Abstract
Genetic relationships between human groups were first studied by comparisons of relative allele frequency at multiple loci. Geographical study of detailed, highly resolved trees of single, non-recombining uniparental loci (mitochondrial DNA: mtDNA and Y chromosome/non-recombining Y: NRY), following specific lineages rather than populations, then revolutionized knowledge of the peopling of the world, although, curiously, the use of geographically highly specific mutations that protect against malaria, found on individual autosomal globin genes, were first in single-locus phylogeography. mtDNA, with its high single nucleotide polymorphism (SNP) mutation rates and relative ease of dating, led the way and gave stronger proof of the recent near replacement of all human species by anatomically modern humans (AMH). AMH left Africa via a single southern exit about 70 000 years ago and rapidly spread around the Indian Ocean towards the Antipodes, long before a small branch left a South Asian colony, earlier on the trail, to populate Europe. The worldwide skeleton phylogeny of mtDNA is fully resolved, but a regional analysis will continue to illuminate subsequent migrations. NRY with a lower SNP mutation rate still has a dating problem relating to use the of single tandem repeats (STRs), but has validated mtDNA results and with more geographical specificity and genomic size, as with the autosomal human genome, has much more detail to offer for the future.
Keywords: Out-of-Africa, migration, genetic phylogeography, climate, chronology, mitochondrial DNA
1. Introduction
Genetic relationships between human groups were first studied by comparisons of relative allele frequencies at multiple loci. This population-based approach had problems of resolution and dating, but results were largely consistent with an African ancestry for anatomically modern humans (AMH). The lineage-based approach—which involved phylogeographic analysis of uniparental non-recombining DNA, based on regionally specific, highly detailed, reconstructed gene trees—has illuminated detailed geography of migrations—in particular, the exit route of AMH from Africa. It should be recalled that the use of geographically highly specific mutations on single-locus phylogenies, to trace migrations, started with autosomes—for instance, the alpha- and beta-globin loci.
Following the discovery of a high rate of α+ (single deletion) thalassaemia in the Pacific [1], protecting against and selected for by malaria [1–5], specific deletional variants were shown to link Melanesia to Polynesia [6], tracing an ancient, staged migration that was only recently fully elucidated using mitochondrial DNA (mtDNA) [7]. Samples collected in the early 1980s for these studies, by this author and others, were used to construct a worldwide beta-globin locus phylogeny, based on population genetic distance analysis of restriction fragment length polymorphism (RFLP) haplotypes, which unambiguously resolved Africans as the deepest single ancestral branch from which all non-African groups arise [8], just scooping the famous ‘mitochondrial Eve’ paper [9] by one year.
The finding of a greater genetic diversity within Africa, when compared with outside, is now abundantly supported by many genetic markers; so Africa is the most likely geographical origin for a modern human dispersal. There still are, however, four basic questions essential in any reconstruction of the African Exodus, in the following logical order: first, how many founding exits of AMH can be seen in the fossil or archaeological record? Second, which of these are evidenced genetically by continuity into modern regional populations (rather than by evanescent interglacial spreads with other fauna in the fossil record)? Third and fourth, which route(s) were taken and when? Two further questions ride on these last: how and why?
2. Regional setting
In geographical terms, there is a growing consensus on a single southern dispersal of AMH extending from Africa via the mouth of the Red Sea moving around the coasts of the Indian Ocean initially to Bali, but ultimately to Melanesia and Australia in the southwest Pacific, and to the Americas on the other side (figure 1). The argument for the single exit [11,12] rather than multiple exits [13,14] is mainly based on the finding of single lineages, initially mtDNA and the Y chromosome non-recombining region (NRY), but now shown in the X chromosome and some autosomes, representing the entire non-African genetic diversity [15]. Recent claims for extra AMH exits at the Eemian interglacial [16–18] need to be viewed in this context.
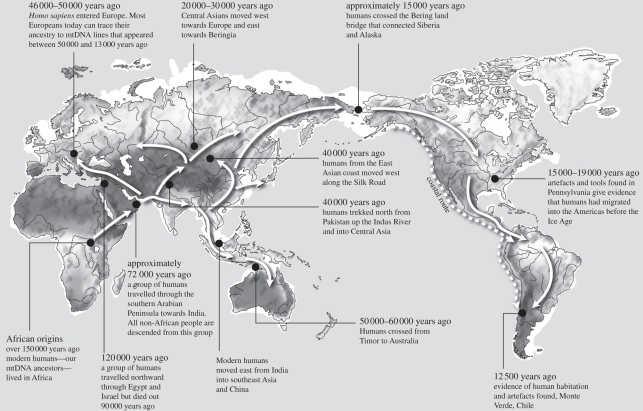
Figure 1.
Narrative map of modern human dispersals, as reconstructed in this review. Adapted from Oppenheimer [10].
The concept that modern human range extension out of Africa proceeded initially along coastlines has depended largely on the rapidity of this movement, as inferred from genetic phylogeography, because only three key founder mtDNA haplotypes (M, N and R) give rise to multiple regionally specific branches en route, in effect, three regional starbursts, found all the way from Arabia to Bali (e.g. fig. 6 in Soares et al. [7]). For this to be possible, these explorers must have been able to exploit coastal marine resources. Shellfishing, starting from at least 160 000 years ago (reviewed by McBrearty & Stringer [19]), was limited to the intertidal zone, and would be over-exploited, necessitating continuous extension along the beach. This beachcombing model provides an immediate and continuous motive for unidirectional, linear migration. Evidence of marine exploitation is found at the very earliest occupation sites in Australia and most significantly at the Australian threshold dating to greater than 42 ka cal. BP in Jerimalai in East Timor [20].
3. Material and methods used in review of genetic phylogeography
(a). Phylogeography
The approach of genetic phylogeography (literally and graphically referring to ‘laying the gene tree on the map’—for a description of the method of founder analysis, see Richards et al. [21]) is particularly useful for answering topographic questions of migration and route but, although used extensively for dating migrations, has less precision for that purpose. Conversely, the disciplines of archaeology, palaeoanthropology and the earth sciences, including palaeoclimatology, are most useful for testing dating regimes and routes suggested by the genetic models. A combined approach should give increased confidence by triangulation.
The study of the geographical spread of gene lines uses three related types of inference: first, a gene tree (figure 2) is built up by mutations in the same stretch of DNA as branch points (preferably non-recombining, e.g. mtDNA and NRY [11,15,22]), from representative population samples. Such trees, if based on non-recombining DNA, are unambiguous and possess great topographic detail and geographical specificity. Second, migrations are inferred, based on the geographical distribution of individual gene branches. Third, the diversity of clusters of gene lines is used to infer time depth of individual branches. These three forms of phylogeographic inference are combined in founder analysis, which is the identification and dating of specific founder lines moving from a source to a target region. A common misunderstanding of dating the arrival of founder lineages in a new region is that carried-over diversity is dated as well, thus giving a falsely older date. On the contrary, founder analysis formally excludes such errors by making a detailed comparison with source populations, thus using only estimates of founding dates on the basis of unique new diversity arising at the target [21].
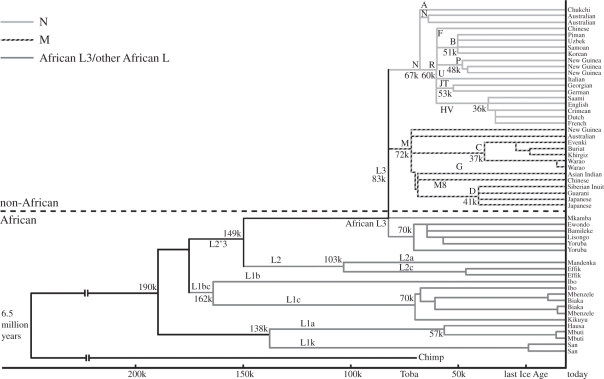
Figure 2.
Phylogeny based on the first mtDNA complete sequence data available in 2000 (from 52 individuals randomly selected around the world). Original branch-date calibration by author. Structure of tree remains essentially the same today. Adapted from fig. 0.3 [10,11]. Note the chimpanzee–human coalescent date derives from analysis of coding region, i.e. not assumed from fossil evidence.
(b). Complete sequence data: sources, phylogeny, calibration and dating method
Dating of genetic phylogenies and founding events has been central to such discussions of models of human migration, and some form of calibration is essential. Most archaeological and genetic models estimate the exit of AMH from Africa at well over 40 ka, thus outside the limits of most carbon dating. There is still rearguard dispute over the accuracy and precision of luminescence studies, which offer to extend geological dating of human activity much further back. The great Toba volcanic eruption in Sumatra (which is very accurately dated at 74 ka), which left its ash-mark around the Indian Ocean and even in the Greenland Icecap, has offered an alternative and tempting older date marker to archaeologists and geneticists and still does.
Genetic dating, if not in its infancy, has only really been of value for less than 30 years and has advanced most for the mitochondrial locus. Formerly, the short hypervariable regions (HVS1–2) of the mtDNA genome offered a large amount of information for relatively little sequencing and, being non-coding, avoided issues of selection. HVS1 sequences still represent over 95 per cent of published mtDNA data and, when combined with key RFLP sites, still give good phylogenetic resolution and phylogeographic information. However, some lack of resolution and network reticulation, owing to homoplasy, still hampers detailed worldwide and regional phylogeography and has raised questions over dating precision. Since the first 52 complete sequences (CS)-mtDNA) were published in 2000 [23], increasingly, CS-mtDNA data with far greater individual sequence information and less overall homoplasy have addressed these gaps [7].
In a review on out-of-Africa by Oppenheimer [12], all available CS-mtDNA data for South Asia, southeast Asia and Sahul were collected in order to estimate relative regional founding dates, using a CS-mtDNA calibration that best approximated the L3 date from the contemporary, most widely used, coding region methods at 83 ka [24,25].
The dates of the relevant non-African lineages, estimated by this method, were interpreted to favour a successful pre-Toba exit by AMH, with the suggestion of a partial extinction bottleneck in India resulting from the Toba event itself [12], but caution was expressed on giving a conclusion either way (i.e. pre- or post-Toba), because of not only the wide error bars on these estimates, which included the date of Toba, but also the questions already hanging over calibration and linearity [26] of the mtDNA clock. The concluding opinion was open: that only accurately dated AMH fossils, found in the appropriate context, could ultimately resolve the Toba issue.
A number of issues have recently been raised over calibration of the mtDNA molecular clock [27,28]. These include (i) uncertainty over the human–chimpanzee fossil split estimates, in terms of both their reliability for calibration and also the mutation rate variation over such long periods, when compared with more recent demographic and geological events within the timescale of AMH colonization of the world; (ii) rate variation in different sectors of the mtDNA molecule; (iii) factors such as the effect of serial, and regionally specific population bottlenecks on calculation of the important Rho statistic used previously in most estimates; (iv) the most important of these concerns: proven nonlinearity, due to both homoplasy, particularly in hypervariable non-coding regions, and gradual purifying selection of non-lethal non-synonymous mutations in the coding regions. Luckily, these nonlinearity effects are regular, predictable and measurable, and can be largely corrected for mathematically, improving precision and linearity, but not of course addressing the direct calibration issues.
A recalibration paper [7] was published in 2009, subsequent to the above-mentioned review [12], using a worldwide phylogeny of greater than 2000 complete mtDNA sequences to measure, predict and correct for nonlinearity effects, calibrating on a chimpanzee–human split of 6.5 Ma. Of the above-mentioned methodological issues, (i) was partially addressed by cross-checking on recent archaeological founder events, (ii) and (iv) were more directly addressed, whereas (iii) was partially addressed by citing age estimates using several different methods (Rho estimation and maximum-likelihood estimates) and making multiple interregional and chronological cross-comparisons.
This author, as a co-worker on that paper, argues that this is now the most accurate and precise human genetic clock calibration available, until another replaces it. An alternative method of calibration was published in 2008, based on the diversity of the Q haplogroup (unique to Melanesia) and a carbon-based colonization date of 45 ka, which produced very similar ages for L3 (61–86 ka), M (53–69 ka) and N (50–64 ka) from a much smaller dataset [29,30]. The authors note that the values could be increased by 11 per cent if the colonization dates subsequently increased to 50 ka, and a similar order increase has in fact happened [31].
The main remaining issues are calibration, the effects of serial bottlenecks and wide confidence intervals of estimates, thus affecting accuracy and precision. Such continuing issues should stimulate efforts to improve methods. This means that genetic dating may not replace good archaeology and precise carbon-based and other dating methods (e.g. evoked luminescence) on identified fossil sites for accurate dating of key events in human migration prehistory. It is, however, important for triangulation to have direct, independent dating of each proxy for the past (archaeology, palaeoanthropology, geology, genetics and even language), because heavy reliance on one method leads to circularity and self-fulfilling models.
The resulting depression of relevant haplogroup ages brought about by the new calibration, and new archaeological evidence pointing in the other direction, inevitably prompts a review of the discourse on AMH exit before or after Toba. Recalibrated mtDNA dates derived from a paper by Soares et al. [7] will be cited subsequently as evidence in this review for consistency, but not as the last word on mtDNA dating.
4. Review of out-of-Africa models
(a). How many exits of anatomically modern human ancestors from Africa? The genetic evidence
(i). Multi-regional versus recent replacement
Darwin [32] first suggested that humans originated in Africa, based solely on the presence of chimps and gorillas there. Since then, there have been two contrasting models of how and when our ancestors left Africa. The multi-regional hypothesis postulated that non-African regional populations each descend largely from separate migrations of Homo erectus, occurring over a million years ago. Multi-regionalists rationalized that the tendency for modern regional populations to resemble each other morphologically, more than any of them resemble their putative H. erectus ancestors, resulted from ‘modern’ gene flow between populations, rather taking the punch out of their own theory, but leaving a testable hypothesis. The other main testable out-of-Africa view, the ‘recent replacement model’, postulates that AMH originated from African stock within the past 200 ka and, having replaced other human groups in Africa, then left within the last 100 ka and replaced all pre-existing human groups outside Africa (reviewed by Oppenheimer [11]).
Limited archaic interbreeding outside Africa: it should be noted that, contrary to a perception common among multi-regionalists, the replacement model does not depend on the concept that AMH was necessarily a new human species rather than, say, a gracile version of Homo heidelbergensis and/or sibling race to other large-brained humans, or that there was a lack of inter-fertility or limited interbreeding initially, between these different regional human populations. The model merely asserts that there was, subsequently, a near-complete, recent physical replacement by AMH.
It is important to clarify the latter perspective and lay to rest the assumption, sometimes made, that any evidence of archaic admixture outside Africa, however small, would necessarily disprove the recent replacement model. The issue could be raised for instance by recent claims of a 1–4% introgression of Neanderthal genome into modern Eurasians [33]. However, these are not necessarily at variance with the view of near-complete replacement, in spite of the absence of evidence of Neanderthal introgression among the many tens of thousands of uniparental genomes studied. It should be remembered that the effective population size of the haploid uniparental genomes would be a quarter that of autosomes, predicting more severe drift and lineage extinction on uniparental loci.
This difference in drift between the two kinds of loci can be directly demonstrated. A good example is the observation that although autosomal loci do show a regular, progressive loss of diversity (e.g. in measures of heterozygosity) with increasing distance east from Ethiopia to the Americas, consistent with a serial founder effect [34], this decay effect is linear and, unlike uniparental loci, shows no sudden step fall on leaving Africa. By inference, this lack of an initial step-drop in diversity means that many autosomal loci probably preserved (i.e. carried-over) multiple alleles on exit from Africa. In contrast, the finding of only single recent African lineages represented on mtDNA and NRY outside Africa (L3 and M168, respectively) itself suggests a very severe initial founding drift effect in those loci immediately after exit. This susceptibility to drift would subsequently have tended to cause the extinction of any minority exotic mtDNA lineages, such as the less than 4 per cent maximum Neanderthal introgression (estimated from the autosomes outside Africa).
Whether such hypothetical extinction of acquired uniparental Neanderthal lineages could have occurred with admixture outside Africa (Green's preferred geographical option) is clearly debatable and depends on where and how soon admixture occurred and what population sizes were involved. However, it is difficult to imagine any scenario in which the ancestors of East Asians could have simultaneously acquired the same dose of Neanderthal autosomal genes as those of Europeans, unless it was soon after the exit and before east–west divergence. As the authors acknowledge, for this to happen: ‘gene flow from Neandertals into the ancestors of non-Africans occurred before the divergence of Eurasian groups from each other’ ([34], p. 710), thus implicitly depending on the single exit hypothesis for AMH, as applied in this case to autosomes. In order for Neanderthals to admix with the common ancestors of all non-Africans, however, they would have had to be in South Arabia at the time. The only evidence for this is the abundant Middle Palaeolithic record there, which certainly raises that possibility.
There is an alternative scenario Green and co-workers wisely did not consider in that publication but, as co-workers, do offer more recently in the context of the Denisova hominin phalanx and tooth found in a Middle Palaeolithic/Upper Palaeolithic transitional context in Southern Siberia [35,36]. This is the possibility of interbreeding with other recent, large-brained, putative H. heidelbergensis descendants in China such as the Maba, Dali (and Jinniushan) types who, on geographical distribution grounds, may have taken the southern route to the Far East.
In this instance, 4–6% of the provisional autosomal sequences from the Denisova finger bone are apparently found in two Melanesians but in no other modern human groups tested so far. MtDNA sequenced from the tooth is very close but not identical to the phalanx found at the same site; and both fossil mtDNAs are deeply separated phylogenetically by an estimated 1.04 Myr from Neanderthals and AMH. Morphologically, the Denisova tooth groups tightly with australopithecines, Homo habilis and African H. erectus. This archaic dental group shows a wide split from a more ‘modern’ group containing Neanderthals and all other European humans (ancient and modern), all AMH and curiously Chinese H. erectus. Indonesian erectus specimens alone took an intermediate position between the two groups. Given the latter observations, the morphologically archaic nature of the Denisova tooth and the Melanesian DNA connection, it is a pity that recent archaic Chinese Homo specimens (e.g. Maba, Dali and Jinniushan), modern Melanesian, Australian and Homo floresiensis teeth could not all be used as relevant types for comparison. In contrast to the erectine morphological qualities of the Denisovan tooth and the divergent mtDNA, comparison of Denisovan autosomal DNA with AMH and Neanderthals was however surprisingly anomalous, in that a closer relationship was inferred between Denisova and Neanderthals than either had with AMH [36]. The strength of evidence for this assertion of more recent common ancestry for the two archaics was, however, less convincing than that for Neanderthal intrusion into AMH. This is because the alternative explanation for the archaic autosomal sequence associations was not tested. This alternative was extended direct interbreeding between Neanderthals and Denisovans due to their clear geographical and temporal overlap in Central Asia.
The possibility of more archaic admixture in Oceania is reinforced by a new paper showing, as predicted above, Denisovan admixture in 100-year-old Australian aboriginal hair, similar in degree to that in Melanesians. Unfortunately, the interpretations made on the findings—which postulate an early archaic Australian wave from Africa 62 000–72 000 years ago and a later Asian wave 25 000–38 000 years ago—are influenced by an old morphologically based two-exit AMH model (see §4a(iii)) and a selective use of autosomal dating of demographic events, unsupported archaeologically. Their analysis and interpretation depend on a statistical test based on only four complete genomes. As their actual results stand, however, they merely extend the influence of ‘Denisovan DNA’ in Oceania and do not falsify the model presented here [37]. Indeed, there is morphological evidence that would reverse the order of the ‘two Australian waves’ because the earliest human crania in Australia are modern and gracile, whereas there is a late appearance of archaic and robust skulls in the Kow Swamp and Coobool Creek sites, radiocarbon-dating to no earlier than 14 300 years ago [38].
Genesis of the recent replacement view: the morphological evidence for recent replacement by AMH actually just predates the genetic evidence [9]. Stringer's [39] definitive paper, whose genesis goes back to a similar one in 1984, was written and presented in 1986. However, the difference in timescale along with the sharply different colonization models offers further proof, in that several predictions can be tested by genetic phylogeography, using non-recombining DNA such as mtDNA and the Y chromosome. Under the multi-regional hypothesis, today's non-African populations should possess some recent lineages and a substantial mixture of ancient lineages (i.e. much more than a few per cent) as old as, or maybe older than, those in Africa, such similarity signalling multiple exits over the past million years. These predictions of multi-regionalism are clearly not fulfilled. Conversely, recent replacement outside Africa should show older lineages within Africa and nearly exclusively young lineages outside, depending on the completeness of replacement as is the case.
When Africa and the rest of the world were first compared using the uniparental mtDNA locus in 1986 [9], the evidence for the recent ‘out-of-Africa’ model was strongly supported, in that the two trees rooted deeply in sub-Saharan Africa (SSA) with derivative branches outside. The first mtDNA tree was low resolution, with a number of African misclassifications. The mtDNA tree was improved and resolved by complete sequence data and a world tree, based on the first 52 complete sequences published, showed that all non-Africans belonged to two sibling twigs (M and N) both arising from L3, one of over a dozen unique African branches (fig. 0.3 in Oppenheimer [11], reproduced here as figure 2).
Two phylogeographic predictions of the recent replacement model are fulfilled in the first resolved, dated worldwide CS-mtDNA tree (figure 2). First, of the dozen or more branches of the tree found only in Africa most, although young (less than 200 ka), are, as expected, much older than L3, M or N. The genetic age of the deepest branch (190 ka) [11] (192 ka in Soares et al. [7]) is consistent with the new dated fossil finds of early AMH in East Africa (164 ka [40]; 195 ka [41]).
(ii). Single recent exit model for anatomically modern human
Second, as mentioned already, the most singular observation for mtDNA is that ‘L3’, the remaining African branch, is not unique to SSA in that it also encompasses all non-Africans. L3 has several branches within Africa, but only two originating outside, namely M and N [11,42,43], and these two are also the only ones found throughout the rest of the world. In effect, this means that the rest of the world was also colonized by descendants of a single ex-African founding mtDNA haplotype—the root type of the L3 haplogroup (more specifically, a founding subtype of the main root holding a nucleotide transition at position 195, shared by M and N). The latter now dates by the molecular clock back 71 600 (CI: 57 100; 86 600) years [7].
N and M outside Africa overall both have similar ages, which are older than their derivative African re-entrants (see §4a(iii)), and on average around 10 kyr younger than their immediate founding ancestor L3-195. Two exceptions to this are N, when dated nearest to Africa, in west South Asia at 71 200 years (CI: 55 800; 87 100), and M dated in India at 49 400 years (CI: 39.0; 60.2). Their inferred origins are located, respectively, in the Gulf region and in India, with widespread co-distribution outside Africa [11,42,43].
This non-African origin of the two L3 branches (M and N) found outside Africa is, ultimately, unambiguous evidence of a single successful L3 exit from Africa, and until recently was paralleled in the male Y chromosome tree [11,15,22], but with two branches, CF and D, both defined by the same single nucleotide polymorphism (M168), originating outside Africa [44] (but see §4a(iii)). That all non-African descendants, for both mtDNA and Y twin lines, each share one and the same close African ancestor is the result of severe drift in uniparental lineages, resulting from a period of isolation in a small founding group, during which all other lineages in the group were lost by genetic drift (see above). But such singularity, in itself, provides serendipitous, unequivocal, proof of the single successful AMH exit, not usually available from autosomes, with their higher effective population size. The chances of two or more founding-exit episodes separated in time and place each giving rise, by drift, to the same two single-source African lineages (or one of each) in all the different parts of the non-African world are vanishingly small, leaving the single exit statistically by far the most likely solution. Such severe single drift at exit is clearly reflected not only in uniparental loci but also in the X chromosome and in several autosomal loci studied [15].
It should be noted that although the evidence and logic for a single successful ex-African AMH mtDNA founder lineage is clear (and forms the basis for the rest of this review), the scenario is surprising and counterintuitive. In the absence of severe drift, a single exit group would be expected to involve multiple founder lineages spreading to different Eurasian locations. A single founder picture is not seen in any other continent-to-continent colonization. For instance, the colonization of the Americas involved five founder mtDNA lineages [11], whereas New Guinea and Australia have, between them, over a dozen ancient founders (see §4c(iv)). For uniparental loci, the most severe bottleneck was thus apparently the first exit, with serial unequal splits, bottlenecks and loss of diversity occurring all the way to the Antipodes and the Americas.
(iii). Models with multiple exits: the Cambridge model
Models including more than one exit have, in the past, been the default (reviewed in Oppenheimer [11]) and are exemplified in publications by Foley and Lahr of Cambridge University and others [13,14,45–47]. These multiple exit views are all essentially prefigured by cranial morphological analyses of the 1990s [13,46]. That work postulated multiple land-locked ‘islands’ of populations of modern humans scattered within the arid Palaeolithic landscape of SSA. Being morphologically different, sub-sets of these ‘population islands’ would then have constituted the discrete sources of subsequent morphological diversity outside SSA, as a result of multiple separate outward migrations. This view was the basis of a later collaborative publication, which effectively matched Y chromosome lineages secondarily to the morphological multi-migration model [13,14], and a book which further elaborated the same model adding genetic data from Central Asia [48]. This apparent dominance of morphological over genetic arguments for numbers and directions of African exits seems odd in retrospect, given the greater specificity of the genetic evidence, but may reflect the early difficulties of getting uniparental evidence to be taken seriously, even by population geneticists.
These morpho-genetic models of the early twentieth century argued for two main exit routes. The first was a southern one across the mouth of the Red Sea 50–45 ka, towards Australia and carrying two of the three non-African Y branches, DE and C (in 2002, Y Chromosome Consortium nomenclature [49]; fig. 3c in Underhill et al. [14]), leaving, as ‘relict’ descendants, small populations around the arc of the Indian Ocean coast such as Andaman islanders, southeast Asian Negritos, Melanesians and Australians, but also with genetic traces in Japan and North America [14,48]. Second and separate from these minority early southern route migrations, it was postulated [14] that the bulk of Eurasian populations, derived from East African F lineage, migrated through northeast Africa to the Levant around 45 ka, and subsequently dispersed variously towards west to Europe and east to the rest of Eurasia between 40 and 20 ka.
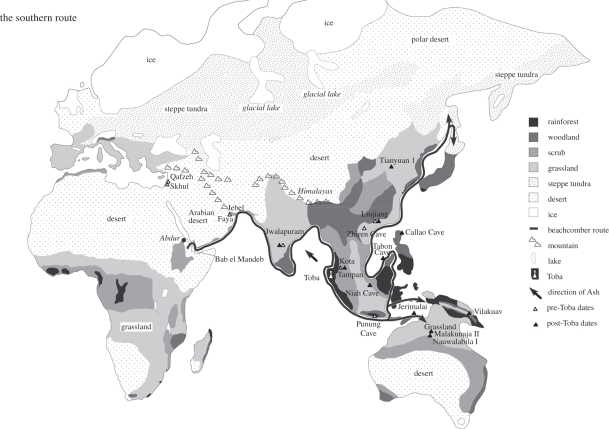
Figure 3.
Map showing single southern route out of Africa and beachcomber arc route from the Red Sea along the Indo-Pacific coast to Australia, including likely extensions to China, Japan and New Guinea, from OIS-4. Vegetation and sea level shown as at Last Glacial Maximum (LGM). Overlay: claimed fossil and cultural evidence for AMH in Eurasia during OIS-5–early OIS-3 (dates in text). Adapted from fig. 1.6 [10,11].
Problems with Y chromosome evidence offered for multiple exits: the foregoing view ignored the fact, implicit in the data, that the Y chromosome tree outside Africa showed a phylogeny similar to that of the mtDNA tree, with one African branch (CDEF) defined by the mutation M168, accounting for all non-Africans [50]. Furthermore, the view of the relicts of earlier exit populations deriving solely from the exit(s) of sub-clades D and C was contradicted by pre-existing phylogeographic evidence [50], which has been confirmed since then [11,22] using DNA obtained from the same so-called relict groups in the Antipodes who possessed ample regionally unique lineages derived from the third Y branch F. Additionally, the claimed, close distributional correspondence of mtDNA haplogroup M with Y sub-clades D and C [14] was undermined by a similar widespread correspondence with the second mtDNA haplogroup N in the same places, including the Antipodes. As will be discussed below, this author, and others, argued logically, both from mtDNA and Y chromosome evidence, for a single definitive exit, probably by the southern route ([11]: ch. 4 and figs. 4.2–4.5) [15,25].
Subsequently, Underhill [22] has implicitly acknowledged these counter-arguments to multiple exits by co-workers in a more recent, combined Y and mtDNA paper. However, there is still a remaining problem of fully reconciling the Y chromosome evidence with that from mtDNA. The problem is found in one of the three Y branches, represented among non-Africans: DE (defined by the YAP+ mutation). This has a disjunct geographical distribution, with the D sub-clade, represented at low levels only in South and southeast Asia and in Japan, while E is represented, on the one hand, locally throughout the Middle East and Europe as a sub-clade E-M35 [51], when on the other hand, the remaining uniquely African E sub-clades make E the commonest Y clade in Africa [50]. Furthermore, while the distribution of D marks it as uniquely East Asian, and one of the original single exit clades, its fraternal sub-clade outside Africa (E-M35) has been argued to have left Africa much later, around or since the Last Glacial Maximum and probably not by the southern route [15,52,53], potentially partially falsifying the single exit hypothesis.
These observations seem to mean that, unlike the mtDNA exit, which is defined by a single branch L3, splitting outside Africa into M and N (see below), up to three Y branches (C, D and F), albeit very closely related, could have split before the initial exit from Africa, rather than after. The argument for this scenario is that the ancestor to D could not have split off after exit, from a single M168-CDF common ancestor outside Africa, leaving E behind, because D and E are both defined by YAP+ and form their own sub-clade [15].
A recent review by Underhill & Kivisild [44] appears partially to solve this conundrum, and to make the single group exit even more convincing, because they use a newer phylogenetic marker, P143 [54], to define a single exit for CF before C and F separated, thus leaving only two Y lineages in the southern out-of-Africa movement: namely D and CF. The unity of C and F at exit incidentally also further renders Underhill's original model, with an earlier exit of C and D before F, impossible and reconciles the finding of C and F together in the Antipodes.
There is yet a further twist to this story that could result in going full circle back to a model first proposed by Hammer et al. [55] in 1998, and previously supported by Oppenheimer [11], known as ‘Out of Africa and Back Again’, whereby the YAP+ mutation occurred outside Africa, forming the DE clade and this dispersed on to East Asia, becoming D, while a second DE branch returned to East Africa, there becoming E.
Fittingly, Underhill (who originally opposed this model) is the senior author on a recent paper offering new phylogeographic evidence that could support it, including a simplifying revision of the E phylogeny, and finding two examples of underived root-types of E in Saudi Arabia [56]. This model would specify, as for mtDNA, a single exit Y lineage and two non-African descendant branches arising outside Africa. To quote: ‘Regions near but external to northeast Africa, like the Levant or the southern Arabian Peninsula could have served as an incubator for the early diversification of non-African uniparental haplogroup varieties like Y chromosome DE-YAP*, CF-P143* and mtDNA M and N molecular ancestors’ ([56], p. 66). The model also predicts that E1b1 (the main trunk of E defined by P2) probably arose in East Africa after DE backflow there.
In line with this prediction, there is now more recent evidence consistent with, firstly, only two primary branches of E1b1 (rather than three as previously thought), but also with their joint East African origin. This includes not only the E-M215 branch carrying the E-M35 sub-clade, for which latter such geographical evidence already exists, but also the other new main E1b1 branch, defined by V8 and V100. This V8/V100 stem now clearly includes the E-M329 branch, observed almost exclusively in East Africa, whereas the rest of the large African branch, previously defined by E-M2, was puzzlingly virtually absent there [51,57]. Remaining, non-P2, minority branches of E, E1b2 and E1a are also found in East Africa (as well as North Africa) [51], consistent with an ultimate E origin in East Africa.
Worldwide tests of prediction: one of the testable predictions of the ‘multiple exit’ model is that various populations including the so-called ‘relict’ groups around the Indian Ocean should show different mtDNA and Y lineages from the rest of the world, as evidence of arising from separate exits. This prediction was first tested formally in 2003 [11], by comparing what was known about uniparental lineages in multiple putative ‘relict’ or aboriginal populations in South Asia, the Indian Ocean, southeast Asia and the Sahul. Multiple exits were found unsupported for mtDNA because all known lineages in these groups belonged to M and/or N haplogroups. In other words, all non-Africans were descended from L3 (figure 2), including all relict non-African groups, thus supporting the single exit ([11]: chapter 4 especially endnote 5 and figs. 4.2 and 4.3). Similarly, all Y lineages in relict groups belonged to C or F. These inferences have been confirmed several times since, with the same single exit conclusion, using larger sample sizes and at higher sequence resolution [25,58], and more relict groups, such as Andaman Islanders, who also held D [59], Melanesians and Aboriginal Australians [22].
(iv). Backflow into Africa
A critical issue in establishing the location of origin of the M and N mtDNA L3 subgroups was explaining the presence of 15 of their branches (1 M; 14N) in a well-conducted study in East Africa, together amounting to 47.8 per cent of mtDNA lineages in Ethiopia, but very little anywhere else in SSA [60]. The equivalent of retention of M and N lineages across the water in Yemen was similar at 47 per cent. Was that evidence of local origin in Ethiopia or backflow from Arabia? Clearly, it was the latter: for N branches, the issue was straightforward, because the Ethiopian branches were not only much younger (12.6–14.4 ka) inside Africa than their equivalent outside, but they were ultimately derived from well-established non-African sub-clades of N. For M, it was slightly different in that the only African M branch, M1, was derived, but apparently had no precedents elsewhere, apart from root-M outside Africa. However, with an age of 41.2 ka (SE17), which is clearly younger than the rest of M outside Africa, M1 was unlikely to represent a solitary African stay-at-home [60].
Male backflow from Asia to Africa has been shown for non-African Y involving F and E-M35 [61], although E-M35 is likely a returnee of East African rather than Middle Eastern origin [52]. Haplogroup F backflow only accounts for 10–40% backflow and is confined to North and East Africa [61]. If, however, as suggested in §4a(iii), DE-YAP* originated just outside Africa, then technically non-African backflow in the Y locus includes E and could ultimately massively outnumber the original African Y throughout Africa, ranging from 59 per cent (rather higher than mtDNA figures cited above) in East Africa and 65–80% in South Africa, to well over 90 per cent in West Africa (data from Cruciani et al. [61]).
Cruciani et al. [62] have recently extended their description of NRY backflow by showing backflow of Eurasian haplogroup R1b1 (ultimately belonging to the F clade and defined by P25) into North and Central Africa. Phylogenetic evidence and coalescence time estimates of a newly identified marker, V88, suggest that these chromosomes may have been carried to Africa by an Asia-to-Africa back migration, through northeast Africa to the central-western part of the African continent (northeast Cameroon), where they can be detected at frequencies as high as 95 per cent in Chadic-speaking peoples from the Central Sahel and several other Afroasiatic-speaking groups from North Africa, while having much lower rates among surrounding Niger-Kordofanian-speaking groups. The R-V88 coalescence time was estimated at 9.2–5.6 ka, in the Early Mid-Holocene [62], suggesting the effects of the interglacial optimum window, greening the Sahara and the Chad Lake basin, thus allowing the spread of people and pastoralism south into SSA.
(b). Which route?
A single successful African exit for AMH has several clear implications. The simplest, but most important of these is that the number of possible subsequent route options was also consequently decreased. Whichever route was taken initially, however, the model thus has to explain how both Europe and Asia could have been colonized ultimately from the same single exit group.
(i). Southern rather than northern exit: genetic evidence
As to which route, there are four potential corridors leading out of SSA, three of which involve some water-crossing. Two of these would be north direct to Europe, to Gibraltar or, via emerged Malta and Sicily landmasses, to Italy. There is no evidence (archaeological or genetic) for early AMH taking either. This leaves the two most commonly suggested: northeast to the Levant across the Sinai from Egypt after crossing the Sahara, and east across the mouth of the Red Sea from Djibouti, thus by-passing the Sahara, to take the coastal route to South Asia.
As far as the genetic evidence is concerned, there are two logical reasons for identifying the southern route across the mouth of the Red Sea as the most likely defining exit route. Firstly, both of the two primary branches of the L3 mtDNA haplogroup outside Africa (M and N; figure 2), are represented in East Eurasia, including South Asia (where they have the highest number of autochthonous M and N lineages in the world), while only N is represented in the Near East/Europe where it is younger than in South Asia (§4c(i–ii)). Second, all the primary branches of N are represented in South Asia [43], whereas the Levantine and European N representatives are less diverse, more derived and younger than those in South Asia (for an in-depth discussion, see earlier studies [11,15]). The single southern exit is also consistent with the earlier archaeological luminescence dates for colonization of the Philippines and Australia than of Europe.
(ii). Climatic constraints and opportunities for an exit route
Another implication of both the single genetic exit and drift is that there must have been great difficulty in establishing a beachhead outside SSA during the Late Pleistocene, presumably as a result of the Saharan–Arabian arid zone creating a barrier to migration out of SSA between 90 and 48 ka [63], but also a barrier to movement between South Asia and the Levant across Arabia (figure 3). The Sahara and the evidence for earlier arrival in Australia than Europe (§4c(ii)) logically points to the southern route.
There is the question of drinking water along a desert salt-sea coastline such as the southern Arabian Peninsula. Sauer [64] has, however, pointed out that low-tide fresh-water seeps occur even on desert beaches. Paradoxically, emerged aquifers may have been the more abundant coastal sources of groundwater, during periods of low sea level [65].
(c). Dating migrations
(i). Possible dates of the definitive exit
Dated archaeological evidence for the exit is lacking at the Bab El Mandeb or on the Arabian coast because sea-level rise has drowned most coastal remains. An exception is some dated cultural evidence at Jebel Faya (see §4c(iii)), which starts far too early for continuity with the genetic evidence, even with the latter's wide confidence intervals. Much of the relevant datable evidence lies either in India or on the eastern side of the Indian Ocean. As mentioned above, a general guide to earliest possible date, and excluding any Eemian exit as lacking progeny, is the genetic age of expansion of L3 at 71 600 years ago (CI: 57 100; 86 600), which crucially includes African L3 [7]. For the lower limit, N, one of the two daughter lineages of L3 outside Africa has been dated near its putative origin in South Asia, where it has a predominantly western distribution, to a similar 71 200 years (CI: 55 800; 87 100). M, the other branch, is surprisingly young in South Asia, where it has a predominantly eastern distribution (see §4c(iii)) at 49 400 years (CI: 39 000; 60 200) [7] (figure 3).
(ii). Delayed migration to the near east and Europe
It is important to explain the issue of the ultimate origins of West Eurasians out of the southern exit route. However, the problem remains as to how and when ancestors of northwest Eurasians crossed the arid zone between South Asia and the Levant/Turkey, and why they were delayed.
Today, there is a green corridor running through the Iraq/Iran arid region, northwest between the western slopes of the Zagros mountains and the Tigris River ending in the Taurus mountains of Anatolia. This would have been closed and arid between ca 90 and 47 ka, i.e. during cold stages OIS-5b 4 and half of 3 [63] (see also Selivanov [66]).
A partial amelioration of climate occurred around 48 ka with Dansgaard–Oeschger Interstadial 14 (e.g. fig. 2 in [67]). With appropriate radiocarbon correction [68], this period would have also coincided with the brief period of raised humidity in the Arabian arid zone 45 ka C14 years ago [63]. A climate permissive model can be reconstructed [12] of AMH populations suddenly being able to move into northwest Eurasia from South Asia only at ca 48 ka (see fig. 1.8 in [11]). This dating would coincide with the earliest appearance of Upper Palaeolithic technology in the Levant and southeast Europe (calibrated to 47/49 and 46 ka, respectively [68]) and also with the age of U8, the oldest mtDNA ‘N’ lineage in the Near East and Europe (at 51.3 ka (44.0–58.8) [69]; see also [11,15]).
(iii). Dating appearance of anatomically modern human outside Africa: Eemian, pre-Toba or post-Toba
Eemian exits and fossils: there is clear archaeological evidence for an earlier AMH exit to the Levant during the moist Eemian interglacial, based on fossil and lithic evidence, dating between 120 and 90 ka [70,71]. Dry glacial conditions returned and Neanderthals then reappeared in the Levant from around 60 ka, but there was a long gap of up to 40 kyr between the last Skhul and Qafzeh AMH skulls, and evidence of the reappearance of AMH in West Eurasia was reported no earlier than 49 ka [68].
Non-African MtDNA and Y chromosome phylogeography and dating evidence are not consistent with an ultimate survival of this northern colony into the modern gene pool, and the Levant exit is presumed abortive [12].
The proven Levantine AMH Eemian exit, however, means that similar Arabian-Far Eastern claims for a southern Eemian AMH exit may well turn out to be correct, although carrying the same lack of genetic evidence for survival. Five such claims currently vie for attention, all with date brackets similar to those of the Levant exit, four with fossils and one with cultural evidence. The latter is at Jebel Faya on the Arabian Peninsula, where lithics with affinities to the Late Middle Stone Age in northeast Africa date between 127 ka and 95 ka [18] (reviewed by Petraglia et al. [16]; Rose [72]).
Similarly, a recent finding of an anterior mandible with two molars at Zhiren Cave in South China was dated by flowstone context to 113–100 ka [17,73]. The AMH attribution was unconfirmed, and even the authors suggest hybridization. The presence of AMH in Java during OIS 5e was recently suggested, on the basis of one hominin tooth [74], first described in 2005 [75], small enough to suggest AMH, and previously found in breccia in Punung cave in southeast Java. The date claim is explicitly tentative and indirect, as must be the attribution. Finally, the famous south China ‘Liujiang skull’ is indubitably AMH, but controversially dated to 111–139 ka by other items in the breccia in which it was found [76], which could put it in the same Eemian age range as the Zhiren Cave and Punung Cave fossils. A lower date of 67 ka based on the covering flowstone seems, however, far safer [12].
The recalibrated age estimates and confidence intervals of the out-of-Africa mtDNA branch L3 of 71 600 years ago (CI: 57 100–86 600 [7]), although wide in this context, exclude the date ranges implied by an Asian Eemian exit (as did previous L3 age estimates [11,12]). So, one has to presume that, such as the Skhul and Qafzeh exit, any Asian exit by AMH during the Eemian must have been evanescent. This view is supported to some extent by the archaeological record in all these cases of the putative early exit—for instance the Jebel faya record, which stops at 95 000 years, far outside the range of L3 age estimates. Autosomal loci with more detail and much larger effective population size may ultimately disprove this perspective, as with Neanderthal admixture, but autosomal dating is currently unreliable.
Exit before the Toba super-eruption (74 ka)? The first out-of-Africa model to suggest the definitive AMH exit before the Toba eruption [11] used the Tampanian pebble tool culture of the Lenggong Valley, Perak, Malaysia as partial cultural evidence, although no fossils were found with the tools covered in Younger Toba Tuff (YTT: ash). The same lithic culture persisted locally, however, latterly associated with AMH, until the Mid-Holocene. Similar lithic cultures from Indonesia dating to around 70 ka are reviewed elsewhere [11,12], but see caveats [77].
Similar claims for pre-Toba exit were made recently for YTT strata in India [16,78], but in this case, the lithic culture above and below the ash was Middle Palaeolithic (MP), with the added argument that local typology better matched South African Middle Stone Age than North African or Levantine MP. Matching such typological comparisons too closely to hominin type does not currently get a good archaeological hearing [77], as was the case here. However, a follow-up detailed description of methods at a recent meeting on Toba in Oxford (February 2010) convinced several, but not all, seasoned archaeologists in the audience (reported in [79]). The meeting held another surprise, in that dating of cultural layers below and above the YTT had not previously been shown to other collaborators. Previously pre- and post-Toba-ash cultural dates for the first site [78] were both very close to the eruption (YTE), but in the new results on nine paired Indian sites, only the pre-Toba dates were close to the YTE at 74 ka BP or earlier, whereas nearly all of the post-Toba dates were about 55 000 years BP or younger [79], thus raising a question mark over the group's key claim of rapid human recovery post-YTE.
Genetics and Toba: a prediction of this author's interpretation of the conjunction of tools and ash in India is that a deep genetically sterile furrow would have split South from southeast Asia, with India eventually recovering from a genetic bottleneck either locally or by re-colonization from either side. Such a furrow was first argued for in the genetic map of South Asia ([11], pp. 180–184). India is characterized by an abundance of unique local diversity as evidenced by a number of uniparental lineages [41,80–82].
Apart from the complete change in representative mtDNA haplogroups between Eastern India and Indo-China, tribal populations on the East Coast of India predominantly exhibit unique M groups and lack N subgroups, consistent with local drift resulting from near extinction. Also consistent with the possibility of local Indian bottleneck, M groups in Eastern India around the Bay of Bengal, although numerous, are systematically younger than elsewhere along the Indian Ocean trail [11,12] (see also table 1 in [80] and table 3 in [7]). However, dates for haplogroups N and R, which locate further west in South Asia, where less ash fell, are relatively higher (see fig. 3 in [12] and table 3 in [7]), consistent with local drift near the east coast of India and a pre-Toba exit.
In summary, the new cultural dates and lithic arguments presented at the recent Toba meeting in Oxford are consistent with this author's long-held speculation on a near pre-Toba AMH colonization of Asia, interrupted by the YTE, and followed by a slow demic recovery in India, but continuity existing elsewhere. However, this is not the final proof and, as mentioned already, the calibrated genetic age (CI) of the exit mtDNA branch L3 is 71 600 years ago (57 100; 86 600), thus bracketing Toba and neither clearly proving nor convincingly excluding a pre-Toba exit.
Post-Toba anatomically modern human fossils in southeast Asia: diagnostic, well-dated fossils are the gold standard in proving presence. Unfortunately, as in Europe, the realities of stone tool production and bone preservation make cultural remains vastly more common than fossils. The oldest dated AMH fossils in southeast Asia show a much older age bracket (67–40 ka) than the oldest such fossils in Europe (35 ka or less [83]). However, with the ‘possible Eemian’ exception of the Punung tooth (§4b(ii)), none predate Toba.
There is currently no firm fossil support for AMH dispersal to SE Asia before Toba (74 ka), let alone of antiquity anywhere near the Eemian, as claimed for the Zhirendong mandible in China (§4b(ii)). Fossil dates however do seem consistent with AMH presence in southeast Asia (and China) by at least 67 ka. The oldest dated human fossil in southeast Asia is now a single human third metatarsal from Callao Cave in Northern Luzon, Philippines [84], with a minimum age of 66 700 ka (±11 ka). There is the inevitable problem of identifying which human the metatarsal belonged to. The bone is gracile, consistent with AMH, but is small, and in the size range of adult local Negritos and that inferred from the Niah femur [85]. However, the size is also consistent with H. habilis and H. floresiensis. The age cannot be assessed accurately owing to the absence of the metatarsal head, although a young adult or adolescent is suggested.
The next oldest human fossil in the region is most likely AMH. A dozen AMH individuals have now been excavated from in-and-around the Tabon cave (Palawan, the Philippines), with fragments including a diagnostic frontal bone and mandible dating from 16 500 to the oldest, a tibia, at 47 ka in age, using Uranium series [86].
Next among these meagre island southeast Asian fossils aged over 40 ka is the site of the Niah Cave ‘Deep skull’ in Sarawak, Borneo. Originally dated at around 40 ka in the early days of the radiocarbon method, the AMH skull site has been re-excavated and securely redated using the ABOX-SC method and bracketed at around 42 ka (45–40 cal. ka BP [87]). The associated cultural sequence goes back at least to 46 ka (46 cal. ka BP [88]). Contemporary to the Niah skull is the aforementioned Timor beachcombing site at the Australian threshold, unfortunately lacking fossils [20].
However, there is a problem with the newly recalibrated estimates of genetic ages for M, N and R haplogroups in East Asia, now clustering at around 60 ka BP or less [7], 5–15 ka lower than estimated before [12], rather than the new archaeological estimates of up to 67 ka BP. Admittedly, the new genetic confidence intervals of all three haplogroups are still sufficiently wide to include 67 ka BP.
(iv). Dating Pleistocene arrivals of humans in Sahul and near Oceania
Archaeology, radiocarbon and luminescence-dating: for Melanesia, the first archaeological evidence of occupation of the island of New Guinea has been radiocarbon dated to 49 ka BP [89] and 40 ka BP to the east, in New Britain island [90]. For Australia, the carbon ceiling has been increased to around 48 ka using the ABOX-SC method [91]. Non-carbon dating techniques, however, suggest an earlier human colonization of Australia by 50–60 ka [92–95].
There is clearly more work required with non-radiocarbon-based techniques. This review is not intended to replace those existing reviews, which are more focused on that controversy. It merely points out that the present ‘oldest non-carbon dates’ are not inconsistent with colonization by 50–60 ka. Bird et al. [95] have reviewed the recent literature, suggesting the arrival of H. sapiens in Australia conservatively by at least 48 ka BP, but that the chronological evidence argues for rather earlier than this. Recent sea-level curves, concentrating on troughs [96], suggest that the best window of opportunity for such a long crossing over the large Sahul shelf would have been 65 ka at a trough of −100 m, with occupation in the Northern Territory only becoming visible on modern littorals from 60–50 ka owing to higher modern sea levels [10].
Sahul genetics: as mentioned already, genetic phylogeography is a better tool for topography than for dating, offering two points. First is the resolution on the descent of all Melanesians and Australian aboriginals on uniparental lineages from the single southern exit [11]. Extensive further work on Australian and Melanesian CS-mtDNA and NRY has not falsified this assertion [22,25,97].
The second point is whether Australia and New Guinea were colonized in parallel and independently, or serially, or by both models. Australians and New Guineans, although genetically more like each other than they are to, for instance, southeast Asians, have deep genetic divisions at several loci [11,98]. However, by 2004 it became clear from complete sequence data and the rare presence of variants of the characteristic Melanesian ‘P’ mtDNA clade, both in southeast Asia and in Australia, that there were at least some common genetic sources and/or direct interchange between the two land masses [98]. Recent CS-mtDNA data [22,99] confirm a much more substantial ‘P’ clade connection and it can be inferred that both these models are correct and that New Guinea was probably colonized in parallel, at a similar time, to Australia, sharing several sources but not others. So, much of the genetic difference between them may derive from subsequent isolation rather than from different sources. Recent autosomal work has independently supported this conclusion [100].
Previous mtDNA genetic dates of arrival of the vanguard in the Sahul estimated by this author [12] were consistent with a pause of around 10 ka at the threshold of the crossing (i.e. from Sundaland/Wallacea). Combining the optically stimulated luminescence (OSL) colonization dates for Australia [92,93] with the recalibrated genetic ages of M, N and R in East Asia would now give no time at all for such leisure before arriving in Australia. This would still infer an extremely rapid transit from Aden at least up to the tip of Sundaland, of around 10 ka, given the recalibrated molecular age of the out-of-Africa L3 lineage at 71 600 years.
The speed of colonization inferred from the Australian archaeological dates above is consistent with the phylogenetic observation that the great arc of dispersal was effected entirely by the three primary root founder haplotypes M, N and R, each giving rise to multiple sets of unique regional daughter branches in every sector around the Indian Ocean. The only exception to this ‘local primary starburst’ rule is the crossing to the Sahul, which was substantially colonized by derivative lineages exemplified by the ‘P’ haplogroup, derived from R and originating in southeast Asia.
For Australia [22], there are nine unique founding lineages (M13, 14 and 42; S; N12, 13 and 14; P5′7 and Q2), implying multiple founder events. Melanesia (New Guinea and islands) has four unique founder lines, with molecular ages varying between 30 and 82 ka (M27, M29′Q, M28 and P2).
Additionally, there are a dozen haplogroups shared between Australia and New Guinea. It is clear that not only did the large ‘P’ haplogroup ultimately originate in southeast Asia, where it is now extremely rare, but that it is represented in both Australia and New Guinea in more than half of its subgroups (P1′6′9, P3a, P3b and P4). Furthermore, this sharing derives in all cases from early splits [12], both before arrival as a result of common sources (e.g. P4) and as exchange between the two landmasses after arrival (e.g. P3a and P3b).
There is also similar sharing of an early source M lineage, M13′14′28′42, probably originating externally to the Sahul, which is mainly present in Australia as subgroups M13, M14 and M42, but has two branches in Melanesia (M28a–b).
The remaining lineage sharing in Australia is a single example of the Melanesian subgroup Q2.
There are examples from the autosomes and the Y chromosome of post-glacial gene flow from New Guinea and southeast Asia to Australia; substantial gene flow between island southeast Asia and Melanesia commenced from the early Holocene (reviewed in previous studies [101–103]).
5. Conclusions
The genetic, climatic and archaeological evidence logically suggests a single southern exit of AMH from Africa, probably during early OIS 4. All non-African groups today are descended from this exit, with the exception of some autosomes (7% or less) apparently derived from admixture with several archaic non-African groups. Whether this exit predated the Toba eruption is currently unclear. A series of founding bottlenecks characterized rapid migration around the Indian Ocean coast to Borneo and Bali at the tip of the Sunda shelf. Then, a sea-level low stand permitted multiple colonizations of the Sahul, followed by prolonged isolation until the post-glacial period, during which maritime gene flow from island southeast Asia recommenced. These last migrations were limited into Australia and substantial into Melanesia. Climate and access to fresh water were crucial determinants of routes and dates for windows of opportunity.
Dates of migration can only be approximated by genetic studies, while palaeoclimatology, palaeoanthropology and archaeology offer potentially firmer options. The possibility that AMH arrived in Asia before the YTE offers both an approach to dating and a source of controversy. Contentious AMH dates include the definitive exit from Africa, as well as first arrivals in India, China, Central Asia, Indo-China, Sunda, Sahul and the Americas.
References
- 1.Oppenheimer S. J., Higgs D. R., Weatherall D. J., Barker J., Spark R. A. 1984. Alpha thalassaemia in Papua New Guinea. Lancet 1, 424–426 10.1016/S0140-6736(84)91754-9 (doi:10.1016/S0140-6736(84)91754-9) [DOI] [PubMed] [Google Scholar]
- 2.Oppenheimer S. J., Hill A. V. S., Gibson F. D., Macfarlane S. B. J., Moody J. B., Pringle J. 1987. The interaction of alpha thalassaemia with malaria. Trans. R. Soc. Trop. Med. Hyg. 81, 322–326 10.1016/0035-9203(87)90253-7 (doi:10.1016/0035-9203(87)90253-7) [DOI] [PubMed] [Google Scholar]
- 3.Flint J., et al. 1986. High frequencies of alpha thalassaemia are the result of natural selection by malaria. Nature 321, 744–750 10.1038/321744a0 (doi:10.1038/321744a0) [DOI] [PubMed] [Google Scholar]
- 4.Senok A. C., Li K., Nelson E. A. S., Yu L. M., Tian L. P., Oppenheimer S. J. 1997. Invasion and growth of Plasmodium falciparum growth is inhibited in fractionated thalassaemic erythrocytes. Trans. R. Soc. Trop. Med. Hyg. 91, 138–143 10.1016/S0035-9203(97)90200-5 (doi:10.1016/S0035-9203(97)90200-5) [DOI] [PubMed] [Google Scholar]
- 5.Senok A. C., Nelson E. A. S., Li K., Oppenheimer S. J. 1997. Thalassaemia trait, red blood cell age and oxidant stress: effects on Plasmodium falciparum growth and sensitivity to artemisinin. Trans. R. Soc. Trop. Med. Hyg. 91, 585–589 10.1016/S0035-9203(97)90037-7 (doi:10.1016/S0035-9203(97)90037-7) [DOI] [PubMed] [Google Scholar]
- 6.Hill A. V. S., Bowden D. K., Trent R. J., Higgs D. R., Oppenheimer S. J., Thein S. L., Mickleson K. N. P., Weatherall D. J., Clegg J. B. 1985. Melanesians and Polynesians share a unique alpha thalassaemia mutation. Am. J. Hum. Genet. 37, 571–580 [PMC free article] [PubMed] [Google Scholar]
- 7.Soares P., Ermini L., Mormina M., Rito T., Röhl A., Oppenheimer S., Macaulay V., Richards M. 2009. Correcting for purifying selection: an improved human mitochondrial molecular clock. Am. J. Hum. Genet. 84, 1–20 10.1016/j.ajhg.2009.05.001 (doi:10.1016/j.ajhg.2009.05.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wainscoat J. S., et al. 1986. Evolutionary relationships of human populations from an analysis of nuclear DNA polymorphisms. Nature 319, 491–493 10.1038/319491a0 (doi:10.1038/319491a0) [DOI] [PubMed] [Google Scholar]
- 9.Cann R. L., Stoneking M., Wilson A. C. 1987. Mitochondrial DNA and human evolution. Nature 325, 31–36 10.1038/325031a0 (doi:10.1038/325031a0) [DOI] [PubMed] [Google Scholar]
- 10.Oppenheimer S. 2004. Out of Eden: the peopling of the world, 2nd edn London, UK: Constable [Google Scholar]
- 11.Oppenheimer S. 2003. Out of Eden: the peopling of the world. London, UK: Constable [Google Scholar]
- 12.Oppenheimer S. 2009. The great arc of dispersal of modern humans: Africa to Australia. Quat. Int. 202, 2–13 10.1016/j.quaint.2008.05.015 (doi:10.1016/j.quaint.2008.05.015) [DOI] [Google Scholar]
- 13.Lahr M., Foley R. 1994. Multiple dispersals and modern human origins. Evol. Anthropol. 3, 48–60 10.1002/evan.1360030206 (doi:10.1002/evan.1360030206) [DOI] [Google Scholar]
- 14.Underhill P., Passarino G., Lin A., Shen P., Lahr M., Foley R., Oefner P., Cavalli-Sforza L. 2001. The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Ann. Hum. Genet. 65, 43–62 10.1046/j.1469-1809.2001.6510043.x (doi:10.1046/j.1469-1809.2001.6510043.x) [DOI] [PubMed] [Google Scholar]
- 15.Richards M., Bandelt H.-J., Kivisild T., Oppenheimer S. 2006. A model for the dispersal of modern humans out of Africa. In Human mitochondrial DNA and the evolution of Homo sapiens (eds Bandelt H.-J., Macaulay V., Richards M.), pp. 227–257 Hamburg, UK: Springer [Google Scholar]
- 16.Petraglia M. D., Haslam M., Fuller D. Q., Boivin N., Clarkson C. 2010. Out of Africa: new hypotheses and evidence for the dispersal of Homo sapiens along the Indian Ocean rim. Ann. Hum. Biol. 37, 288–311 10.3109/03014461003639249 (doi:10.3109/03014461003639249) [DOI] [PubMed] [Google Scholar]
- 17.Liu W., et al. 2010. Human remains from Zhirendong, South China, and modern human emergence in East Asia. Proc. Natl Acad. Sci. USA 107, 19 201–19 206 10.1073/pnas.1014386107 (doi:10.1073/pnas.1014386107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Armitage S. J., Jasim S. A., Marks A. E., Parker A. G., Usik V. I., Uerpmann H. P. 2011. The southern route ‘Out of Africa’: evidence for an early expansion of modern humans into Arabia. Science 331, 453–456 10.1126/science.1199113 (doi:10.1126/science.1199113) [DOI] [PubMed] [Google Scholar]
- 19.McBrearty S., Stringer C. 2007. The coast in colour. Nature 449, 793–794 10.1038/449793a (doi:10.1038/449793a) [DOI] [PubMed] [Google Scholar]
- 20.O'Connor S. 2007. New evidence from East Timor contributes to our understanding of earliest modern human colonisation east of the Sunda shelf. Antiquity 81, 523–535 [Google Scholar]
- 21.Richards M., et al. 2000. Tracing European founder lineages in the near Eastern mtDNA pool. Am. J. Hum. Genet. 67, 1251–1276 [PMC free article] [PubMed] [Google Scholar]
- 22.Hudjashov G., et al. 2007. Revealing the prehistoric settlement of Australia by Y chromosome and mtDNA analysis. Proc. Natl Acad. Sci. USA 104, 8726–8730 10.1073/pnas.0702928104 (doi:10.1073/pnas.0702928104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingman M., Kaessmann H., Pääbo S., Gyllensten U. 2000. Mitochondrial genome variation and the origin of modern humans. Nature 408, 708–713 10.1038/35047064 (doi:10.1038/35047064) [DOI] [PubMed] [Google Scholar]
- 24.Mishmar D., et al. 2003. Natural selection shaped regional mtDNA variation in humans. Proc. Natl Acad. Sci. USA 100, 171–176 10.1073/pnas.0136972100 (doi:10.1073/pnas.0136972100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macaulay V., et al. 2005. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science 308, 1034–1036 10.1126/science.1109792 (doi:10.1126/science.1109792) [DOI] [PubMed] [Google Scholar]
- 26.Ho S. Y., Phillips M. J., Cooper A., Drummond A. J. 2005. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 22, 1561–1568 10.1093/molbev/msi145 (doi:10.1093/molbev/msi145) [DOI] [PubMed] [Google Scholar]
- 27.Endicott P., Ho S. W. Y., Metspalu M., Stringer C. 2009. Evaluating the mitochondrial timescale of human evolution. Trends Ecol. Evol. 24, 515–521 10.1016/j.tree.2009.04.006 (doi:10.1016/j.tree.2009.04.006) [DOI] [PubMed] [Google Scholar]
- 28.Henn B. M., Gignoux C. R., Feldman M. W., Mountain J. L. 2009. Characterizing the time dependency of human mitochondrial DNA mutation rate estimates. Mol. Biol. Evol. 26, 217–230 10.1093/molbev/msn244 (doi:10.1093/molbev/msn244) [DOI] [PubMed] [Google Scholar]
- 29.Atkinson Q. D., Gray R. D., Drummond A. J. 2008. mtDNA variation predicts population size in humans and reveals a major southern Asian chapter in human prehistory. Mol. Biol. Evol. 25, 468–474 10.1093/molbev/msm277 (doi:10.1093/molbev/msm277) [DOI] [PubMed] [Google Scholar]
- 30.Atkinson Q. D., Gray R. D., Drummond A. J. 2009. Bayesian coalescent inference of major human mitochondrial DNA haplogroup expansions in Africa. Proc. R. Soc. B 276, 367–373 10.1098/rspb.2008.0785 (doi:10.1098/rspb.2008.0785) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Summerhayes G. R., Leavesley M., Fairbairn A., Mandui H., Field J., Ford A., Fullagar R. 2010. Human adaptation and plant use in highland New Guinea 49,000 to 44,000 years ago. Science 330, 78–81 10.1126/science.1193130 (doi:10.1126/science.1193130) [DOI] [PubMed] [Google Scholar]
- 32.Darwin C. R. 1871. The descent of man, and selection in relation to sex, ch. 6 London, UK: John Murray [Google Scholar]
- 33.Green R. E., et al. 2010. A draft sequence of the Neanderthal genome. Science 328, 710–722 10.1126/science.1188021 (doi:10.1126/science.1188021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramachandran S., Deshpande O., Roseman C. C., Rosenberg N. A., Feldman M. W., Cavalli-Sforza L. L. 2005. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl Acad. Sci. USA 102, 15 942–15 947 10.1073/pnas.0507611102 (doi:10.1073/pnas.0507611102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krause J., Fu Q., Good J. M., Viola B., Shunkov M. V., Derevianko A. P., Pääbo S. 2010. The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature 464, 894–897 10.1038/nature08976 (doi:10.1038/nature08976) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reich D., et al. 2010. Genetic history of an archaic hominin group from Denisova cave in Siberia. Nature 468, 1053–1060 10.1038/nature09710 (doi:10.1038/nature09710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasmussen M., et al. 2011. An aboriginal Australian genome reveals separate human dispersals into Asia. Science 334, 94–98 10.1126/science.1211177 (doi:10.1126/science.1211177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown P. 1992. Recent human evolution in East Asia and Australasia. Phil. Trans. R. Soc. Lond. B 337, 235–242 10.1098/rstb.1992.0101 (doi:10.1098/rstb.1992.0101) [DOI] [PubMed] [Google Scholar]
- 39.Stringer C. B. 1989. Documenting the origin of modern humans. In The emergence of modern humans (ed. Trinkaus E.), pp. 67–96 Cambridge, UK: Cambridge University Press [Google Scholar]
- 40.White T. D., Asfaw B., DeGusta D., Gilbert H., Richards G. D., Suwa G., Howell F. C. 2003. Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature 423, 742–747 10.1038/nature01669 (doi:10.1038/nature01669) [DOI] [PubMed] [Google Scholar]
- 41.McDougall I., Brown F. H., Fleagle J. G. 2005. Stratigraphic placement and age of modern humans from Kibish, Ethiopia. Nature 433, 733–736 10.1038/nature03258 (doi:10.1038/nature03258) [DOI] [PubMed] [Google Scholar]
- 42.Oppenheimer S. 2002. The first exodus: journey out of Africa. Geogr. Mag. 74, 32–35 [Google Scholar]
- 43.Metspalu M., et al. 2004. Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans. BMC Genet. 5, 26–50 10.1186/1471-2156-5-26 (doi:10.1186/1471-2156-5-26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Underhill P. A., Kivisild T. 2007. Use of Y chromosome and mitochondrial DNA population structure in tracing human migrations. Annu. Rev. Genet. 41, 539–564 10.1146/annurev.genet.41.110306.130407 (doi:10.1146/annurev.genet.41.110306.130407) [DOI] [PubMed] [Google Scholar]
- 45.Lahr M., Foley R. 1998. Towards a theory of modern human origins: geography, demography, and diversity in recent human evolution. Yearb. Phys. Anthropol. 41, 137–176 [DOI] [PubMed] [Google Scholar]
- 46.Lahr M. M. 1996. The evolution of modern human diversity: a study of cranial variation. Cambridge, UK: Cambridge University Press [Google Scholar]
- 47.Stringer C. B. 2000. Coasting out of Africa. Nature 405, 24–27 10.1038/35011166 (doi:10.1038/35011166) [DOI] [PubMed] [Google Scholar]
- 48.Wells S. 2002. The journey of man: a genetic odyssey. Allen Lane, UK: The Penguin Press [Google Scholar]
- 49.The Y Chromosome Consortium. 2002. A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 12, 339–348 10.1101/gr.217602 (doi:10.1101/gr.217602) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Underhill P. A., et al. 2000. Y chromosome sequence variation and the history of human populations. Nat. Genet. 26, 358–361 10.1038/81685 (doi:10.1038/81685) [DOI] [PubMed] [Google Scholar]
- 51.Semino O., et al. 2004. Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am. J. Hum. Genet. 74, 1023–1034 10.1086/386295 (doi:10.1086/386295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cruciani F., et al. 2004. Phylogeographic analysis of haplogroup E3b (E-M215) Y chromosomes reveals multiple migratory events within and out of Africa. Am. J. Hum. Genet. 74, 1014–1022 10.1086/386294 (doi:10.1086/386294) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luis J. R., Rowold D. J., Regueiro M., Caeiro B., Cinnioglu C., Roseman C., Underhill P. A., Cavalli-Sforza L. L., Herrera R. J. 2004. The Levant versus the horn of Africa: evidence for bidirectional corridors of human migrations. Am. J. Hum. Genet. 74, 532–544 10.1086/382286 (doi:10.1086/382286) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karafet T. M., Mendez F. L., Meilerman M. B., Underhill P. A., Zegura S. L., Hammer M. F. 2008. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 18, 830–838 10.1101/gr.7172008 (doi:10.1101/gr.7172008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hammer M. F., Karafet T., Rasanayagam A., Wood E. T., Altheide T. K., Jenkins T., Griffiths R. C., Templeton A. R., Zegura S. L. 1998. Out of Africa and back again: nested cladistic analysis of human Y chromosome variation. Mol. Biol. Evol. 15, 427–441 [DOI] [PubMed] [Google Scholar]
- 56.Abu-Amero K. K., Hellani A., González A. M., Larruga J. M., Cabrera V. M., Underhill P. A. 2009. Saudi Arabian Y-chromosome diversity and its relationship with nearby regions. BMC Genet. 10, 59–68 10.1186/1471-2156-10-59 (doi:10.1186/1471-2156-10-59) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trombetta B., Cruciani F., Sellitto D., Scozzari R. 2011. A new topology of the human Y chromosome haplogroup E1b1 (E-P2) revealed through the use of newly characterized binary polymorphisms. PLoS ONE 6, e16073. 10.1371/journal.pone.0016073 (doi:10.1371/journal.pone.0016073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hill C., et al. 2006. Phylogeography and ethnogenesis of aboriginal southeast Asians. Mol. Biol. Evol. 23, 1–12 10.1093/molbev/msl124 (doi:10.1093/molbev/msl124) [DOI] [PubMed] [Google Scholar]
- 59.Thangaraj K., Chaubey G., Kivisild T., Reddy A. G., Singh V. K., Rasalkar A. A., Singh L. 2005. Reconstructing the origin of Andaman Islanders. Science 308, 996. 10.1126/science.1109987 (doi:10.1126/science.1109987) [DOI] [PubMed] [Google Scholar]
- 60.Kivisild T., et al. 2004. Ethiopian mitochondrial DNA heritage: tracking gene flow across and around the Gate of Tears. Am. J. Hum. Genet. 75, 752–770 10.1086/425161 (doi:10.1086/425161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cruciani F., et al. 2002. A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am. J. Hum. Genet. 70, 1197–1214 10.1086/340257 (doi:10.1086/340257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cruciani F., et al. 2010. Human Y chromosome haplogroup R-V88: a paternal genetic record of early mid Holocene trans-Saharan connections and the spread of Chadic languages. Eur. J. Hum. Genet. 18, 800–807 10.1038/ejhg.2009.231 (doi:10.1038/ejhg.2009.231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yan Z., Petit-Maire N. 1994. The last 140 ka in the Afro-Asian arid/semi-arid transitional zone. Palaeogeogr. Palaeoclimatol. Palaeoecol. 110, 217–233 10.1016/0031-0182(94)90085-X (doi:10.1016/0031-0182(94)90085-X) [DOI] [Google Scholar]
- 64.Sauer C. O. 1963. Seashore–primitive home of man? In Land and life: a selection from the writings of Carl Ortwin Sauer (ed. John L.), pp. 312–330 Berkeley, CA: University of California Press [Google Scholar]
- 65.Faure H., Walter R. C., Grant D. R. 2002. The coastal oasis: ice age springs on emerged continental shelves. Glob. Planet. Change 33, 47–56 10.1016/S0921-8181(02)00060-7 (doi:10.1016/S0921-8181(02)00060-7) [DOI] [Google Scholar]
- 66.Selivanov Y. I. 1982. The Dasht-E Lut desert of Iran. Problemy Osvoeniya Pustyn (Problems of Desert Development) (1), 14–19 (in Russian) [Google Scholar]
- 67.Schultz H., von Rad U., Erlenkeuser H. 1998. Correlation between Arabian Sea and Greenland climate oscillations of the past 110,000 years. Nature 393, 54–57 10.1038/31750 (doi:10.1038/31750) [DOI] [Google Scholar]
- 68.Mellars P. 2006. A new radiocarbon revolution and the dispersal of modern humans in Eurasia. Nature 439, 931–935 10.1038/nature04521 (doi:10.1038/nature04521) [DOI] [PubMed] [Google Scholar]
- 69.Soares P., Achilli A., Semino O., Davies W., Macaulay V., Bandelt H.-J., Torroni A., Richards M. B. 2010. The Archaeogenetics of Europe. Curr. Biol. 20, R174–R183 10.1016/j.cub.2009.11.054 (doi:10.1016/j.cub.2009.11.054) [DOI] [PubMed] [Google Scholar]
- 70.Grün R., Stringer C. B. 1991. Electron spin resonance dating and the evolution of modern humans. Archaeometry 33, 153–199 10.1111/j.1475-4754.1991.tb00696.x (doi:10.1111/j.1475-4754.1991.tb00696.x) [DOI] [Google Scholar]
- 71.Stringer C. B., Grün R., Schwarcz H. P., Goldberg P. 1989. ESR dates for the hominid burial site of Es Skhul in Israel. Nature 338, 756–758 10.1038/338756a0 (doi:10.1038/338756a0) [DOI] [PubMed] [Google Scholar]
- 72.Rose J. I. 2010. New light on human prehistory in the Arabo-Persian Gulf Oasis. Curr. Anthropol. 51, 849–883 10.1086/657397 (doi:10.1086/657397) [DOI] [Google Scholar]
- 73.Dennell R. 2010. Early Homo sapiens in China. Research news and views. Nature 468, 512–513 10.1038/468512a (doi:10.1038/468512a) [DOI] [PubMed] [Google Scholar]
- 74.Westaway K. E., et al. 2007. Age and biostratigraphic significance of the Punung Rainforest Fauna, East Java, Indonesia, and implications for Pongo and Homo. J. Hum. Evol. 53, 709–717 10.1016/j.jhevol.2007.06.002 (doi:10.1016/j.jhevol.2007.06.002) [DOI] [PubMed] [Google Scholar]
- 75.Storm P., Aziz F., de Vos J., Kosasih D., Baskoro S., Ngaliman, van den Hoek Ostende L. W. 2005. Late Pleistocene Homo sapiens in a tropical rainforest fauna in East Java. J. Hum. Evol. 49, 536–545 10.1016/j.jhevol.2005.06.003 (doi:10.1016/j.jhevol.2005.06.003) [DOI] [PubMed] [Google Scholar]
- 76.Shen G., Wang W., Wang Q., Zhao J., Collerson K., Zhou C., Tobias P. V. 2002. U-series dating of Liujiang hominid site in Guanxi, Southern China. J. Hum. Evol. 43, 817–829 10.1006/jhev.2002.0601 (doi:10.1006/jhev.2002.0601) [DOI] [PubMed] [Google Scholar]
- 77.Moore M. W., Brumm A. 2007. Stone artifacts and hominins in island southeast Asia: new insights from Flores, eastern Indonesia. J. Hum. Evol. 52, 85–102 10.1016/j.jhevol.2006.08.002 (doi:10.1016/j.jhevol.2006.08.002) [DOI] [PubMed] [Google Scholar]
- 78.Petraglia M., et al. 2007. Middle Paleolithic assemblages from the Indian subcontinent before and after the Toba super-eruption. Science 317, 114–116 10.1126/science.1141564 (doi:10.1126/science.1141564) [DOI] [PubMed] [Google Scholar]
- 79.Balter M. 2010. News of the week, archaeology: of two minds about Toba's impact. Science 327, 1187–1188 10.1126/science.327.5970.1187-a (doi:10.1126/science.327.5970.1187-a) [DOI] [PubMed] [Google Scholar]
- 80.Sun C., et al. 2006. The dazzling array of basal branches in the mtDNA macrohaplogroup M from India as inferred from complete genomes. Mol. Biol. Evol. 23, 683–690 10.1093/molbev/msj078 (doi:10.1093/molbev/msj078) [DOI] [PubMed] [Google Scholar]
- 81.Palanichamy M. G., et al. 2004. Phylogeny of mitochondrial DNA macrohaplogroup N in India, based on complete sequencing: implications for the peopling of South Asia. Am. J. Hum. Genet. 75, 966–978 10.1086/425871 (doi:10.1086/425871) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thangaraj K., Chaubey G., Singh V. K., Vanniarajan A., Thanseem I., Reddy A. G., Singh L. 2006. In situ origin of deep rooting lineages of mitochondrial Macrohaplogroup ‘M’ in India. BMC Genomics 7, 151–156 10.1186/1471-2164-7-151 (doi:10.1186/1471-2164-7-151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trinkaus E., et al. 2003. An early modern human from the Pesteracu Oase, Romania. Proc. Natl Acad. Sci. USA 100, 11 231–11 236 10.1073/pnas.2035108100 (doi:10.1073/pnas.2035108100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mijares A. S., et al. 2010. New evidence for a 67000-year-old human presence at Callao Cave, Luzon Philippines. J. Hum. Evol. 59, 123–132 10.1016/j.hevol.20104.008 (doi:10.1016/j.hevol.20104.008) [DOI] [PubMed] [Google Scholar]
- 85.Bulbeck D., O'Connor S. 2011. The Watinglo mandible: a second terminal Pleistocene Homo sapiens fossil from tropical Sahul with a test on existing models for the human settlement of the region. HOMO J. Comp. Hum. Biol. 62, 1–29 10.1016/j.jchb.2010.10.002 (doi:10.1016/j.jchb.2010.10.002) [DOI] [PubMed] [Google Scholar]
- 86.Détroit F., Dizon E., Falguères C., Hameau S., Ronquillo W., Sémah F. 2004. Upper Pleistocene Homo sapiens from the Tabon cave (Palawan, the Philippines): description and dating of new discoveries. CR Palevol. 3, 705–712 10.1016/j.crpv.2004.06.004 (doi:10.1016/j.crpv.2004.06.004) [DOI] [Google Scholar]
- 87.Higham T. F. G., Barton H., Turnery C. S. M., Barker G., Ramsey C. B., Brock F. 2009. Radiocarbon dating of charcoal from tropical sequences: results from the Niah Great Cave, Sarawak, and their broader implications. J. Quat. Sci. 24, 189–197 10.1002/jqs.1197 (doi:10.1002/jqs.1197) [DOI] [Google Scholar]
- 88.Barker G., et al. 2007. The ‘human revolution’ in lowland tropical southeast Asia: the antiquity and behavior of anatomically modern humans at Niah Cave (Sarawak, Borneo). J. Hum. Evol. 52, 243–261 10.1016/j.jhevol.2006.08.011 (doi:10.1016/j.jhevol.2006.08.011) [DOI] [PubMed] [Google Scholar]
- 89.Summerhayes G. R., Leavesley M., Fairbairn A., Mandui H., Field J., Ford A., Fullagar R. 2010. Human adaptation and plant use in highland New Guinea 49,000 to 44,000 years ago. Science 330, 78–80 10.1126/science.1193130 (doi:10.1126/science.1193130) [DOI] [PubMed] [Google Scholar]
- 90.Leavesley M. G., Bird M. I., Fifield L. K., Dos Santos G. M., Hausladen P. A., Di Tada M. L. 2002. Buang Merabak: 40,000 radiocarbon years of human occupation in the Bismarck Archipelago, Papua New Guinea. Aust. Archaeol. 55, 55–57 [Google Scholar]
- 91.Turney C., et al. 2001. Early human occupation at Devil's Lair, southwestern Australia 50,000 years ago. Quat. Res. 55, 3–13 10.1006/qres.2000.2195 (doi:10.1006/qres.2000.2195) [DOI] [Google Scholar]
- 92.Roberts R. G., Jones R., Spooner N. A., Head M. J., Murray A. S., Smith M. A. 1994. The human colonization of Australia: optical dates of 53,000 and 60,000 years bracket human arrival at the Deaf Adder Gorge, Northern Territory. Quat. Sci. Rev. 13, 575–583 10.1016/0277-3791(94)90080-9 (doi:10.1016/0277-3791(94)90080-9) [DOI] [Google Scholar]
- 93.Roberts R. G., Jones R. 2001. Chronologies of carbon and of silica: evidence concerning the dating of the earliest human presence in Northern Australia. In Humanity from African naissance to coming millennia (ed. Tobias P. V.), pp. 239–248 Firenze, Italy: Florence University Press [Google Scholar]
- 94.Bird M. I., Turney C. S. M., Fifield L. K., Jones R., Palmer A., Cresswell R., Robertson S. 2002. Radiocarbon analysis of the early archaeological site of Nauwalabila I, Arnhemland, Australia: implications for sample suitability and stratigraphic integrity. Quat. Sci. Rev. 21, 1061–1075 10.1016/S0277-3791(01)00058-0 (doi:10.1016/S0277-3791(01)00058-0) [DOI] [Google Scholar]
- 95.Bird M. I., Hope G., Taylor D. 2004. Populating PEP II: the dispersal of humans and agriculture through Austral-Asia and Oceania. Quat. Int. 118–119, 145–163 10.1016/S1040-6182(03)00135-6 (doi:10.1016/S1040-6182(03)00135-6) [DOI] [Google Scholar]
- 96.Siddall M., Rohling E. J., Almogi-Labin A., Hemleben C. H., Meischner D., Schmelzer I., Smeed D. A. 2003. Sea-level fluctuations during the last glacial cycle. Nature 423, 853–858 10.1038/nature01690 (doi:10.1038/nature01690) [DOI] [PubMed] [Google Scholar]
- 97.Oppenheimer S. 2005. The Significance of the Orang Asli and their possible relationship to the Pleistocene peoples of the Malay Peninsula and to the colonization of the World. In The Perak man and other prehistoric skeletons of Malaysia (ed. Majid Z.), pp. 447–462 Penang, Malaysia: University of Science Malaysia [Google Scholar]
- 98.Oppenheimer S. J. 2005. Arguments for and logical consequences of a single successful exit of anatomically modern humans (AMH) from Africa. Proceedings of the Australasian Society for Human Biology (2004): human contacts in the past: origins, adaptations, and health implications. HOMO J. Comp. Hum. Biol. 56, 291–292 [Google Scholar]
- 99.Friedlaender J. S., Friedlaender F. R., Hodgson J. A., Stoltz M., Koki G., Horvat G., Zhadanov S., Schurr T. G., Merriwether D. A. 2007. Melanesian mtDNA Complexity. PLoS ONE 2, e248. 10.1371/journal.pone.0000248 (doi:10.1371/journal.pone.0000248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McEvoy B. P., Lind J. M., Wang E. T., Moyzis R. K., Visscher P. M., van Holst Pellekaan S. M., Wilton A. N. 2010. Whole-genome genetic diversity in a sample of Australians with deep aboriginal ancestry. Am. J. Hum. Genet. 87, 297–305 10.1016/j.ajhg.2010.07.008 (doi:10.1016/j.ajhg.2010.07.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Oppenheimer S. 2004. Austronesian spread into southeast Asia and Oceania: where from and when. Pacific archaeology: assessments and prospects: Proc. Int. Conf. for the 50th Anniversary of the first Lapita excavation, Kone, Noumea, 2002. Les Cahiers de l'Archeologie en Nouvelle Caledonie, 15, 54–70 [Google Scholar]
- 102.Hill C., et al. 2007. A mitochondrial stratigraphy for Island southeast Asia. Am. J. Hum. Genet. 80, 29–43 10.1086/510412 (doi:10.1086/510412) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Soares P., et al. 2011. Ancient voyaging and Polynesian origins. Am. J. Hum. Genet. 88, 1–9 10.1016/j.ajhg.2011.01.009 (doi:10.1016/j.ajhg.2011.01.009) [DOI] [PMC free article] [PubMed] [Google Scholar]