Abstract
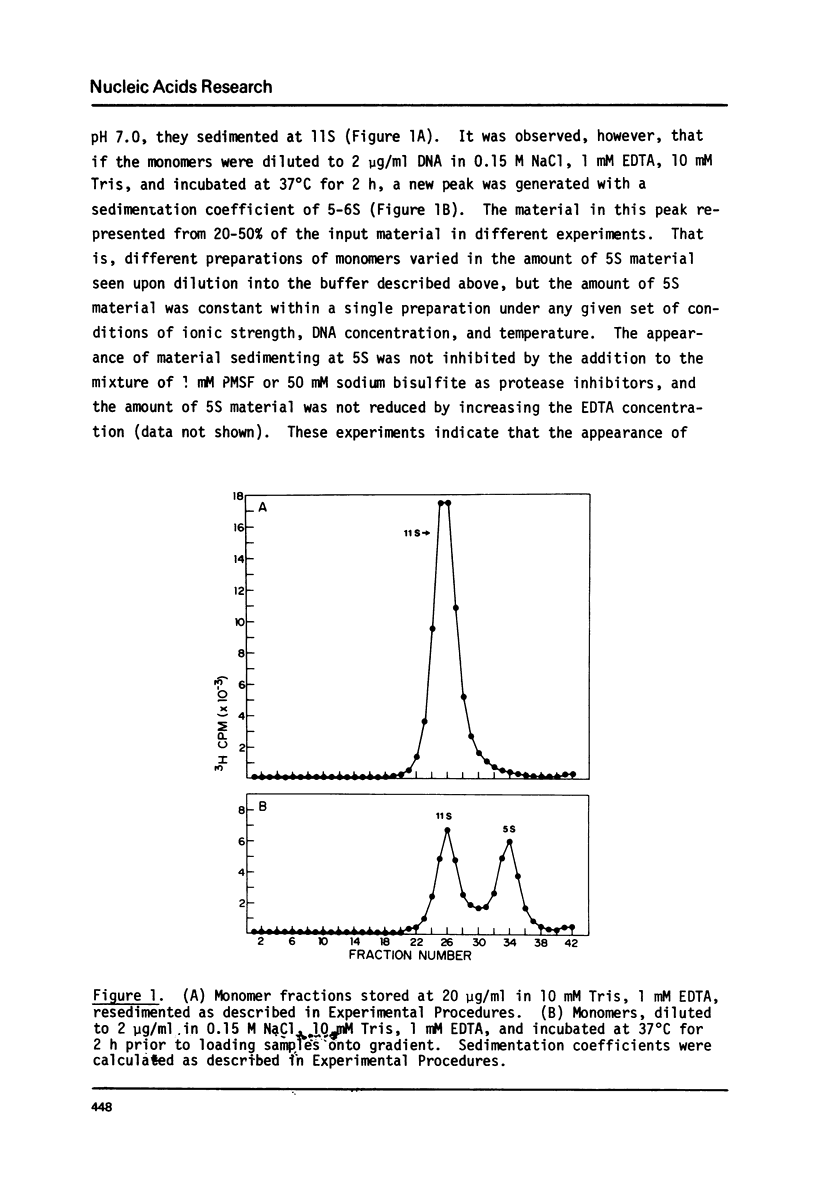
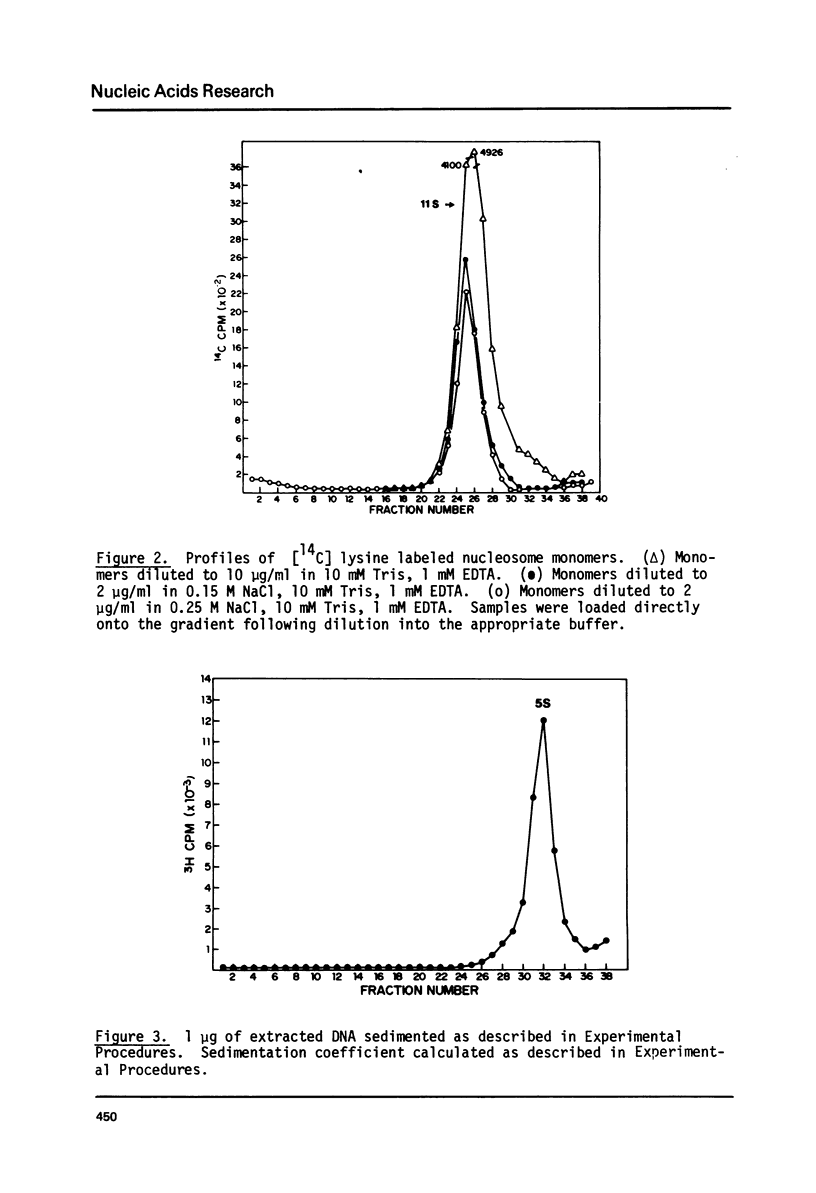
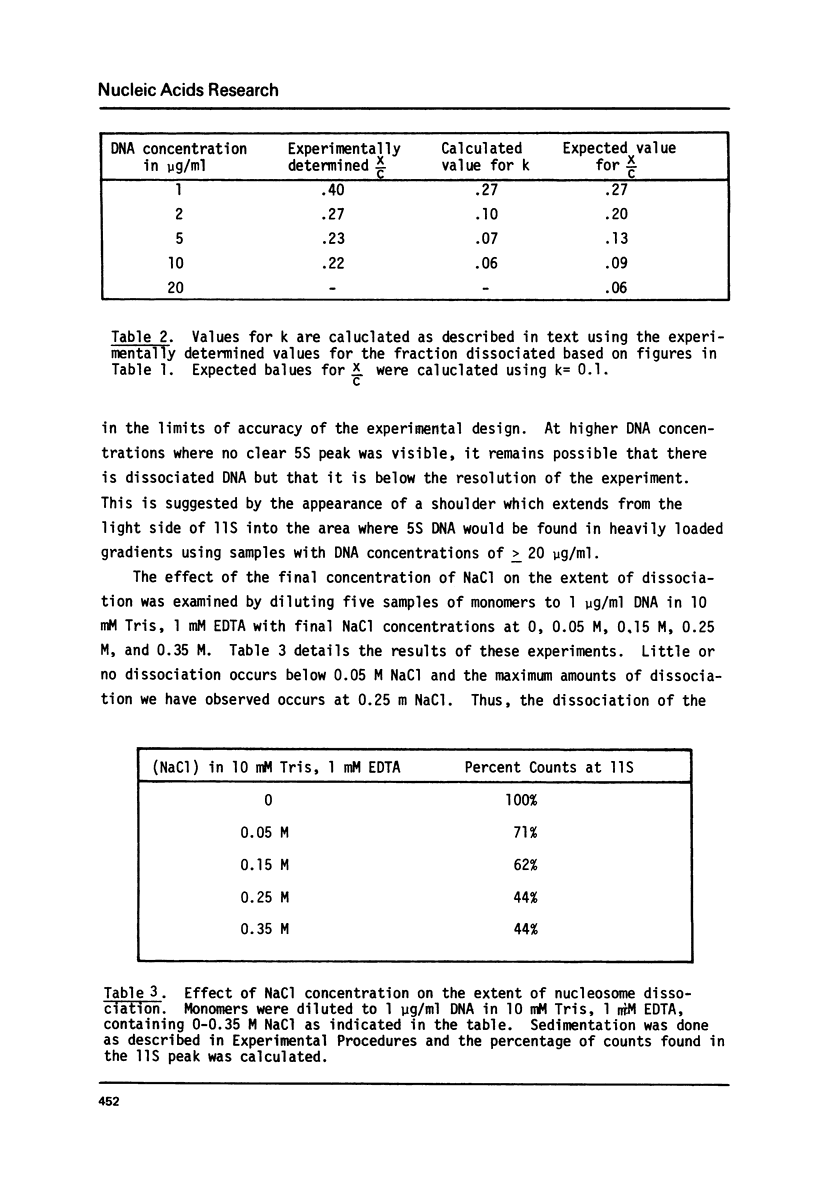
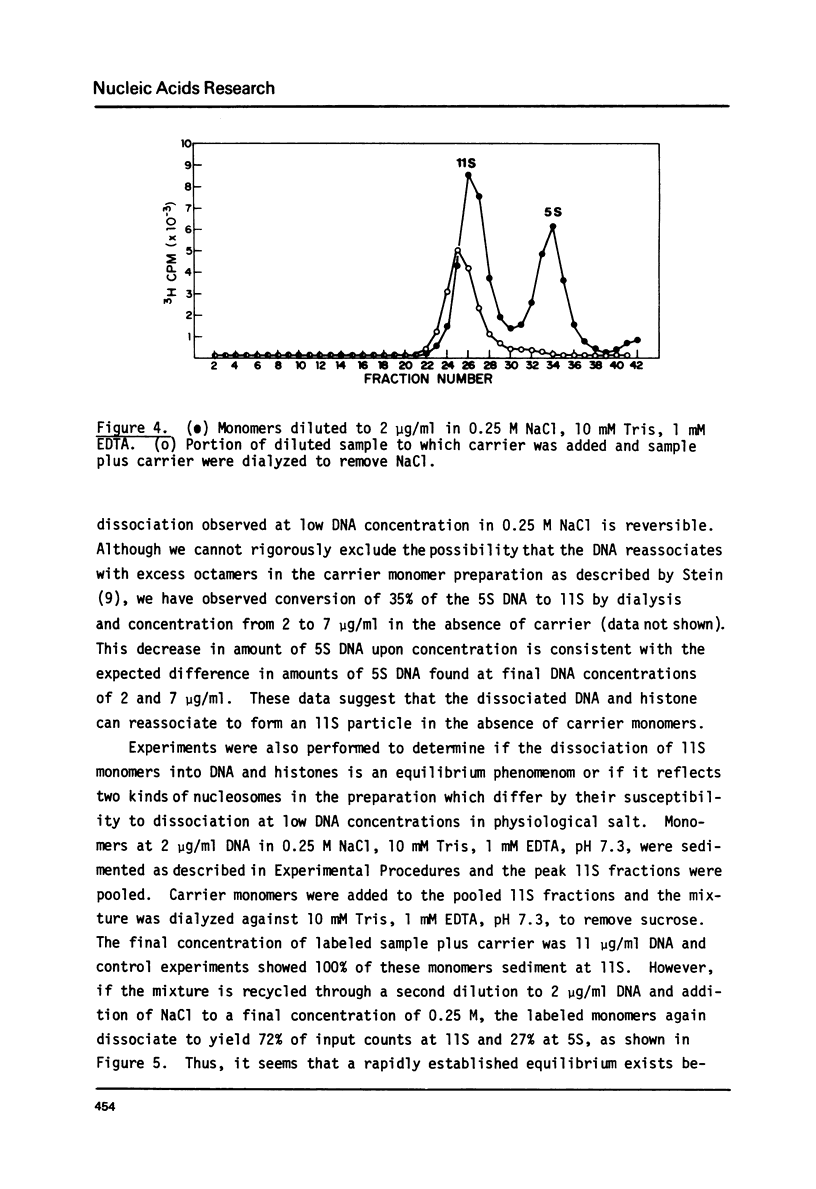
Monomer nucleosomes purified on isokinetic sucrose gradients are shown to dissociate into component DNA and histones at physiological ionic strength upon dilution to a DNA concentration below 20 microgram/ml. The starting material is 11S, contains 145-190 BP DNA, and equimolar amounts of the four core histones with slightly less H1. Dilution of monomers in the presence of 0.14 M NaCl results in the rapid conversion of 10-40% of the 3H thymidine labeled material from 11S to 5S (5S is coincident with the S value of monomer length DNA). The proportion of nucleosomes which dissociate increases with increasing NaCl concentration between 0.15 M and 0.35 M and decreases with increasing DNA concentration above 1 microgram/ml. Recycling 11S monomers, which remain after dissociation, through a second dilution in salt generates an equivalent proportion of 5S material as seen after the initial dilution. Thus, the dissociation does not result from special properties of a subset of nucleosomes. An equilibrium between intact monomer and free DNA and histones appears to be rapidly established under the conditions described and the dissociated DNA will reassociate with histones to form 11S monomers if conditions of high DNA concentration and low ionic strength are established.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carter C. W., Jr, Levinger L. F. Self redigestion of native chromatin. Biochem Biophys Res Commun. 1977 Feb 7;74(3):955–960. doi: 10.1016/0006-291x(77)91611-4. [DOI] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Isenberg I. Histones. Annu Rev Biochem. 1979;48:159–191. doi: 10.1146/annurev.bi.48.070179.001111. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Jacobs M. F., Houghton M. The nature of the interaction of nucleosomes with a eukaryotic RNA polymerase II. Nucleic Acids Res. 1979 Sep 25;7(2):377–399. doi: 10.1093/nar/7.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty K. S., Jr, Vollmer R. T., McCarty K. S. Improved computer program data for the resolution and fractionation of macromolecules by isokinetic sucrose density gradient sedimentation. Anal Biochem. 1974 Sep;61(1):165–183. doi: 10.1016/0003-2697(74)90343-1. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Stein A. DNA folding by histones: the kinetics of chromatin core particle reassembly and the interaction of nucleosomes with histones. J Mol Biol. 1979 May 15;130(2):103–134. doi: 10.1016/0022-2836(79)90421-2. [DOI] [PubMed] [Google Scholar]



