Abstract
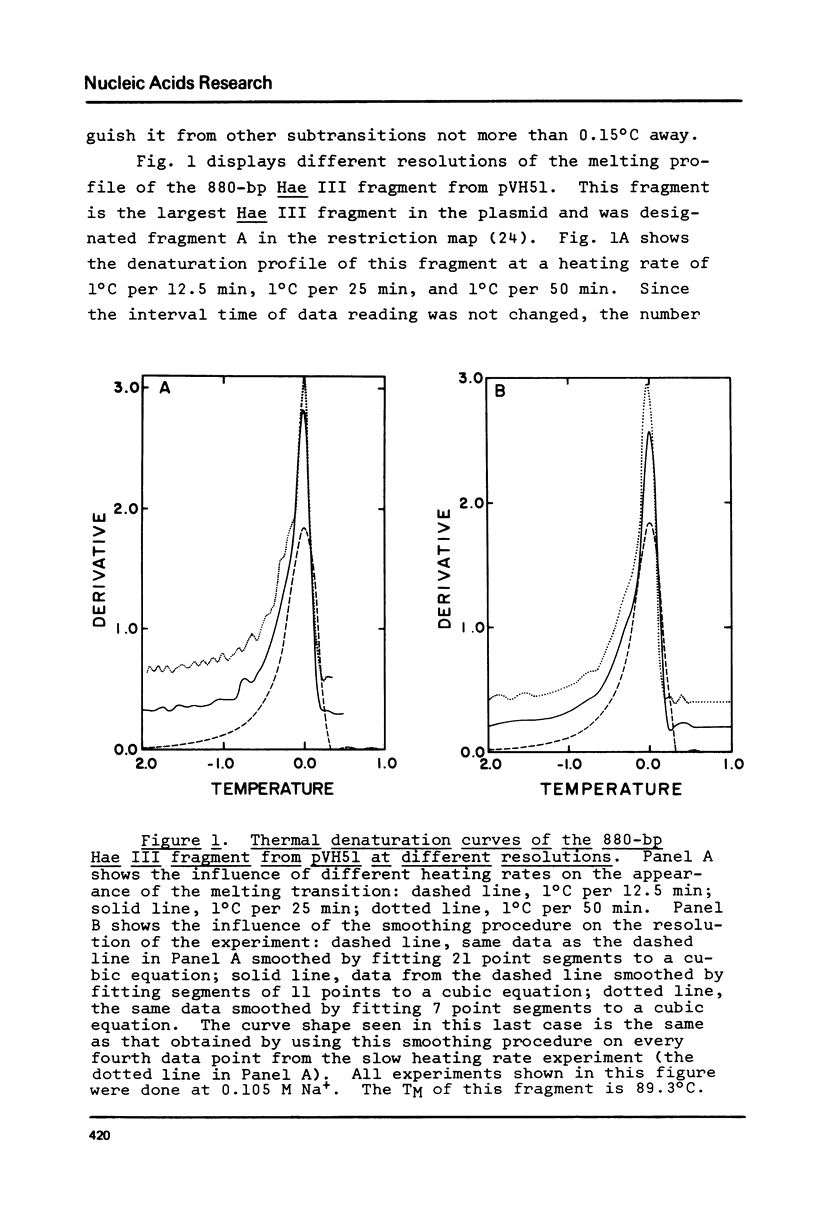
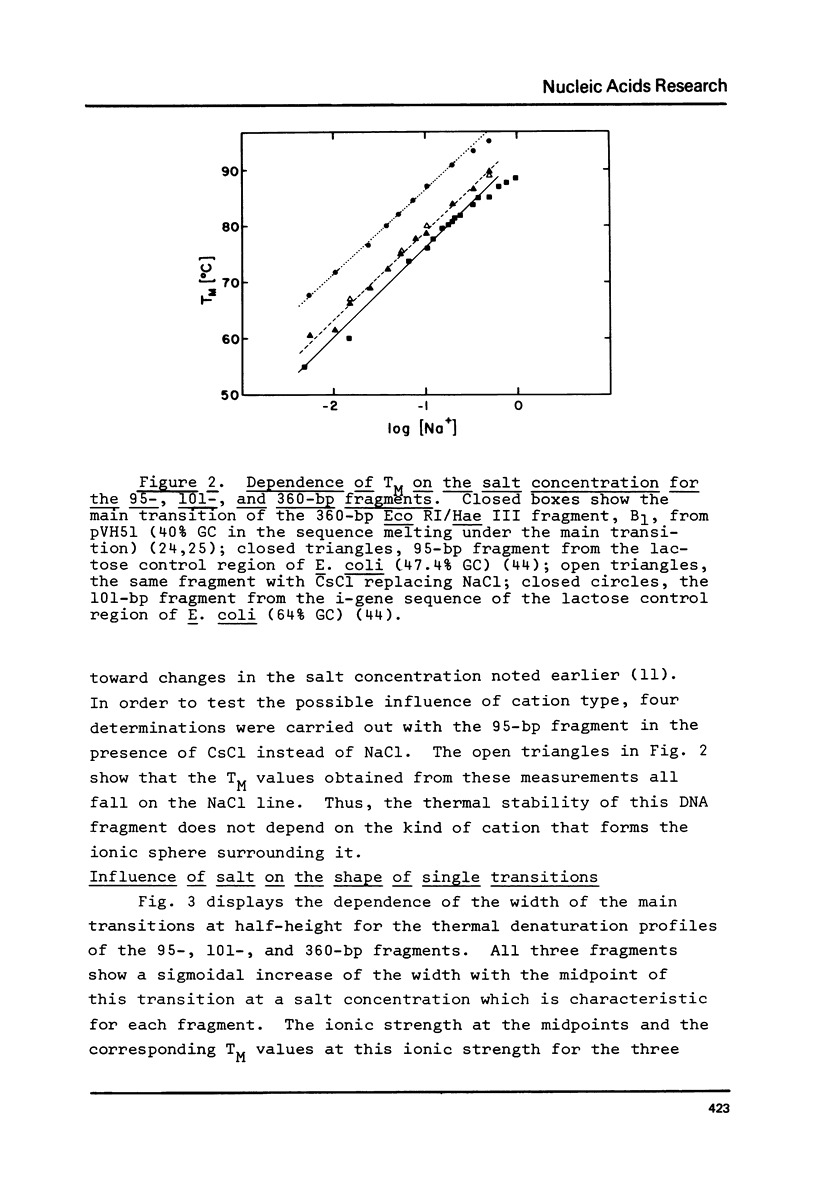
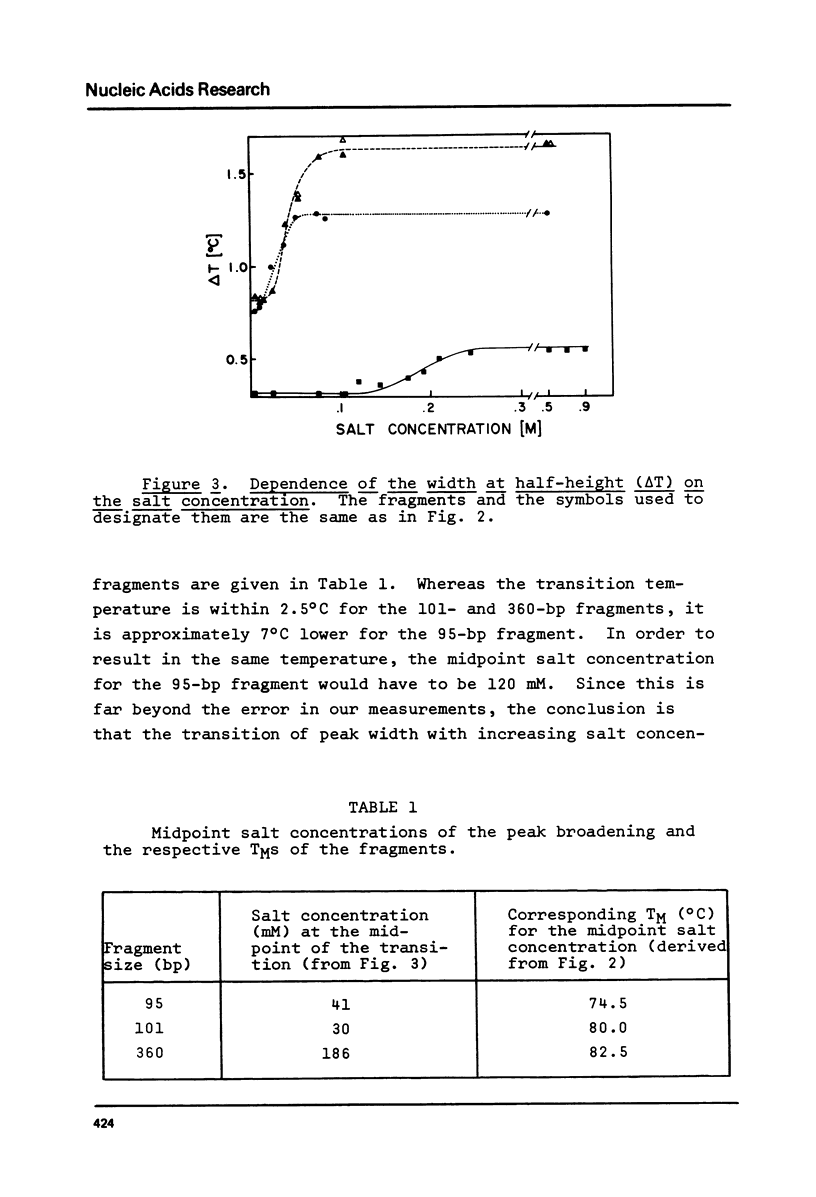
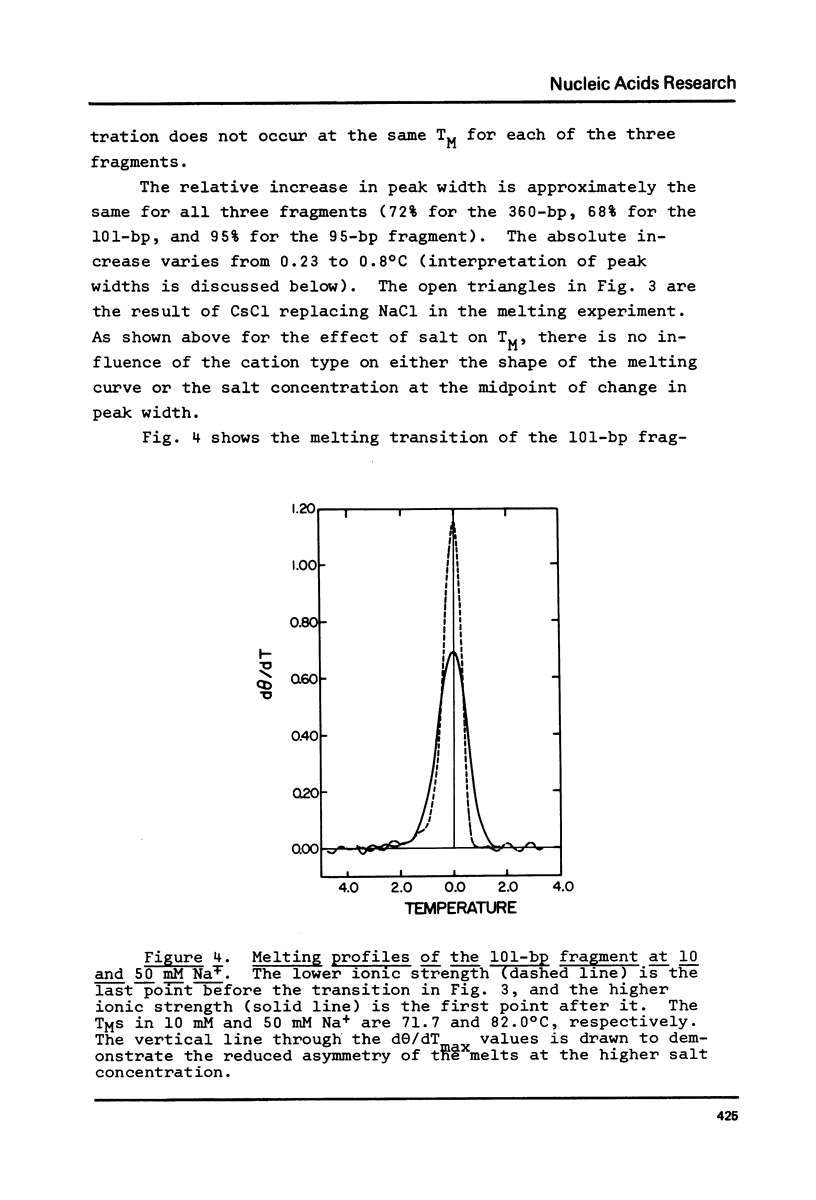
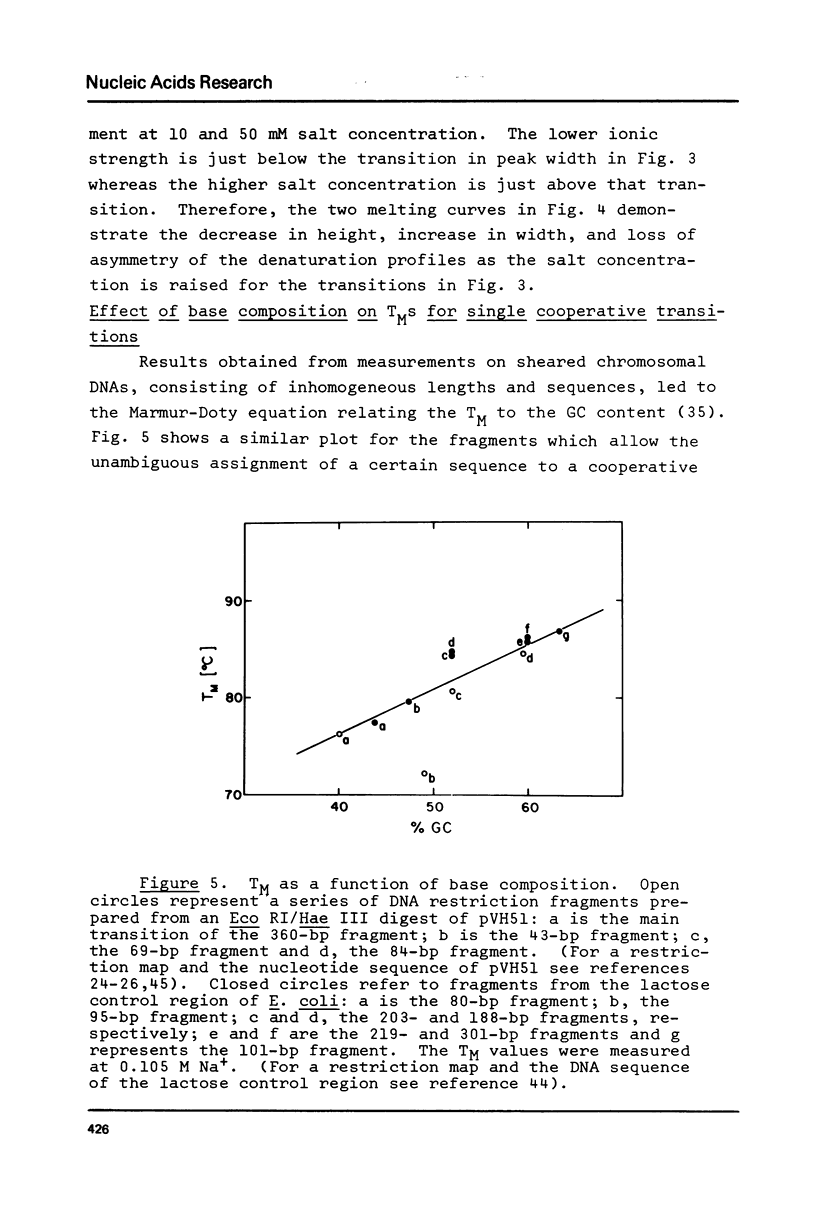
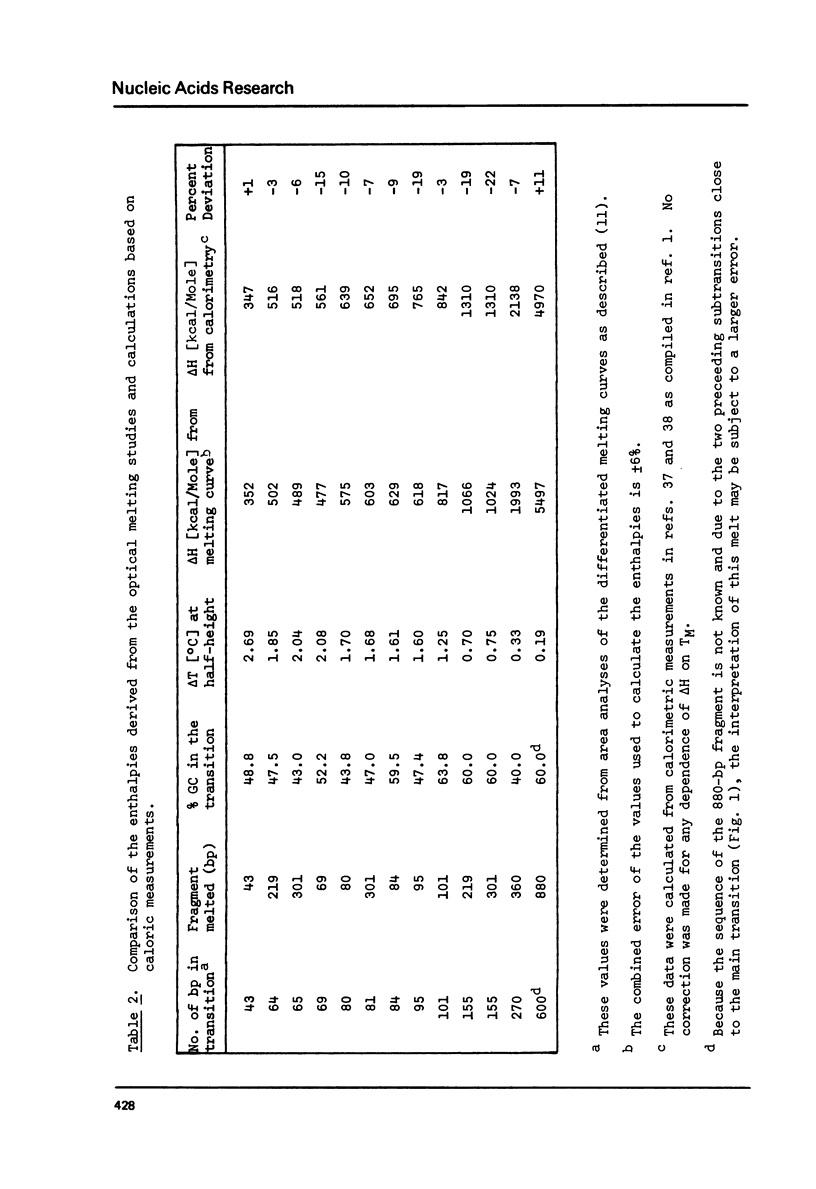
The influence of cation concentration on the thermal denaturation of DNA restriction fragments from the E. coli lac regulatory region and from pVH51, ranging in size from 43- to 880- bp, is described. Upon increasing the ionic strength, the melting transitions broaden in a cooperative manner at salt concentrations characteristic for the specific fragment. For three fragments studied in detail, the salt concentration dependence at the midpoint varied between 0.03 and 0.19 M Na+. Along with the broadening, the melting transitions become more symmetrical. This result is discussed with respect to the irreversibility of melting transitions at low ionic strength. After a cooperative broadening, the shape of the melting curves remains constant up to salt concentrations of 0.5 M Na+. The dTM/dlog[Na+] values for three fragments fall between 15.7 and 16.7. An easily applicable approximation of the van't Hoff equation is used to evaluate the enthalpies of 13 transitions arising from the denaturation of 43 to 600 bp. The results of this analysis are compared to calculations of the expected enthalpies based on calorimetric measurements. The TMs of most transitions were directly related to the base composition, but several deviations from the predicted behavior were observed. The possible influences of fragment length and sequence on the thermal stability are discussed. The experimental and mathematical procedure to resolve a thermal denaturation transition with a width f 0.17 +/- 0.01 degrees and its distinction from another preceeding transition only approximately 0.15 degrees away in an 880-bp Hae III fragment from pVH51 is described. This transition is about half as wide as the smallest one reported to date.
Full text
PDF















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ansevin A. T., Vizard D. L., Brown B. W., McConathy J. High-resolution thermal denaturation of DNA. I. Theoretical and practical considerations for the resolution of thermal subtransitions. Biopolymers. 1976 Jan;15(1):153–174. doi: 10.1002/bip.1976.360150111. [DOI] [PubMed] [Google Scholar]
- Azbel M. Y. DNA sequencing and melting curve. Proc Natl Acad Sci U S A. 1979 Jan;76(1):101–105. doi: 10.1073/pnas.76.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crothers D. M. Calculation of melting curves for DNA. Biopolymers. 1968 Oct;6(10):1391–1404. doi: 10.1002/bip.1968.360061003. [DOI] [PubMed] [Google Scholar]
- Dix D. E., Straus D. B. DNA helix stability. I. Differential stabilization by counter cations. Arch Biochem Biophys. 1972 Sep;152(1):299–310. doi: 10.1016/0003-9861(72)90219-6. [DOI] [PubMed] [Google Scholar]
- Eichinger B. E., Fixman M. Helix-coil transition in heterogeneous chains. II. DNA model. Biopolymers. 1970 Feb;9(2):205–221. doi: 10.1002/bip.1970.360090206. [DOI] [PubMed] [Google Scholar]
- Frank-Kamenetskii M. D., Vologodskii A. V. The nature of the fine structure of DNA melting curves. Nature. 1977 Oct 20;269(5630):729–730. doi: 10.1038/269729a0. [DOI] [PubMed] [Google Scholar]
- Gotoh O., Husimi Y., Yabuki S., Wada A. Hyperfine structure in melting profile of bacteriophage lambda DNA. Biopolymers. 1976 Apr;15(4):655–670. doi: 10.1002/bip.1976.360150406. [DOI] [PubMed] [Google Scholar]
- Gotoh O., Wada A., Yabuki S. Salt-concentration dependence of melting profiles of lambda phage DNAs: evidence for long-range interactions and pronounced end effects. Biopolymers. 1979 Apr;18(4):805–824. doi: 10.1002/bip.1979.360180406. [DOI] [PubMed] [Google Scholar]
- Grant R. C., Kodama M., Wells R. D. Enzymatic and physical studies on (dI-dC) n -(dI-dC) n and (dG-dC) n -(dG-dC) n . Biochemistry. 1972 Feb 29;11(5):805–815. doi: 10.1021/bi00755a020. [DOI] [PubMed] [Google Scholar]
- Gómez B., Lang D. Denaturation map of bacteriophage T7 DNA and direction of DNA transcription. J Mol Biol. 1972 Sep 28;70(2):239–251. doi: 10.1016/0022-2836(72)90536-0. [DOI] [PubMed] [Google Scholar]
- Hardies S. C., Hillen W., Goodman T. C., Wells R. D. High resolution thermal denaturation analyses of small sequenced DNA restriction fragments containing Escherichia coli lactose genetic control loci. J Biol Chem. 1979 Oct 25;254(20):10128–10134. [PubMed] [Google Scholar]
- Hardies S. C., Patient R. K., Klein R. D., Ho F., Reznikoff W. S., Wells R. D. Construction and mapping of recombinant plasmids used for the preparation of DNA fragments containing the Escherichia coli lactose operator and promoter. J Biol Chem. 1979 Jun 25;254(12):5527–5534. [PubMed] [Google Scholar]
- Hardies S. C., Wells R. D. Preparation and characterization of large amounts of restriction fragments containing the E. coli lac control elements. Gene. 1979 Sep;7(1):1–14. doi: 10.1016/0378-1119(79)90039-8. [DOI] [PubMed] [Google Scholar]
- Hershfield V., Boyer H. W., Chow L., Helinski D. R. Characterization of a mini-ColC1 plasmid. J Bacteriol. 1976 Apr;126(1):447–453. doi: 10.1128/jb.126.1.447-453.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff A. J., Roos A. L. Hysteresis of denaturation of DNA in the melting range. Biopolymers. 1972;11(6):1289–1294. doi: 10.1002/bip.1972.360110612. [DOI] [PubMed] [Google Scholar]
- Klump H., Ackermann T. Experimental thermodynamics of the helix--random coil transition. IV. Influence of the base composition of DNA on the transition enthalpy. Biopolymers. 1971;10(3):513–522. doi: 10.1002/bip.360100307. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Karlström O. Heat-induced depyrimidination of deoxyribonucleic acid in neutral solution. Biochemistry. 1973 Dec 4;12(25):5151–5154. doi: 10.1021/bi00749a020. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974 Jul 30;13(16):3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972 Sep 12;11(19):3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- Lyubchenko Y. L., Vologodskii A. V., Frank-Kamenetskii M. D. Direct comparison of theoretical and experimental melting profiles for RF II phiX174 DNA. Nature. 1978 Jan 5;271(5640):28–31. doi: 10.1038/271028a0. [DOI] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori H., Tomizawa J. Nucleotide sequence of the region required for maintenance of colicin E1 plasmid. Mol Gen Genet. 1979 Oct 3;176(2):161–170. doi: 10.1007/BF00273210. [DOI] [PubMed] [Google Scholar]
- Oka A., Nomura N., Morita M., Sugisaki H., Sugimoto K., Takanami M. Nucleotide sequence of small ColE1 derivatives: structure of the regions essential for autonomous replication and colicin E1 immunity. Mol Gen Genet. 1979 May 4;172(2):151–159. doi: 10.1007/BF00268276. [DOI] [PubMed] [Google Scholar]
- Scheffler I. E., Sturtevant J. M. Thermodynamics of the helix-coil transition of the alternating copolymer of deoxyadenylic acid and deoxythymidylic acid. J Mol Biol. 1969 Jun 28;42(3):577–580. doi: 10.1016/0022-2836(69)90244-7. [DOI] [PubMed] [Google Scholar]
- Tachibana H., Wada A., Gotoh O., Takanami M. Location of the cooperative melting regions in bacteriophage fd DNA. Biochim Biophys Acta. 1978 Feb 16;517(2):319–328. doi: 10.1016/0005-2787(78)90198-3. [DOI] [PubMed] [Google Scholar]
- Vizard D. L., Ansevin A. T. High resolution thermal denaturation of DNA: thermalites of bacteriophage DNA. Biochemistry. 1976 Feb 24;15(4):741–750. doi: 10.1021/bi00649a004. [DOI] [PubMed] [Google Scholar]
- Wada A., Tachibana H., Gotoh O., Takanami M. Long range homogeneity of physical stability in double-stranded DNA. Nature. 1976 Sep 30;263(5576):439–440. doi: 10.1038/263439a0. [DOI] [PubMed] [Google Scholar]
- Wada A., Tachibana H., Ueno S., Husimi Y., Machida Y. Melting fine structure of DNA fragments of known base sequence from theta X174. Nature. 1977 Sep 22;269(5626):352–353. doi: 10.1038/269352a0. [DOI] [PubMed] [Google Scholar]
- Wada A., Ueno S., Tachibana H., Husimi Y. Stability mapping along the DNA double strand and its relation to the genetic map. J Biochem. 1979 Mar;85(3):827–832. [PubMed] [Google Scholar]
- Wells R. D., Blakesley R. W., Hardies S. C., Horn G. T., Larson J. E., Selsing E., Burd J. F., Chan H. W., Dodgson J. B., Jensen K. F. The role of DNA structure in genetic regulation. CRC Crit Rev Biochem. 1977;4(3):305–340. doi: 10.3109/10409237709102561. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Goodman T. C., Hillen W., Horn G. T., Klein R. D., Larson J. E., Müller U. R., Neuendorf S. K., Panayotatos N., Stirdivant S. M. DNA structure and gene regulation. Prog Nucleic Acid Res Mol Biol. 1980;24:167–267. doi: 10.1016/s0079-6603(08)60674-1. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Hardies S. C., Horn G. T., Klein B., Larson J. E., Neuendorf S. K., Panayotatos N., Patient R. K., Selsing E. RPC-5 column chromatography for the isolation of DNA fragments. Methods Enzymol. 1980;65(1):327–347. doi: 10.1016/s0076-6879(80)65043-5. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Larson J. E., Grant R. C., Shortle B. E., Cantor C. R. Physicochemical studies on polydeoxyribonucleotides containing defined repeating nucleotide sequences. J Mol Biol. 1970 Dec 28;54(3):465–497. doi: 10.1016/0022-2836(70)90121-x. [DOI] [PubMed] [Google Scholar]



