Abstract
The 5'-terminal of Saccharomyces cerevisiae 18 S and 25 S rRNA are precisely mapped within the sequence of the rDNA repeating unit. The 3'-terminal of 25 S rRNA and 37 S pre-rRNA are located within a 548 bp segment of the rDNA repeating unit by the use of a DNA polymerase I extension technique. The analysis of the rDNA sequences at the structural gene boundaries reveals the presence of oligonucleotide repeats which may be involved in transcription or processing control mechanisms. The sequence of rDNA in the transcription termination region is determined and possible mechanisms shaping the 3'-end of 25 S rRNA are discussed.
Full text
PDF

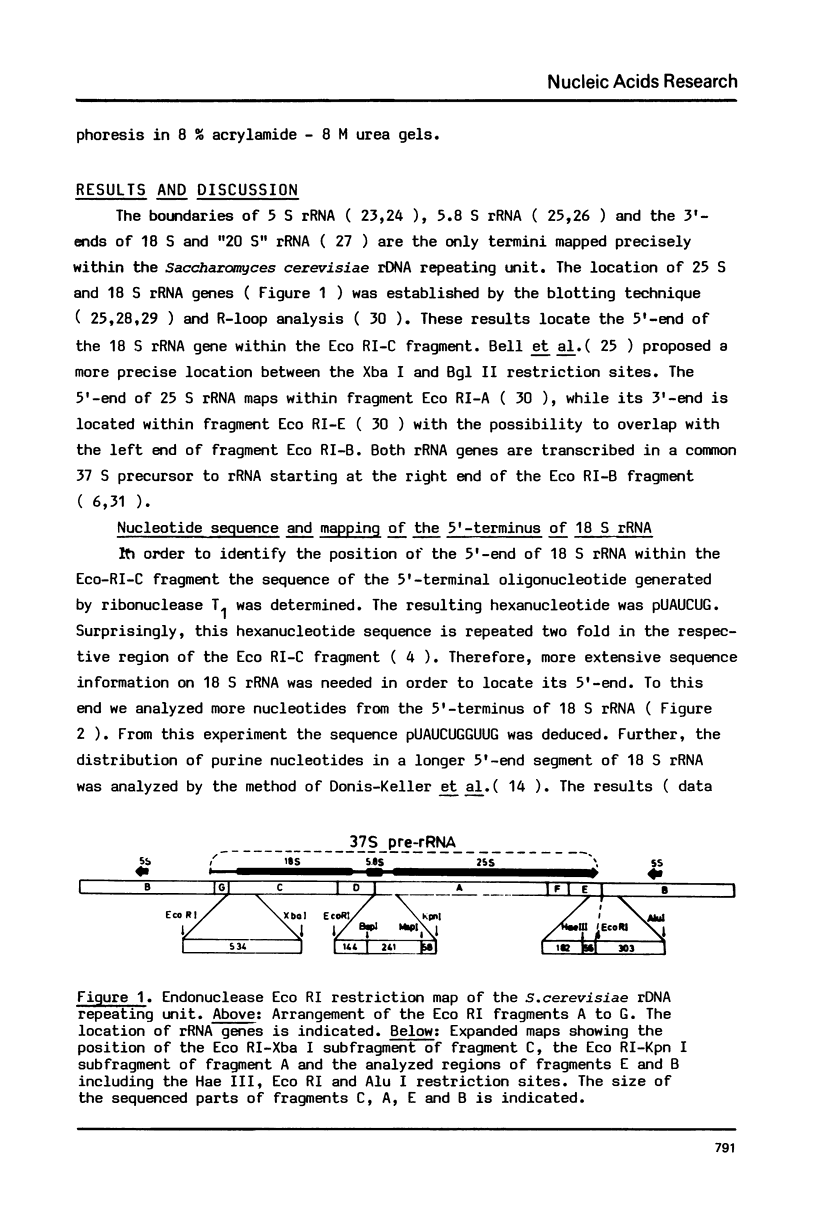


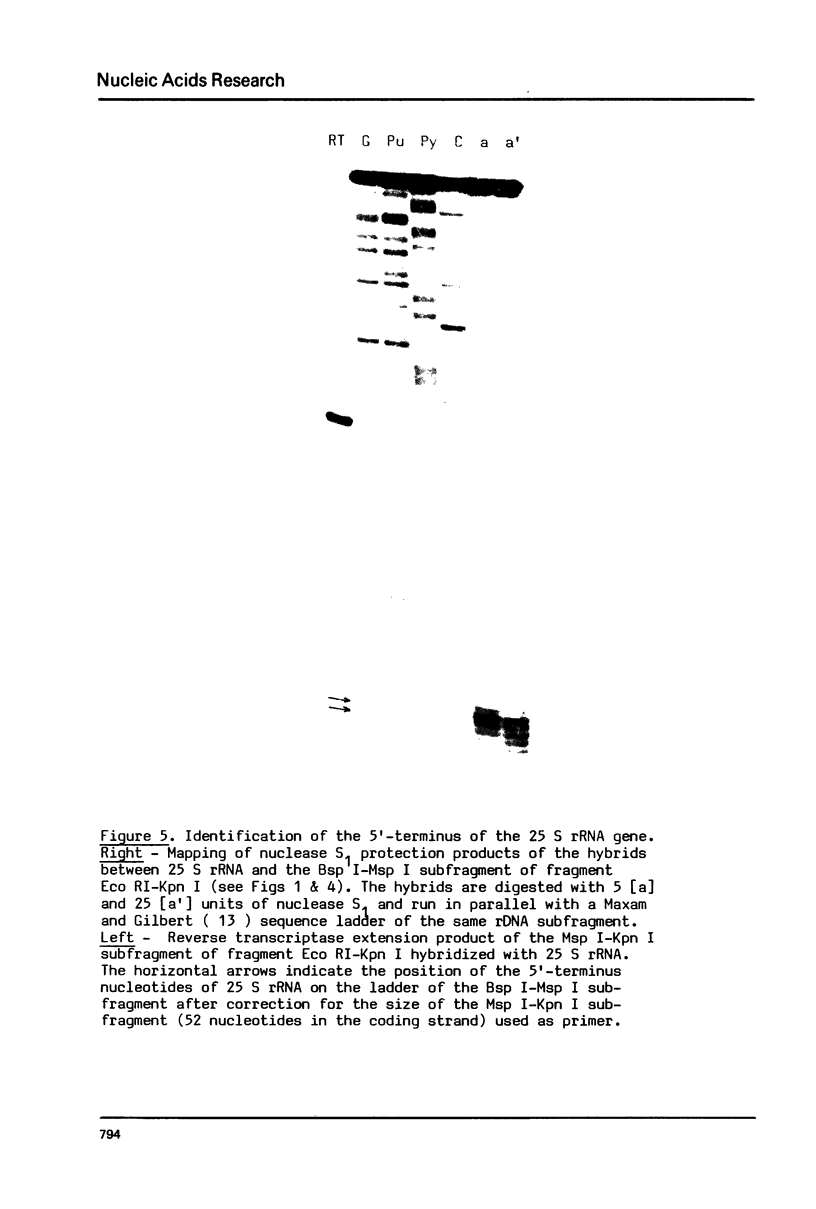





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Bayev A. A., Georgiev O. I., Hadjiolov A. A., Kermekchiev M. B., Nikolaev N., Skryabin K. G., Zakharyev V. M. The structure of the yeast ribosomal RNA genes. 2. The nucleotide sequence of the initiation site for ribosomal RNA transcription. Nucleic Acids Res. 1980 Nov 11;8(21):4919–4926. doi: 10.1093/nar/8.21.4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. I., DeGennaro L. J., Gelfand D. H., Bishop R. J., Valenzuela P., Rutter W. J. Ribosomal RNA genes of Saccharomyces cerevisiae. I. Physical map of the repeating unit and location of the regions coding for 5 S, 5.8 S, 18 S, and 25 S ribosomal RNAs. J Biol Chem. 1977 Nov 25;252(22):8118–8125. [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- De Jonge P., Klootwijk J., Planta R. J. Terminal nucleotide sequences of 17-S ribosomal RNA and its immediate precursor 18-S RNA in yeast. Eur J Biochem. 1977 Jan;72(2):361–369. doi: 10.1111/j.1432-1033.1977.tb11260.x. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn I. M., Chappell J. B. A simple method for the preparation of 32-P-labelled adenosine triphosphate of high specific activity. Biochem J. 1964 Jan;90(1):147–149. doi: 10.1042/bj0900147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolov A. A. Biogenesis of ribosomes in eukaryotes. Subcell Biochem. 1980;7:1–80. doi: 10.1007/978-1-4615-7948-9_1. [DOI] [PubMed] [Google Scholar]
- Hadjiolov A. A., Nikolaev N. Maturation of ribosomal ribonucleic acids and the biogenesis of ribosomes. Prog Biophys Mol Biol. 1976;31(2):95–144. doi: 10.1016/0079-6107(78)90006-8. [DOI] [PubMed] [Google Scholar]
- Jay E., Bambara R., Padmanabhan R., Wu R. DNA sequence analysis: a general, simple and rapid method for sequencing large oligodeoxyribonucleotide fragments by mapping. Nucleic Acids Res. 1974 Mar;1(3):331–353. doi: 10.1093/nar/1.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemenz R., Geiduschek E. P. The 5' terminus of the precursor ribosomal RNA of Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Jun 25;8(12):2679–2689. doi: 10.1093/nar/8.12.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S., Sures I., Kedes L. H. Sequence of the 5'-end of Strongylocentrotus purpuratus H2b histone mRNA and its location within histone DNA. Nature. 1979 Jun 21;279(5715):737–739. doi: 10.1038/279737a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Tizard R., Skryabin K. G., Gilbert W. Promotor region for yeast 5S ribosomal RNA. Nature. 1977 Jun 16;267(5612):643–645. doi: 10.1038/267643a0. [DOI] [PubMed] [Google Scholar]
- Moss T., Birnstiel M. L. The putative promoter of a Xenopus laevis ribosomal gene is reduplicated. Nucleic Acids Res. 1979 Aug 24;6(12):3733–3743. doi: 10.1093/nar/6.12.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath K., Bollon A. P. Generation of discrete yeast DNA fragments by endonuclease RI. Nature. 1975 Sep 11;257(5522):155–157. doi: 10.1038/257155a0. [DOI] [PubMed] [Google Scholar]
- Nikolaev N., Georgiev O. I., Venkov P. V., Hadjiolov A. A. The 37 S precursor to ribosomal RNA is the primary transcript of ribosomal RNA genes in Saccharomyces cerevisiae. J Mol Biol. 1979 Jan 25;127(3):297–308. doi: 10.1016/0022-2836(79)90331-0. [DOI] [PubMed] [Google Scholar]
- Peters G. G., Hayward R. S. Extension of RNA molecules with DNA. An approach to the study of termination sequences recognised by RNA polymerase. Eur J Biochem. 1972 Dec 4;31(2):360–366. doi: 10.1111/j.1432-1033.1972.tb02541.x. [DOI] [PubMed] [Google Scholar]
- Petes T. D., Hereford L. M., Skryabin K. G. Characterization of two types of yeast ribosomal DNA genes. J Bacteriol. 1978 Apr;134(1):295–305. doi: 10.1128/jb.134.1.295-305.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen P., Thomas M., Kramer R. A., Davis R. W. Unique arrangement of coding sequences for 5 S, 5.8 S, 18 S and 25 S ribosomal RNA in Saccharomyces cerevisiae as determined by R-loop and hybridization analysis. J Mol Biol. 1978 Aug 15;123(3):387–404. doi: 10.1016/0022-2836(78)90086-4. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Rubtsov P. M., Musakhanov M. M., Zakharyev V. M., Krayev A. S., Skryabin K. G., Bayev A. A. The structure of the yeast ribosomal RNA genes. I. The complete nucleotide sequence of the 18S ribosomal RNA gene from Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Dec 11;8(23):5779–5794. doi: 10.1093/nar/8.23.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Brownlee G. G., Barrell B. G. A two-dimensional fractionation procedure for radioactive nucleotides. J Mol Biol. 1965 Sep;13(2):373–398. doi: 10.1016/s0022-2836(65)80104-8. [DOI] [PubMed] [Google Scholar]
- Shine J., Hunt J. A., Dalgarno L. Studies on the 3'-terminal sequences of the large ribosomal ribonucleic acid of different eukaryotes and those associated with "hidden" breaks in heart-dissociable insects 26S ribonucleic acid. Biochem J. 1974 Sep;141(3):617–625. doi: 10.1042/bj1410617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skriabin K. G., Kraev A. S., Rubtsov P. M., Baev A. A. Polnaia posledovatel'nost' nukleotidov speisernoi oblasti, raspolozhennoi mezhdu genami 18S i 5.8S RNK drozhzhei. Dokl Akad Nauk SSSR. 1979;247(3):761–765. [PubMed] [Google Scholar]
- Skriabin K. G., Zakhar'ev V. M., Rubtsov P. M., Baev A. A. Posledovatel'nost' nukleotidov predpolagaemoi oblasti initsiatsii transkriptsii ribosomnogo operona drozhzhei. Dokl Akad Nauk SSSR. 1979;247(5):1275–1277. [PubMed] [Google Scholar]
- Skryabin K. G., Maxam A. M., Petes T. D., Hereford L. Location of the 5.8S rRNA gene of Saccharomyces cerevisiae. J Bacteriol. 1978 Apr;134(1):306–309. doi: 10.1128/jb.134.1.306-309.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Sugiura M., Takanami M. Analysis of the 5'-terminal nucleotide sequences of ribonucleic acids. II. Comparison of the 5'-terminal nucleotide sequences of ribosomal RNA's from different organisms. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1595–1602. doi: 10.1073/pnas.58.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Weisblum B. Construction of a colicin E1-R factor composite plasmid in vitro: means for amplification of deoxyribonucleic acid. J Bacteriol. 1975 Jan;121(1):354–362. doi: 10.1128/jb.121.1.354-362.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela P., Bell G. I., Venegas A., Sewell E. T., Masiarz F. R., DeGennaro L. J., Weinberg F., Rutter W. J. Ribosomal RNA genes of Saccharomyces cerevisiae. II. Physical map and nucleotide sequence of the 5 S ribosomal RNA gene and adjacent intergenic regions. J Biol Chem. 1977 Nov 25;252(22):8126–8135. [PubMed] [Google Scholar]
- Veldman G. M., Brand R. C., Klootwijk J., Planta R. Some characteristics of processing sites in ribosomal precursor RNA of yeast. Nucleic Acids Res. 1980 Jul 11;8(13):2907–2920. doi: 10.1093/nar/8.13.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkov P. V., Hadjiolov A. A., Battaner E., Schlessinger D. Saccharomyces cerevisiae: sorbitol-dependent fragile mutants. Biochem Biophys Res Commun. 1974 Feb 4;56(3):599–604. doi: 10.1016/0006-291x(74)90646-9. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Flügel R. M., Larson J. E., Schendel P. F., Sweet R. W. Comparison of some reactions catalyzed by deoxyribonucleic acid polymerase from avian myeloblastosis virus, Escherichia coli, and Micrococcus luteus. Biochemistry. 1972 Feb 15;11(4):621–629. doi: 10.1021/bi00754a025. [DOI] [PubMed] [Google Scholar]