis of the methylation sites in yeast ribosomal RNA. Eur J Biochem. 1973 Nov 15;39(2):325–333. doi: 10.1111/j.1432-1033.1973.tb03130.x. [DOI] [PubMed] [Google Scholar]
- Lambowitz A. M., Luck D. J. Letter: Methylation of mitochondrial RNA species in the wild-type and poky strains of Neurospora crassa. J Mol Biol. 1975 Jul 25;96(1):207–214. doi: 10.1016/0022-2836(75)90192-8. [DOI] [PubMed] [Google Scholar]
- Lambowitz A. M., Luck D. J. Studies on the poky mutant of eurospora crassa. Fingerprint analysis of mitochondrial ribosomal RNA. J Biol Chem. 1976 May 25;251(10):3081–3095. [PubMed] [Google Scholar]
- Maden B. E., Reeder R. H. Partial mapping of methylated sequences in Xenopus laevis ribosomal RNA by preparative hybridization to cloned fragments of ribosomal DNA. Nucleic Acids Res. 1979 Mar;6(3):817–830. doi: 10.1093/nar/6.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E., Tartof K. Nature of the ribosomal RNA transcribed from the X and Y chromosomes of Drosophila melanogaster. J Mol Biol. 1974 Nov 25;90(1):51–64. doi: 10.1016/0022-2836(74)90255-1. [DOI] [PubMed] [Google Scholar]
- Nierhaus D., Nierhaus K. H. Identification of the chloramphenicol-binding protein in Escherichia coli ribosomes by partial reconstitution. Proc Natl Acad Sci U S A. 1973 Aug;70(8):2224–2228. doi: 10.1073/pnas.70.8.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubtsov P. M., Musakhanov M. M., Batchikova N. V., Skriabin K. S., Baev A. A. Opredelenie pervichnoi struktury fragmentov ribosomnogo operona pekarskikh drozhzhei, kodiruiushchikh 18 S rRNK. Dokl Akad Nauk SSSR. 1979;248(3):760–762. [PubMed] [Google Scholar]
- Santer M., Shane S. Area of 16S ribonucleic acid at or near the interface between 30S and 50S ribosomes of Escherichia coli. J Bacteriol. 1977 May;130(2):900–910. doi: 10.1128/jb.130.2.900-910.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. The use of nuclease P1 in sequence analysis of end group labeled RNA. Nucleic Acids Res. 1977 Dec;4(12):4091–4108. doi: 10.1093/nar/4.12.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert G., Fiers W. A micromethod for base analysis of 32P-labeled oligoribonulcleotides. Anal Biochem. 1977 Nov;83(1):222–227. doi: 10.1016/0003-2697(77)90530-9. [DOI] [PubMed] [Google Scholar]
- Volckaert G., Fiers W. Micro thin-layer techniques for rapid sequence analysis of 32P-labeled RNA: double digestion and pancreatic ribonuclease analyses. Anal Biochem. 1977 Nov;83(1):228–239. doi: 10.1016/0003-2697(77)90531-0. [DOI] [PubMed] [Google Scholar]
- Volckaert G., Jou W. M., Fiers W. Analysis of 32P-labeled bacteriophage MS2 RNA by a mini-fingerprinting procedure. Anal Biochem. 1976 May 7;72:433–446. doi: 10.1016/0003-2697(76)90551-0. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Fox G. E., Zablen L., Uchida T., Bonen L., Pechman K., Lewis B. J., Stahl D. Conservation of primary structure in 16S ribosomal RNA. Nature. 1975 Mar 6;254(5495):83–86. doi: 10.1038/254083a0. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Magrum L. J., Gupta R., Siegel R. B., Stahl D. A., Kop J., Crawford N., Brosius J., Gutell R., Hogan J. J. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980 May 24;8(10):2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zablen L., Woese C. R. Procaryote phylogeny IV: concerning the phylogenetic status of a photosynthetic bacterium. J Mol Evol. 1975 Jun 9;5(1):25–34. doi: 10.1007/BF01732011. [DOI] [PubMed] [Google Scholar]




