Abstract
The human leucocyte antigen (HLA) system shows extensive variation in the number and function of loci and the number of alleles present at any one locus. Allele distribution has been analysed in many populations through the course of several decades, and the implementation of molecular typing has significantly increased the level of diversity revealing that many serotypes have multiple functional variants. While the degree of diversity in many populations is equivalent and may result from functional polymorphism(s) in peptide presentation, homogeneous and heterogeneous populations present contrasting numbers of alleles and lineages at the loci with high-density expression products. In spite of these differences, the homozygosity levels are comparable in almost all of them. The balanced distribution of HLA alleles is consistent with overdominant selection. The genetic distances between outbred populations correlate with their geographical locations; the formal genetic distance measurements are larger than expected between inbred populations in the same region. The latter present many unique alleles grouped in a few lineages consistent with limited founder polymorphism in which any novel allele may have been positively selected to enlarge the communal peptide-binding repertoire of a given population. On the other hand, it has been observed that some alleles are found in multiple populations with distinctive haplotypic associations suggesting that convergent evolution events may have taken place as well. It appears that the HLA system has been under strong selection, probably owing to its fundamental role in varying immune responses. Therefore, allelic diversity in HLA should be analysed in conjunction with other genetic markers to accurately track the migrations of modern humans.
Keywords: histocompatibility, HLA, migrations, selection, convergent evolution, diversification
1. Genetic and functional variation of major histocompatibility complex genes and products
The major histocompatibility complex (MHC) was initially identified because differences in proteins from different individuals that are encoded in this genetic system play a major role in the rejection of tissues and organs. Two types of histocompatibility molecules, class I and II, are expressed in nucleated cells or antigen-presenting cells, respectively. The class I and class II MHC genes encode cell surface heterodimers; they play an important role in antigen presentation, tolerance and self/non-self recognition [1–3]. The MHC class I molecules form a stable tri-molecular complex composed of an MHC-encoded heavy chain, beta-2 microglobulin and small peptides. In live cells, the peptides presented in these complexes derive from intracellular proteins; this complex is the ligand of the antigen receptor of cytotoxic T-lymphocytes. In these molecules, some sub-structures, called peptide-binding specificity pockets, accommodate the side chains of the bound peptides [3–7]. The MHC class II molecules are also tri-molecular complexes, composed of a peptide and two subunits (alpha and beta) that are encoded in the MHC; in the case of the class II molecules, the peptides presented derive from extracellular proteins that are endocytosed in the antigen-presenting cells. The class II molecules are the ligands of the T-cell receptor of the helper T-lymphocytes. In spite of the fact that the class I and class II molecules are structurally different from each other, they present similar spatial conformations.
The MHC of humans is located in the short arm of chromosome 6; this is called the human leucocyte antigen (HLA) system and spans approximately 3.5 megabases. In this system, three regions can be identified according to the gene type content. The class II region is centromeric; it includes the genes that encode for three isotypes (DR, DQ, DP) of class II molecules. The genes encoding for the heavy chain of the class I molecules reside in the most telomeric region. The intervening region between the class I and class II regions is denominated the class III region. In this region, there are many genes involved in immune function; these include the genes encoding for the C2 and C4 proteins of the complement cascade as well as heat-shock proteins and tumour necrosis factors.
The HLA system is rich in highly homologous genes, many of them pseudogenes that do not encode for any functional protein. The alleles of different contiguous loci may be found together in the same individual more often than expected by random distribution according to their gene frequencies. The genetic phenomenon of this association at the population level is denominated linkage disequilibrium (LD).
Since its discovery, one of the most striking features of the HLA system has been the observation of an extensive degree of variation in both the number of loci and the number of alleles at those loci. These loci are the most polymorphic ones in the human genome. Over 6400 alleles have been identified, and more than 2000 of these are at a single locus (HLA-B). More than 1000 alleles have been observed at each of the HLA-A, -C and –DRB1 loci. The first hint of this diversity was obtained with the use of serological and cellular reagents. It has been speculated that this degree of diversity is a correlate of the biological functions of the molecules encoded in the MHC region. The extensive population polymorphism of the MHC genes may have resulted from selective pressures and functional adaptations [8–10]. It has been shown that the highest degree of variability of MHC proteins is found in residues pointing toward the peptide-binding region [11,12]. Cornerstone discoveries in the 1980s demonstrated that the main function of both class I and class II MHC molecules is to bind and present antigenic peptides to T-lymphocytes. The three-dimensional structure of both the class I and class II molecules shows that their distal membrane domains fold in a manner that defines a cavity, called the antigen recognition site (ARS), which accommodates peptides with notable precision. It has been shown that the antigenic peptides are bound by these molecules through interactions between the side chains of their amino acids and sub-structures (peptide-binding pockets) of the MHC molecules. The peptides eluted from different HLA alleles show distinctive patterns and at certain positions (e.g. for many class I alleles, the eluted peptides present predominant amino acids at position 2 and the carboxyl-terminus position). The specificity for peptide preferences for each HLA allele correlates directly with the composition of amino acid residues pointing toward the peptide-binding pockets.
The amino acid sequences of HLA alleles reveal that many alleles differ from each other only by substitutions in residues that contribute to the structure of the peptide-binding pockets. Therefore, this variation may lead to differences in immune responses among individuals. It is thus thought that the distinguishing allelic polymorphism is functional because the alleles with different amino acid sequences may have a differential peptide-binding capability. The immune responsiveness through peptide binding may therefore be considered as a dominant trait. If this is the case, then distribution of MHC alleles in different populations may be a consequence of functional polymorphisms. In many instances, the immune response to a particular protein of a pathogen may depend on the MHC alleles carried by an individual. Individuals heterozygous for MHC alleles have a wider peptide-binding repertoire and therefore have the capability to respond to various pathogens. However, the HLA system displays a functional redundancy in that there are several homologous expressed loci (e.g. HLA-A, -B and -C) that may compensate for the deficits presented by homozygosity at a single locus. The heterozygous advantage may be demonstrated in some species (e.g. chicken), which do not have a redundant MHC in which it has been clearly demonstrated that heterozygous individuals do have an advantage in responding to pathogens and are able to survive different infectious epidemics [13].
In addition to their natural biological function, i.e. to bind and present peptides, the class I and class II histocompatibility antigens play an important role in allogeneic transplantation. Matching for the alleles at the class I and class II MHC loci impacts the outcome of both solid organ [14,15] and haematopoietic stem cell [15–17] allogeneic transplants.
2. Allelic diversity of HLA loci in various populations
The distribution of HLA alleles defined at the serological level was initially examined in various outbred populations. It was observed that the HLA-A, -B -C and -DRB1 loci display levels of homozygosity below those expected for populations evolving under neutral conditions (e.g. genetic drift). When these loci are examined using molecular typing methods, it is found that many serologically indistinguishable subtypes can be observed in the same population. On the other hand, some alleles of the same serotype or allelic lineage that display limited structural differences are observed with distinctive frequency distributions in different populations.
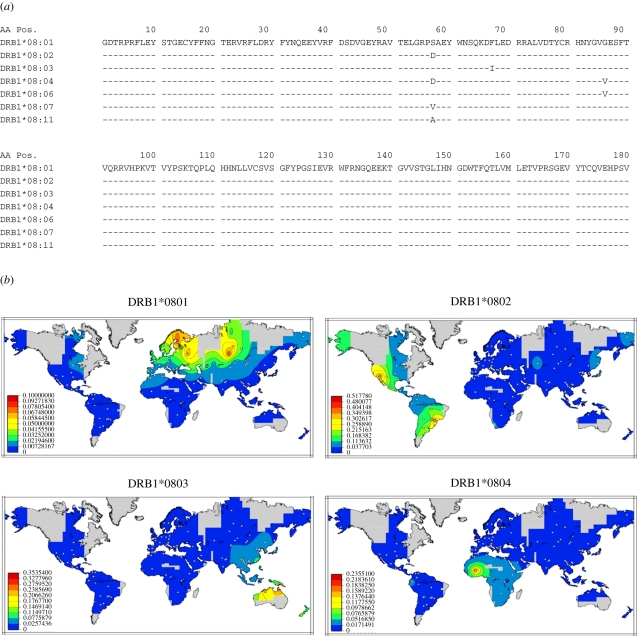
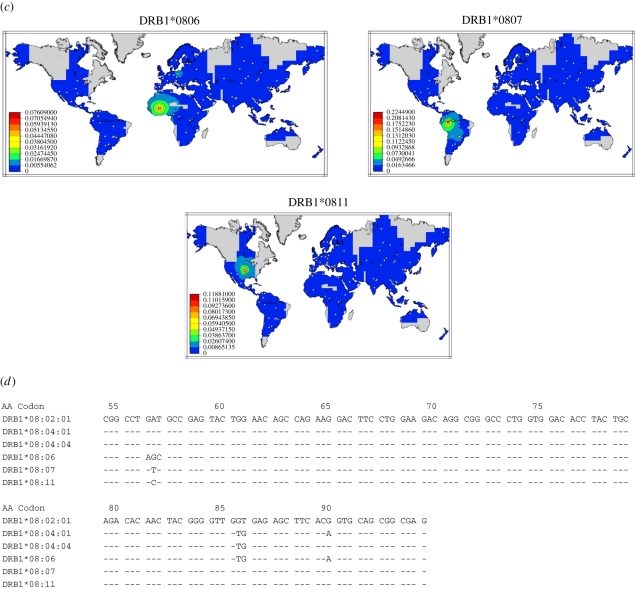
The HLA nomenclature has evolved over time in an attempt to capture the definitions achieved by methodological advances while trying to maintain or correlate with historical definitions. According to the current nomenclature [18], alleles of a specific locus are annotated using the name of the locus followed by an asterisk that separates the name from four different field types that are separated by colons. Under this nomenclature, the first field describes the allele family, which often corresponds to the serological antigen carried by the allotype. The second field is assigned in the order in which the sequences have been determined. Alleles whose numbers differ in the first two fields differ in one or more nucleotide substitutions that change the amino acid sequence of the encoded protein. Alleles that differ only by synonymous nucleotide substitutions within the coding sequence are distinguished by the use of the third field. Alleles that only differ by sequence polymorphisms in introns or in the 5′ and 3′ untranslated regions that flank the exons and introns are distinguished by the use of the fourth field. Figure 1a shows the protein sequences of the most common alleles of the HLA-DR8 serotype; figure 1b,c displays the gene frequency distributions of the most common subtypes of this serotype in various world populations. Each of these alleles is common in a specific region of the world and may be absent from other populations. This example illustrates how technical resolution limitations may lead to erroneous inferences of genetic relatedness between populations. Table 1 shows the different haplotype fragments (blocks) that include alleles of HLA-DR8 alleles and the associated alleles of the contiguous DQA1 and DQB1 loci. Some alleles have identical protein sequences and only differ in their nucleotide sequence by silent substitutions. The analysis of both nucleotide sequence homology and haplotype constitution of alleles at contiguous loci may help elucidate the evolutionary relationships between alleles; figure 1d shows the nucleotide sequences that distinguish several alleles of this group. The alleles DRB1*08:04:02, DRB1*08:04:04, DRB1*08:07 and DRB1*08:11, which are found only in populations from the American continent, may be evolutionarily related and derive from the allele DRB1*08:02:01 which has a high frequency in almost all Native American populations. DRB1*08:02:01 is also found in Asian populations; the presence of this allele may identify the founder migrations from Asia to America through the Bering Strait. In contrast, the alleles DRB1*08:04:01 and DRB1*08:06, found most often in Africans, may be related. The evolutionary relations are proposed because even a single mutation/gene conversion may lead to the generation of a novel allele.
Figure 1.
(a) Amino acid sequences of the most common subtypes of HLA-DR8. The amino acid sequences of the extracellular domains of the alleles of the DRB1*08 group are compared, and indicate amino acid identity in a specific position with the allele DRB1*08:01. (b,c) Distribution of alleles of the HLA-DRB1*08 was inferred from 224 population samples, indicated by the plotted points. (d) Distinguishing nucleotide substitutions of common subtypes of HLA-DR8 of Native Americans and Africans. The graphical representations were obtained using PyPop software from a compilation of a meta-analytic review [19]. The coloured regions indicate interpolated allele frequency; grey areas are too far from a sampled population to permit meaningful frequency estimation. Only non-migrant populations were used to generate these maps. Frequency maps for all alleles at all loci are available at http://www.pypop.org/popdata/. Adapted from Solberg et al. [20].
Table 1.
DR-DQ blocks bearing alleles of the DRB1*08 group. Alleles at DQA1 and DQB1 that associate tightly with alleles of the DRB1*08 group are shown. These haplotype fragments were classified according to their frequencies in different populations [17–31]. Differences in shading denote different DQA1-DQB1 blocks that may associate with different alleles of the HLA-DRB1*08 group.
| DRB1 | DQA1 | DQB1 | frequency overall | exclusive for an ethnic group |
|---|---|---|---|---|
| 08:06 | 01:02 | 06.02 | common | African |
| 08:06 | 05:05 | 03.19 | less common | African (narrow distribution, North African?) |
| 08:04:01 | 04:01 | 03:19 | very common | African |
| 08:04:01 | 05:05 | 03:19 | common | African |
| 08:04:01 | 01:02 | 06:02 | rare | African |
| 08:04:01 | 04:01 | 04:02 | common | African/European |
| 08:01 | 04:01 | 04:02 | very common | European |
| 08:01 | 03:01 | 0302 | rare | European |
| 08:03 | 06:01 | 03:01 | common | European |
| 08:03 | 01:03 | 06:01 | very common | Asian |
| 08:02:01 | 04:01 | 04:02 | very common | American Indian/Asian |
| 08:02:01 | 03:01 | 0302 | common | Asian |
| 08:07 | 04:01 | 04:02 | common | South American Indian |
| 08:11 | 04:01 | 04:02 | common | North American Indian |
| 08:04:04 | 04:01 | 04:02 | common | North American Indian (narrow distribution) |
| 08:04:02 | 04:01 | 04:02 | common | South American Indian (narrow distribution) |
3. HLA alleles in outbred populations
In major outbred populations living in the USA (European Americans, African Americans, Hispanic or Latino Americans, Native Americans and Asian Pacific Islanders), we observed more than 25 HLA-A, 40 HLA-B, 15 HLA-C, 25 DRB1, 17 DQB1 and 15 DPB1 alleles with gene frequencies higher than 0.05 [21–24]. The allele distributions are fairly evenly distributed in most HLA loci analysed (A, B, C, DRB1, DQA1 and DQB1) with the exception of DPB1, in which only four alleles account for the majority of the genes of this locus. This even allele distribution results in low levels of homozygosity, again with the exception of DPB1. This distribution suggests overdominant selection (heterozygous advantage or frequency-dependent selection are indistinguishable).
In these studies, we were able to identify HLA alleles that were common and uniquely found in one group, but that were virtually absent in other groups; several ethnic-specific HLA alleles were identified in Asians, Africans and Native Americans, while in the Europeans we observed only a few common ethnic-specific alleles (DRB1*08:01:01 and DRB1*16:01:01). Among the loci with more alleles, HLA-A presented the lowest levels of diversity in Asians, Native Americans and Europeans, with a few alleles predominating and higher levels of homozygosity than HLA-B, C and DRB1 loci. In contrast, HLA-A in African Americans did not present any highly predominant allele; the level of homozygosity of HLA-A was only lower than that observed for HLA-B. The findings in the outbred populations of the USA are consistent with those from studies on large groups of individuals from the US National Marrow Donor Program [25]. Similar or larger levels of diversity were identified in outbred populations from the South American continent [26].
The so-called Hispanic/Latino groups are defined on the basis of the use of the Spanish language of the country of origin of the ancestors in the American continent. In the United States, the main contribution to the Hispanic/Latino groups comes from migrations from the Caribbean (Cuba, the Dominican Republic and Puerto Rico) or from Central America and Mexico. There are significant regional variations. The Hispanic subjects from states bordering with Mexico present specific haplotypes that are found in Spain, Native Americans from Mexico and Southern USA and, strikingly, in the Middle East. The top 10 full haplotypes of Hispanics and Mexicans include the top two haplotypes found in Lebanon and non-Ashkenazi Jews, four haplotypes that are common in all European populations and four haplotypes including alleles that are uniquely found in natives from Mexico. This observation indicates recent migrations and admixture. The presence of Middle Eastern haplotypes may represent the contribution of the Sephardic Diaspora migrating to the New World after being expelled from Spain at the end of the fifteenth century.
4. HLA variation in sub-saharan Africans
We investigated the allelic and haplotypic diversity of the HLA system in sub-Saharan African populations. In these populations, the distributions of genotypes at all loci and in all populations fit Hardy–Weinberg equilibrium expectations [27]. Similar to the outbred populations from the USA, most of the sub-Saharan African populations did not display a single predominant allele at any of the loci. In addition, all HLA allelic lineages from each of the class I and class II loci were observed in these populations. Interestingly, large numbers of alleles of HLA-A and B loci and fewer alleles of HLA-C and DRB1 loci that have intermediate or high frequencies were found virtually only in the African populations. Most of the African-only alleles are widely distributed in the African continent and their origin may predate the separation of linguistic groups. The sub-Saharan African populations individually present levels of diversity in HLA loci that are comparable but do not exceed those observed in other populations with the exception of HLA-A, which has lower levels of homozygosity in the African populations. The Luo population from Kenya presents the highest levels of allele and haplotypic diversity; this population shows the lowest genetic distance with other sub-Saharan populations. This finding is consistent with the hypotheses that this population is older or that there was a significant gene flow from other populations.
5. HLA profile of some Middle Eastern populations
We analysed the distribution of HLA alleles in Jewish subjects living in Israel [28]; these were classified into 31 groups defined by contemporary country-of-origin information, and their similarities and differences on the basis of HLA haplotypes were studied. In these groups, we observed significant allelic overlap with European populations; Jewish populations presented a few ethnic-specific alleles. The Ashkenazi and non-Ashkenazi groups presented distinctive HLA allele frequencies; even more clear distinctions arose through the analyses of haplotypes and haplotype fragments that identified even more clear differences between these groups. For example, the extended haplotype A*26:01-C*12:03-B*38:01-DRB1*04:02-DQB1*03:02 is typically found, often reaching very high haplotype frequency (greater than 0.1000), in groups with Ashkenazi descent; in contrast this haplotype is absent or rare in the Jewish populations with non-Ashkenazi ancestry.
In a recent study of Lebanese families [29], we observed high levels of diversity but no alleles that are unique to this population. Most of the alleles observed in this group are found in either Europeans or Far East Asians; the distribution of HLA alleles in Lebanese is significantly different from those observed in Europeans, Africans and Far East Asians. This population presents striking differences from other populations in the distribution of alleles of HLA-B; some alleles that are common in Lebanese are rare or have low frequency in most world populations. The allele B*73:01, which is structurally divergent from other alleles of HLA-B, is found frequently in the Lebanese population. The allele B*73:01 presents with its highest world frequency in the Lebanese population (gf = 0.0173). Contrasting with other populations in which this allele associates tightly only with C*15:05:01, in Lebanese the allele B*73:01 associates with C*15:05:01 and C*12:02:02. This observation suggests that the presence of B*73:01 in the Middle East may be older than in other world populations, and thus that this allele arose in this region and spread to other populations in Africa, Europe and Asia. A recent report showed that this allele was indeed identified in DNA from archaic humans [30], called Denisovans, and suggests that admixture between modern humans and archaic humans may have occurred in West Asia. In the Lebanese population, the two most common haplotypes are extended (A*33:01-C*08:02-B*14:02-DRB1*01:02-DQB1*05:01 and A*24:02-C*04:01-B*35:02-DRB1*11:04-DQB1*03:01). These two haplotypes are also the most common ones in non-Ashkenazi Jewish populations and are found often among Ashkenazi groups. These observations indicate that while some alleles and HLA haplotypes are found often in many populations from the Middle East, significant differences are identified when analysing the distribution of extended haplotypes.
6. HLA studies in Native American populations
We studied isolated populations including subjects of American Indian tribes from Mexico and South America [19,31–33]. We also studied subjects self-identified as Native Americans from the USA [21,23]. In all populations, the number of allelic lineages was significantly reduced when compared with other populations. In spite of the finding of a restricted number of alleles, we observed high levels of heterozygosity, with exception of the DPB1 locus.
The examination of ethnic-specific alleles indicated most of the findings in both inbred and outbred populations belonged to HLA-A, B and DRB1 loci. In the American Indian tribes, we observed very few allelic lineages (4 HLA-A, 7 -B, 7 -C, 4 -DRB1, 2 DQA1, 2 DQB1 and 5 DPB1). In spite of the limited number of lineages, we observed several alleles of the same lineage present in each tribe. Many of the alleles found in these tribes were not observed in other outbred populations or tribes. It can be postulated that these alleles were generated in the Americas and are novel alleles. Gene conversion events could be invoked as the mechanism for their generation. In fact, all putative novel alleles may derive from a few founder alleles (those alleles of each lineage found in other populations) and all the nucleotide sequences donated in the gene conversion events may have come from other founder alleles. Almost all novel alleles identified differ from other alleles in the same lineages by amino acid substitutions in residues pointing toward the peptide-binding groove, and may potentially have new peptide-binding capabilities. Most of the postulated gene conversion events could have involved alleles of the same locus. The HLA-B locus presented a relatively degree of diversity and the majority of the putative novel alleles found in these populations were from this locus, and it has been postulated that HLA-B has diversified more rapidly in the South American tribes. Interestingly, in many tribes the novel alleles are present at the highest gene frequencies, suggesting that the novel alleles generated in America were positively selected in these populations probably because they provided selective advantages. It is conceivable that with a limited founder polymorphism any novel allele that arose enlarged the peptide-binding repertoire of these populations. Perhaps the HLA-B locus diverged more than the HLA-A locus in the South American tribes, simply as a result of a higher number of opportunities for intra-locus gene conversions because this locus presented a larger number of founder alleles. However, it should be noted that the HLA-B locus displays high levels of allele diversity across all populations, and across all HLA loci; it may be that this locus is less constrained functionally than others, and is more tolerant of allelic diversity in general.
These studies identified large genetic distances between populations from the American continent; the distances are significantly reduced when replacing alleles by their corresponding serotypes. In contrast, the genetic distances in populations from other continents are in general smaller and correlate well with geographical distances. Furthermore, the genetic distances in populations from other continents do not differ significantly when evaluated by distribution of alleles or their corresponding broad serotypes.
7. HLA studies in other populations
The distribution of HLA alleles in different world populations has been the subject of collaborative studies conducted through the course of many years in the context of the International Histocompatibility Workshops. The Fifth International Histocompatibility Workshop in 1972 first conducted systematic anthropological HLA studies under the guidance of Prof. Jean Dausset and Prof. Walter Bodmer. Since then, the distribution of HLA diversity in human populations has been under close scrutiny using the typing tools that were available at the time. A recent meta-analysis of HLA distributions included data from 497 population samples [20]. Most of the datasets examined in this study included data from studies published in journals and additional datasets included in the International Histocompatibility Workshops and from a web-based compilation (AlleleFrequencies.net [34]). These studies found similar allele distribution patterns for most populations and loci. As with Native American populations (described above), the degree of differentiation was higher among populations from southeast Asia, Polynesia, Melanesia and Australia [35–40] than the rest of the world, and populations from these areas display reduced diversity in allelic lineages. The distribution of HLA alleles in ‘island type’ populations also resembled the ones described above for Native Americans; in the populations from Oceania, the DRB1 locus has more allelic lineages and appears to present higher degrees of differentiation. The findings described in the present report are concordant with those described thoroughly and in detail in the recent report by Sanchez-Mazas et al. [41].
8. HLA haplotypes and haplotype fragments (blocks) in different populations
LD patterns between alleles of various HLA loci may provide significant insight with regard to the history of a particular allele. The examination of both LD and structural features may help elucidate possible evolutionary relations between alleles. Population studies have revealed that the alleles of the DRB1 locus display tight associations, in some examples they were absolute, with DQA1 and DQB1 alleles. Some DRB1 alleles with high sequence homology have associations with the same DQA1 and DQB1 alleles. These shared block associations may mark the evolutionary relationship between some DRB1 alleles. This may be due to a rapid or recent diversification of an allelic lineage, or it may be due to selection for specific cis combinations of DRB1 and DQ alleles. The analysis of the linkage disequilibria between alleles of the class I loci showed tight associations between alleles of HLA-B and HLA-C and somewhat weaker between HLA-B and HLA-A. These data suggest that in the class I region, the strength of the associations between alleles of different loci correlates with the physical distances separating the loci.
Within B-C haplotypes, we observe two distinct patterns of LD; these may represent distinct modes of evolution. In one case, HLA-B alleles with similar nucleotide sequences (displaying a range of frequencies) were in LD with the same HLA-C allele. As with the DRB1-DQ haplotypes discussed above, this suggests the diversification of HLA-B allelic lineages in the context of specific B-C haplotypes, and may represent haplotype-level selection or rapid allelic diversification. In the second case, HLA-B alleles related in nucleotide sequence and observed at similar or balanced frequencies were in LD with different HLA-C alleles. This may be due to ancient HLA-B allelic diversification that has been recombined onto different B-C haplotypes, or alternatively to strong selection for B-C haplotype diversity, maximizing the available peptide-binding repertoire.
The current haplotypic composition of the HLA class I loci may result from two distinct effects. On one hand, LD between neighbouring loci may mark the evolutionary relationship between parental and novel alleles. On the other hand, the similar frequencies between novel and parental alleles may have resulted from selective advantages to respond to different pathogens and may be related to their differential peptide-binding abilities. Since molecules encoded by different class I loci have an overlapping peptide-binding function, differences in the haplotypic composition may result from the complementary/compensatory abilities of alleles in the same haplotype to bind peptides from different pathogens that have exerted selection.
These findings indicate that strong selection has operated at various levels on the HLA system. The current level of diversity and the variation in observed allelic distributions for different populations probably result from evolutionary forces that have changed as human populations have encountered new environments in their spread around the globe.
9. Convergent evolution in HLA
Some alleles have identical protein sequences and are distinguished at the nucleotide sequence level by silent substitutions or substitutions in non-coding segments. In many cases, these alleles are related by descent from a common ancestral sequence, but there are some examples of alleles that appear to have arisen independently. Figure 1 and table 1 include the DRB1*08:04 alleles that have distinct nucleotide sequences and distinctive associations with DQA1 and DQB1 alleles. A similar observation was made for other alleles differing only by silent substitutions. Figure 2a includes the nucleotide sequences of the B*52:01:01 and B*52:01:02 alleles; these alleles differ by one silent substitution at the third nucleotide of codon 23. The allele B*52:01:02 carries the same codon found in B*51:01:01; B*51:01:01 is present in the same populations in which B*52:01:02 is found, and it can be postulated that B*52:01:02 may derive from B*51:01:01, having arisen from a gene conversion event introducing a segment present in B*40:02:01. In sub-Saharan Africans, both B*52:01:02 and B*51:01:01 are in LD with C*16:01, while the B*52:01:01 allele is in LD with C*12:02:02 in Asians and Europeans (figure 2b). A de novo generation of HLA-B*52:01:02 may be also postulated in Native American populations. Convergent evolution indicates that the same allele can be generated in two or more independent events (table 2). Once generated, the novel structurally identical allele may be selected on the basis of its functional capabilities. It is postulated that additional convergent evolution events may have taken place through the evolution of the human MHC. The occurrence of the same allele in LD with different alleles at neighbouring loci in different, geographically distant populations suggests that these events may have occurred. Alternatively, these alleles may be ancient, and diverged through recombination that generated new haplotypes. Undetected convergent evolution events may be confounding in the investigation of population relationships, leading to erroneously close relations between populations.
Figure 2.
Possible evidence for convergent evolution. (a) Nucleotide sequence alignment of exon-2 of the most common alleles of HLA-B*52:01 and B*51:01 groups. The alleles B*52:01:01 and B*52:01:02 differ only by one nucleotide replacement at the third nucleotide of codon 23. The allele B*52:01:02 shares the same nucleotide with B*52:01:02. A putative distinct origin for B*52:01:01 and B*52:01:02 can be postulated; B*52:01:02 may have originated from a gene conversion event in which B*51:01:01 was the recipient of a DNA fragment of a minimal size of 14 nucleotides spanning codons 63–67. The donor of this fragment may be B*40:02:01; many other alleles of HLA-B have identical sequence in the donor segment and could be alternative donor candidates. (b) Distribution of B-C haplotypes bearing the alleles B*52:01:01 and B*52:01:02. Frequency maps for all alleles at all loci are available at http://www.pypop.org/popdata/. Adapted from Solberg et al. [20].
Table 2.
Frequency of HLA-C-B blocks in different populations. Frequencies were extracted from references [21,25,27]. The block B*52:01-C*12:02 is common in Europeans and Asians; this block includes the allele B*52:01:01; the blocks B*52:01-C*16:01 and B*52:01-C*03:03 are common in Africans and Native Americans; these haplotypes include B*52:01:02. The distinctive associations with alleles of HLA-C lend support to the hypothesis that B*52:01:01 and B*52:01:02 have distinctive origins. Differences in shading denote different DQA1-DQB1 blocks that may associate with different alleles of the HLA-DRB1*08 group.
| C | B | API_freq. | EUR_freq. | HIS_freq. | AFA_freq. |
|---|---|---|---|---|---|
| 1202 | 5201 g | 0.03354 | 0.00928 | 0.01229 | 0.00145 |
| 1601 | 5201 g | 0.00000 | 0.00019 | 0.00053 | 0.01226 |
| 0303 g | 5201 g | 0.00000 | 0.00000 | 0.00748 | 0.00000 |
10. Selection for diversification and convergent evolution may be confounding when tracking migrations
In the present and other reports [20,41], it is readily noticed that the genetic distances between open populations correlate well with their geographical locations, and for migrant populations with their regions of origin. In contrast, the genetic distance measurements are larger than expected between inbred populations of the same region. These larger than expected distances may derive from a large number of unique alleles in a small number of lineages as the result of limited founder polymorphism. In these populations, any novel allele may have been positively selected to enlarge the communal peptide-binding repertoire. Conversely, some alleles are found in multiple populations with distinctive haplotypic associations, suggesting that convergent evolution events may have taken place as well. Owing to its fundamental role in the vertebrate immune response, the HLA system has been under strong selection for millions of years. Therefore, allelic diversity in HLA should be analysed in the context of HLA haplotypes and blocks and in conjunction with other genetic markers to accurately track the migrations of modern humans.
References
- 1.Davis M. M., Bjorkman P. J. 1989. A model for T cell receptor and MHC/peptide interaction. Adv. Exp. Med. Biol. 254, 13–16 [DOI] [PubMed] [Google Scholar]
- 2.Marrack P., Kappler J. 1986. The antigen specific, major histocompatibility complex restricted receptor on T cells. Adv. Immunol. 38, 1–30 10.1016/S0065-2776(08)60005-X (doi:10.1016/S0065-2776(08)60005-X) [DOI] [PubMed] [Google Scholar]
- 3.Marrack P., Bender J., Jordan M., Rees W., Robertson J., Schaefer B. C., Kappler J. 2001. Major histocompatibility complex proteins and TCRs: do they really go together like a horse and carriage? J. Immunol. 167, 617–621 [DOI] [PubMed] [Google Scholar]
- 4.Marrack P., Kappler J. 1988. The T cell repertoire for antigen and MHC. Immunol. Today 9, 308–315 10.1016/0167-5699(88)91324-2 (doi:10.1016/0167-5699(88)91324-2) [DOI] [PubMed] [Google Scholar]
- 5.Saper M. A., Bjorkman P. J., Wiley D. C. 1991. Refined structure of the human histocompatibility antigen HLA A2 at 2.6A resolution. J. Mol. Biol. 219, 277–319 10.1016/0022-2836(91)90567-P (doi:10.1016/0022-2836(91)90567-P) [DOI] [PubMed] [Google Scholar]
- 6.Brown J. H., Jardetzky T. S., Gorga J. C., Stern L. J., Urban R. G., Strominger J. L., Wiley D. C. 1993. Three dimensional structure of the human class II histocompatibility antigen HLA DR1. Nature 364, 33–39 10.1038/364033a0 (doi:10.1038/364033a0) [DOI] [PubMed] [Google Scholar]
- 7.McFarland B. J., Beeson C. 2002. Binding interactions between peptides and proteins of the class II major histocompatibility complex. Med. Res. Rev. 22, 168–203 10.1002/med.10006 (doi:10.1002/med.10006) [DOI] [PubMed] [Google Scholar]
- 8.Lawlor D. A., Zemmour J., Ennis P. D., Parham P. 1990. Evolution of class I MHC genes and proteins: from natural selection to thymic selection. Annu. Rev. Immunol. 8, 23–63 10.1146/annurev.iy.08.040190.000323 (doi:10.1146/annurev.iy.08.040190.000323) [DOI] [PubMed] [Google Scholar]
- 9.Hughes A. L., Ota T., Nei M. 1990. Positive Darwinian selection promotes charge profile diversity in the antigen binding cleft of class I major histocompatibility complex molecules. Mol. Biol. Evol. 7, 515–524 [DOI] [PubMed] [Google Scholar]
- 10.Gustafsson K., Wiman K., Emmoth E., Larhammar D., Böhme J., Hyldig-Nielsen J. J., Ronne H., Peterson P. A., Rask L. 1984. Mutations and selection in the generation of class II histocompatibility antigen polymorphism. EMBO J. 3, 1655–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes A. L., Nei M. 1992. Maintenance of MHC polymorphism. Nature 355, 402–403 10.1038/355402b0 (doi:10.1038/355402b0) [DOI] [PubMed] [Google Scholar]
- 12.Hughes A. L., Nei M. 1989. Nucleotide substitution at major histocompatibility complex class II loci: evidence for overdominant selection. Proc. Natl Acad. Sci. USA 86, 958–962 10.1073/pnas.86.3.958 (doi:10.1073/pnas.86.3.958) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaufman J. 2000. The simple chicken major histocompatibility complex: life and death in the face of pathogens and vaccines. Phil. Trans. R. Soc. Lond. B 355, 1077–1084 10.1098/rstb.2000.0645 (doi:10.1098/rstb.2000.0645) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Opelz G., Wujciak T., Dohler B., Scherer S., Mytilineos J. 1999. HLA compatibility and organ transplant survival. Collaborative Transplant Study. Rev. Immunogenet. 1, 334–342 [PubMed] [Google Scholar]
- 15.Erlich H. A., Opelz G., Hansen J. 2001. HLA DNA typing and transplantation. Immunity 14, 347–356 10.1016/S1074-7613(01)00115-7 (doi:10.1016/S1074-7613(01)00115-7) [DOI] [PubMed] [Google Scholar]
- 16.Hansen J. A., Yamamoto K., Petersdorf E., Sasazuki T. 1999. The role of HLA matching in hematopoietic cell transplantation. Rev. Immunogenet. 1, 359. [PubMed] [Google Scholar]
- 17.Petersdorf E. W., Anasetti C., Martin P. J., Hansen J. A. 2003. Tissue typing in support of unrelated hematopoietic cell transplantation. Tissue Antigens 61, 1–11 10.1034/j.1399-0039.2003.610101.x (doi:10.1034/j.1399-0039.2003.610101.x) [DOI] [PubMed] [Google Scholar]
- 18.Marsh S. G., et al. 2010. Nomenclature for factors of the HLA system, 2010. Tissue Antigens 75, 291–455 10.1111/j.1399-0039.2010.01466.x (doi:10.1111/j.1399-0039.2010.01466.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez-Viña M., Lazaro A. M., Sun Y., Miller S., Forero L., Stastny P. 1995. Population diversity of B-locus alleles observed by high-resolution DNA typing. Tissue Antigens 45, 153–168 10.1111/j.1399-0039.1995.tb02435.x (doi:10.1111/j.1399-0039.1995.tb02435.x) [DOI] [PubMed] [Google Scholar]
- 20.Solberg O. D., Mack S. J., Lancaster A. K., Single R. M., Tsai Y., Sanchez-Mazas A., Thomson G. 2008. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum. Immunol. 69, 443–464 10.1016/j.humimm.2008.05.001 (doi:10.1016/j.humimm.2008.05.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao K., Hollenbach J., Shi X., Shi W., Chopek M., Fernandez-Vina M. A. 2001. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum. Immunol. 62, 109–130 10.1016/S0198-8859(01)00298-1 (doi:10.1016/S0198-8859(01)00298-1) [DOI] [PubMed] [Google Scholar]
- 22.Fernandez Vina M. A., Moraes J. R., Moraes M. E., Miller S., Stastny P. 1991. HLA class II haplotypes in Amerindians and in black North and South Americans. Tissue Antigens 38, 235–240 10.1111/j.1399-0039.1991.tb01904.x (doi:10.1111/j.1399-0039.1991.tb01904.x) [DOI] [PubMed] [Google Scholar]
- 23.Fernandez-Viña M. A., et al. 1991. Alleles at four HLA class II loci determined by oligonucleotide hybridization and their associations in five ethnic groups. Immunogenetics 34, 299–312 10.1007/BF00211994 (doi:10.1007/BF00211994) [DOI] [PubMed] [Google Scholar]
- 24.Fernandez-Vina M., Moraes M. E., Stastny P. 1991. DNA typing for class II HLA antigens with allele-specific or group-specific amplification. III. Typing for 24 alleles of HLA-DP. Hum. Immunol. 30, 60–68 10.1016/0198-8859(91)90072-H (doi:10.1016/0198-8859(91)90072-H) [DOI] [PubMed] [Google Scholar]
- 25.Maiers M., Gragert L., Klitz W. 2007. High-resolution HLA alleles and haplotypes in the United States population. Hum. Immunol. 68, 779–788 [Erratum in: Hum. Immunol. 2008 69,141.] [DOI] [PubMed] [Google Scholar]
- 26.Moraes M. E., Fernandez-Viña M., Salatiel I., Tsai S., Moraes J. R., Stastny P. 1993. HLA class II DNA typing in two Brazilian populations. Tissue Antigens 41, 238–242 10.1111/j.1399-0039.1993.tb02012.x (doi:10.1111/j.1399-0039.1993.tb02012.x) [DOI] [PubMed] [Google Scholar]
- 27.Cao K., et al. 2004. Differentiation between African populations is evidenced by the diversity of alleles and haplotypes of HLA class I loci. Tissue Antigens 63, 293–325 10.1111/j.0001-2815.2004.00192.x (doi:10.1111/j.0001-2815.2004.00192.x) [DOI] [PubMed] [Google Scholar]
- 28.Klitz W., Gragert L., Maiers M., Fernandez-Viña M., Ben-Naeh Y., Benedek G., Brautbar C., Israel S. 2010. Genetic differentiation of Jewish populations. Tissue Antigens 76, 442–458 10.1111/j.1399-0039.2010.01549.x (doi:10.1111/j.1399-0039.2010.01549.x) [DOI] [PubMed] [Google Scholar]
- 29.Cano P., Testi M., Khoriaty E., Monsef J. B., Troiano M., Inati A., Fernandez-Vina M. A., Andreani M. Submitted HLA diversity in the Lebanese population defined by the analysis of HLA haplotypes defined by segregation analysis. [Google Scholar]
- 30.Abi-Rached L., et al. 2011. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science 334, 89–94 10.1126/science.1209202 (doi:10.1126/science.1209202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerna M., Falco M., Friedman H., Raimondi E., Maccagno A., Fernandez-Viña M., Stastny P. 1993. Differences in HLA class II alleles of isolated South American Indian populations from Brazil and Argentina. Hum. Immunol. 37, 213–220 10.1016/0198-8859(93)90504-T (doi:10.1016/0198-8859(93)90504-T) [DOI] [PubMed] [Google Scholar]
- 32.Lázaro A. M., Moraes M. E., Marcos C. Y., Moraes J. R., Fernández-Viña M. A., Stastny P. 1999. Evolution of HLA-class I compared to HLA-class II polymorphism in Terena, a South-American Indian tribe. Hum. Immunol. 60, 1138–1149 10.1016/S0198-8859(99)00092-0 (doi:10.1016/S0198-8859(99)00092-0) [DOI] [PubMed] [Google Scholar]
- 33.Hollenbach J. A., Thomson G., Cao K., Fernandez-Vina M., Erlich H. A., Bugawan T. L., Winkler C., Winter M., Klitz W. 2001. HLA diversity, differentiation, and haplotype evolution in Mesoamerican Natives. Hum. Immunol. 62, 378–390 10.1016/S0198-8859(01)00212-9 (doi:10.1016/S0198-8859(01)00212-9) [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez-Galarza F. F., Christmas S., Middleton D., Jones A. R. 2011. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 39 (Suppl. 1), D913–D919 10.1093/nar/gkq1128 (doi:10.1093/nar/gkq1128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao X., Veale A., Serjeantson S. W. 1992. HLA class II diversity in Australian aborigines: unusual HLA-DRB1 alleles. Immunogenetics 36, 333–337 10.1007/BF00215663 (doi:10.1007/BF00215663) [DOI] [PubMed] [Google Scholar]
- 36.Gao X., Zimmet P., Serjeantson S. W. 1992. HLA-DR, DQ sequence polymorphisms in Polynesians, Micronesians, and Javanese. Hum. Immunol. 34, 153–161 10.1016/0198-8859(92)90107-X (doi:10.1016/0198-8859(92)90107-X) [DOI] [PubMed] [Google Scholar]
- 37.Gao X., Bhatia K., Trent R. J., Serjeantson S. W. 1992. HLA-DR, DQ nucleotide sequence polymorphisms in five Melanesian populations. Tissue Antigens 40, 31–37 10.1111/j.1399-0039.1992.tb01954.x (doi:10.1111/j.1399-0039.1992.tb01954.x) [DOI] [PubMed] [Google Scholar]
- 38.Mack S. J., et al. 2000. Evolution of Pacific/Asian populations inferred from HLA class II allele frequency distributions. Tissue Antigens 55, 383–400 10.1034/j.1399-0039.2000.550501.x (doi:10.1034/j.1399-0039.2000.550501.x) [DOI] [PubMed] [Google Scholar]
- 39.Main P., Attenborough R., Chelvanayagam G., Bhatia K., Gao X. 2001. The peopling of New Guinea: evidence from class I human leukocyte antigen. Hum. Biol. 73 365–383 [Erratum in: Hum Biol. 2001 73, 782] [DOI] [PubMed] [Google Scholar]
- 40.Bugawan T. L., Mack S. J., Stoneking M., Saha M., Beck H. P., Erlich H. A. 1999. HLA class I allele distributions in six Pacific/Asian populations: evidence of selection at the HLA-A locus. Tissue Antigens 53, 311–319 10.1034/j.1399-0039.1999.530401.x (doi:10.1034/j.1399-0039.1999.530401.x) [DOI] [PubMed] [Google Scholar]
- 41.Sanchez-Mazas A., et al. 2011. Immunogenetics as a tool in anthropological studies. Immunology 133, 143–164 10.1111/j.1365-2567.2011.03438.x (doi:10.1111/j.1365-2567.2011.03438.x) [DOI] [PMC free article] [PubMed] [Google Scholar]