Abstract
Although the taxonomy of the ca 18 000 species of butterflies and skippers is well known, the family-level relationships are still debated. Here, we present, to our knowledge, the most comprehensive phylogenetic analysis of the superfamilies Papilionoidea, Hesperioidea and Hedyloidea to date based on morphological and molecular data. We reconstructed their phylogenetic relationships using parsimony and Bayesian approaches. We estimated times and rates of diversification along lineages in order to reconstruct their evolutionary history. Our results suggest that the butterflies, as traditionally understood, are paraphyletic, with Papilionidae being the sister-group to Hesperioidea, Hedyloidea and all other butterflies. Hence, the families in the current three superfamilies should be placed in a single superfamily Papilionoidea. In addition, we find that Hedylidae is sister to Hesperiidae, and this novel relationship is supported by two morphological characters. The families diverged in the Early Cretaceous but diversified after the Cretaceous–Palaeogene event. The diversification of butterflies is characterized by a slow speciation rate in the lineage leading to Baronia brevicornis, a period of stasis by the skippers after divergence and a burst of diversification in the lineages leading to Nymphalidae, Riodinidae and Lycaenidae.
Keywords: Lepidoptera, Papilionoidea, diversification, phylogeny, DNA, morphology
1. Introduction
With 18 363 described species [1], butterflies and skippers are a highly diverse group with a species-level taxonomy that is the best known among invertebrates. However, the evolutionary history of the group is not well known, including when the group evolved, and how it diversified, although recent studies on subgroups within the butterflies are suggesting intriguing hypotheses about where, when and how the clades originated and diversified [2–6].
Despite being one of the most familiar and best-studied groups of insects, the higher phylogeny of the true butterflies (superfamily Papilionoidea) has been a tough nut to crack. The family divisions of this alluring superfamily are well established and traditionally agreed upon to include the families Papilionidae, Pieridae, Nymphalidae, Lycaenidae [7] and recently Riodinidae [8,9]. It is also rather apparent that the family Hesperiidae (skippers) and Hedylidae (butterfly moths) are affiliated with the butterflies [10,11]. However, the inter-relationships of these families have long been contradictory, and results of even recent studies have not made the matter any easier [9,12–15]. Solving this systematic puzzle would allow better understanding of the evolutionary history of these popular organisms that serve as biological models in many fields of research.
The inability of molecular and morphological data to resolve phylogenetic relationships is often attributed to ancient, rapid diversification of lineages [16–18]. Such patterns are important signals of evolutionary history: could they be linked with geological or climatic events in the Earth's history such as the Cretaceous–Palaeogene (K–Pg) mass extinction [3,6]? Or perhaps, are they linked to biological innovations, such as colonizing novel host plants [4]? Knowledge of when major lineages have diverged would help in understanding patterns of diversification and discovering the links to the events that were responsible for the current diversity of butterflies and skippers.
Through time-calibrated phylogenies, it is also possible to investigate whether the high number of extant groups of butterflies and skippers is the result of rapid diversifications over a short period of time, or whether the numbers we see now are the expected numbers for an old lineage with even diversification rates (since a constant rate of diversification over long periods will certainly produce a very species-rich lineage) [19]. This can be achieved by estimating past rates of speciation (birth: b) and extinction (death: d) along a phylogeny and for the lineages of interest. Estimating a phylogenetic hypothesis for all the ca 18 000 species of butterflies and skippers is impractical, and thus methods that are able to correct for incomplete phylogenies are needed (taking into account, the species richness of lineages) [19]. Such methods have been recently developed and allow us to understand the patterns of ancient diversifications that are inferred based on estimated dates of diversifications.
The aim of this paper is to perform, to our knowledge, the most comprehensive phylogenetic analysis of the superfamilies Papilionoidea, Hesperioidea and Hedyloidea to date, in order to assess the relationships among their constituent families. We also investigated whether any lineage underwent unusually rapid diversification compared with the net diversification speed (r = b – d) across the phylogeny of butterflies and skippers. We present a morphological data matrix, which includes extensive data: 191 characters derived from adults, larvae and pupae, as well as a molecular data matrix that comprised eight protein-coding gene regions (one from the mitochondrial genome and seven from the nuclear genome). We use this data to investigate the evolutionary history of the butterflies and skippers.
2. Methods
(a). Molecular data sampling
Taxa for this study were selected based on previous analyses of families [3,4,6,9,20] and covered all subfamilies in each family (except Pseudergolinae in Nymphalidae and Aphnaeinae in Lycaenidae; electronic supplementary material, table S1). In a few cases (especially for Lycaenidae), specimens were not available for sequencing and thus previously published sequences were used [9]. Twenty-five outgroups were initially chosen (electronic supplementary material, table S1) based on a recent study on all Lepidoptera [14], but for most analyses, the closest outgroups, i.e. five species of Thyrididae and two species of Callidulidae, were used. We sequenced one mitochondrial (COI) and up to seven protein-coding nuclear gene regions (EF-1α, Wingless, RpS5, MDH, GAPDH, CAD and IDH) accounting for a total of 6165 bp with gaps. PCR conditions and sequencing have been described in detail previously [21]. Sequencing was performed mainly with an ABI 3730 capillary sequencer (Oulu) and an ABI PRISMR 3130xl capillary sequencer (Turku).
(b). Morphology
The morphological dataset includes characters derived from the scale vestiture and sclerotized structures of Papilionoidea (electronic supplementary material, tables S2 and S3). Characters were sought from adults, larvae and pupae by studying actual specimens from various museum collections, and supplemented by re-examining characters used in previous studies [12]. For the study of adult morphology, dissections prepared from dry mounted specimens were used. The study of the larval characters was restricted to the last instar larva, some preserved in ethanol and others dry-inflated. Pupal characters were examined, whenever possible, from both entire preserved pupae and from exuviae from which the adult had emerged. Detailed methods used in the study of morphology are given in electronic supplementary material, table S2.
In total, 54 ingroup species were examined, mostly corresponding to the species that were sequenced (electronic supplementary material, table S1). Both male and female specimens were available for most taxa, and either larva, pupa or both available for 36 of these species. Of the outgroup, larvae and pupae were obtained only for Scardia boletella and Pterodecta felderi. The character list comprises 191 characters (143 binary states, 48 multi-states; electronic supplementary material, table S2). Of these characters, 45 are larval, 32 pupal and 114 are adult characters. Reasoning for major modifications in traditional character coding is reported in the electronic supplementary material, table S2. The data of the character coding are presented in the matrix (electronic supplementary material, table S3).
(c). Phylogenetic analyses
The data were analysed using maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference (BI). MP and ML analyses were run on individual gene regions, combined molecular data, all data, molecular data with third codon positions removed and on a reduced dataset containing only taxa with six or more gene regions sequenced. MP was performed using Tree analysis using New Technology (TNT) [22]. Morphological characters were treated as unordered [23] gaps in molecular characters as a fifth state. Bootstrap and Bremer support values for nodes were obtained using a script written for TNT [24]. ML was performed with RAxML [25], with 100–500 bootstrap replicates. Each gene was assigned the GTR + Γ model, based on Akaike information criterion (AIC) values calculated using FindModel [26].
BI was performed with MrBayes v. 3.1 [27] on the combined dataset. Missing nucleotides were coded as ‘?’. Parameter values were estimated separately for each gene region and the morphological data using the ‘unlink’ command and the rate prior (ratepr) set to ‘variable’. As in the ML analyses, each gene was assigned the GTR + Γ model. The morphological data were modelled under the rate variable model [28].
Bayesian inference of phylogeny and times of divergence were carried out using the program BEAST v. 1.5.4 [29] using only the ingroup taxa. The dataset was partitioned into morphology and the gene regions and analysed simultaneously under the GTR + Γ model for each gene partition separately, and with a relaxed clock allowing branch lengths to vary according to an uncorrelated lognormal distribution [30]. The tree prior was set to the birth–death process. Three fossils were used as calibration points. The split between Hypanartia and Vanessa (Nymphalidae) was constrained to be 34 Ma (s.d. ± 3 Ma) based on two Florissant fossils Prodryas persephone and Vanessa amerindica [31] as well as results from previous studies [2]. Similarly, two other Florissant fossils, Stolopsyche libytheoides and Oligodonta florissantensis [31,32], were used to constrain the age of the first divergence in Pierini (Pieridae) in a similar manner. The third calibration point was taken from a previous study [6], where the divergence between Parnassiinae and Papilioninae was constrained to be between 50 and 80 Ma (uniform distribution). Initial analyses showed that there were serious problems with inferring relationships (the topology was found to be inverted with the root going between Lycaenidae and Riodinidae); thus we placed two constraints on the topology based on the combined analyses using TNT and MrBayes, i.e. all except Papilionidae were constrained to form a monophyletic group and Pieridae, Nymphalidae, Lycaenidae and Riodinidae were constrained to be a monophyletic group. All other priors were left to the defaults in BEAST. Parameters were estimated using four independent runs of 10 million generations each, with parameters sampled every 1000 generations. Convergence was checked in Tracer v. 1.4.6 and summary trees were generated using TreeAnnotator v. 1.5.3, both part of the BEAST package.
(d). Rates of diversification
We analysed patterns of diversification along the tree of butterflies and skippers using algorithms for modelling evolutionary diversification using stepwise AIC (MEDUSA) as implemented in R [19,33] in the package Geiger [34]. MEDUSA fits alternative birth–death models to a phylogenetic tree taking into account taxonomic data in order to estimate changes in net diversification rates along the branches. MEDUSA obtains values of b and d based on the best-fit of the models according to comparisons of AIC scores [19]. First, the MEDUSA algorithm estimates likelihood and AIC scores for the simplest birth–death model (with two parameters, b and d). Next, MEDUSA compares the AIC scores of the two-parameter model with an incrementally more complex model, which includes five parameters (for two birth parameters and two death parameters owing to one break point and a shift-location parameter). MEDUSA continues this process until the addition of parameters does not improve the AIC scores.
We obtained information about species numbers for major lineages of butterflies and skippers from several sources [1,35]. This information was assigned to 28 lineages in our phylogeny after pruning terminals belonging to the same monophyletic groups (electronic supplementary material, table S4). We let MEDUSA estimate up to 10 turnover points in our tree but present the pattern of diversification obtained using a moderate corrected AIC threshold (cut-off value = 4) [19]. MEDUSA produces corrected AIC scores (AICc) [36], which is a bias correction to the AIC when sample size is small. We selected the model with the lowest AICc score, which in this case was the same when using a moderate cut-off value 4 for normal AIC scores, which means rejecting models that do not improve the AIC scores by more than 4 [19].
3. Results
(a). Systematic implications
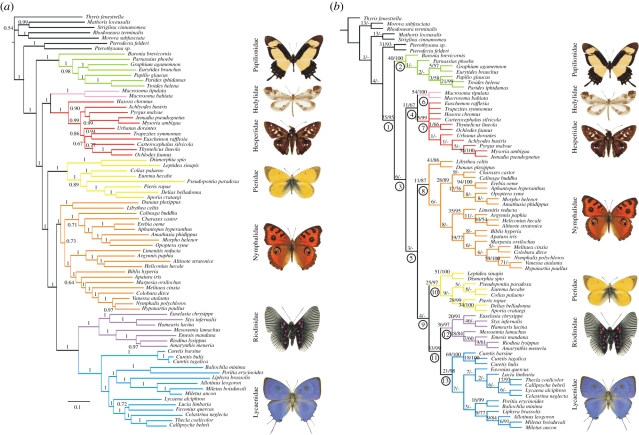
Two major results are apparent from our analyses: the paraphyly of butterflies, with Papilionidae being sister to the rest of the butterflies, skippers and butterfly moths, and the sister-group relationship between Hedylidae and Hesperiidae (figure 1 and electronic supplementary material, figure S1). We recovered the position of Papilionidae as sister-group to Hedylidae + Hesperiidae + the rest of butterflies in all the analyses except the parsimony analysis of morphology alone, where Hedylidae, Hesperiidae and the rest of Papilionoidea form an unresolved trichotomy (electronic supplementary material, figure S1a).
Figure 1.
Phylogenetic hypotheses generated by combined analysis of morphological data and data from eight gene regions. (a) Tree resulting from the Bayesian analysis of combined molecular and morphological datasets. Posterior probabilities under the GTR + G model are shown above branches. (b) Parsimony analysis of combined molecular and morphological datasets. The consensus tree of two equally parsimonious trees (length 31 384). Bremer support/bootstrap values for nodes above branches. Numbers in circles refer to electronic supplementary material, table S5 showing partitioned Bremer support values and partitioned congruence index values for the respective nodes.
The position of Pieridae was highly unstable, it being found to be the sister-group to Lycaenidae + Riodinidae (figure 1b and electronic supplementary material, figure S1c,e), to Nymphalidae + Lycaenidae + Riodinidae (figure 1a and electronic supplementary material, figure S1b,f), to Papilionidae (electronic supplementary material, figure S1a) and finally to the clade excluding Papilionidae (electronic supplementary material, figure S1d), depending on the data and the method of analysis. All the analyses found Riodinidae and Lycaenidae to be sister lineages.
(b). Ages of lineages and rates of diversification
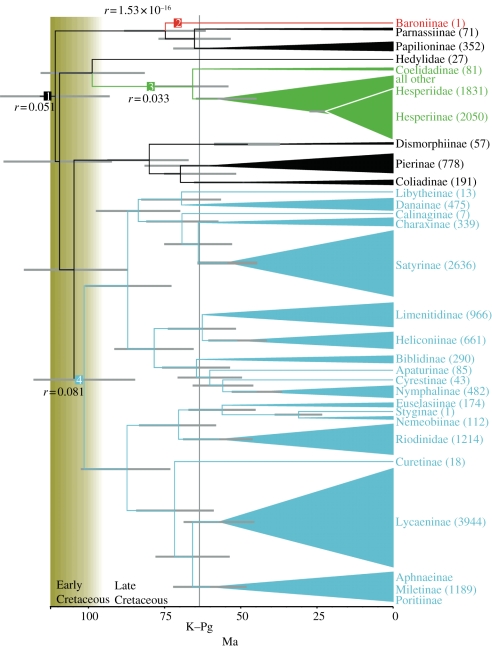
The analysis of times of divergence suggests that the lineages leading to Papilionidae, Hesperiidae + Hedylidae and the rest of the butterflies diverged quite rapidly from each other in the Early Cretaceous, some 110 million years ago (Ma; figure 2). Lineages leading to extant families all diverged from each other by 90 Ma, with Pieridae diverging from the common ancestor to the other butterflies at about 105 Ma, nymphalids from lycaenids and riodinids about 102 Ma, hedylids diverged from hesperiids about 99 Ma and finally riodinids diverging from lycaenids about 88 Ma.
Figure 2.
Ages of major lineages and their rates of diversification. Values to the right of taxon names give estimated numbers of extant species for the given taxon. Changes in rates of diversification (r) are indicated at the appropriate node.
These initial divergences appear to be followed by a period of no family-level divergences of extant lineages (figure 2). The first family that began diversifying was Nymphalidae, with the initial divergence happening at about the same time as the divergence of the Riodinidae and Lycaenidae lineages, i.e. in the Late Cretaceous. Strikingly, most within-family divergences leading to extant subfamily lineages occurred after the K–Pg boundary (also known as the K–T or Cretaceous–Tertiary boundary), the exceptions being the aforementioned Nymphalidae [3] and possibly Pieridae [4].
The diversification analysis suggests that the current diversity of butterflies and skippers is best explained by three shifts in the rates of diversification during their evolutionary history (figure 2). The best-fit model had a log-likelihood value of −288.05 and 11 parameters (four birth rates, four death rates and three shift-location parameters), and it had the best-corrected AIC score (AICc = 604.24). The diversification analysis found the net rate of diversification (r) of butterflies and skippers to be 0.051 lineages per million years (table 1). The lineage leading to the subfamily Baroniinae (Papilionidae) has strikingly low rates of speciation and extinction, leading to a very low net rate (r = 1.528 × 10e−16). Another decrease in the rate of speciation is inferred to have happened in Hesperiidae (r = 0.0327). The clade including Nymphalidae, Lycaenidae and Riodinidae is inferred to have an elevated net rate of diversification (r = 0.081) compared with the background diversification rates in butterflies and skippers.
Table 1.
Estimates of diversification rates for clades as noted in figure 2 (cut-off value = 4). (r, net rate of diversification; b, birth rate; d, extinction rate; ΔAIC, improvement in AIC scores after the addition of a turnover in diversification rates; ΔAICc, corrected AIC scores for each model that added a turnover in diversification rate in the respective clade.)
| clade | r | b | d | ΔAIC | ΔAICc |
|---|---|---|---|---|---|
| 1 | 0.051 | 0.390 | 0.340 | — | 615.540 |
| 2 | 1.53 × 10e−16 | 1.54 × 10e−16 | 1.22 × 10e−18 | 8.523 | 608.006 |
| 3 | 0.033 | 8.441 | 8.408 | 4.634 | 605.268 |
| 4 | 0.081 | 0.341 | 0.259 | 4.047 | 604.240 |
4. Discussion
(a). Systematic implications
The idea that Papilionoidea do not form a natural group has never been entertained in the history of the classification of these well-studied insects. Only recently have two studies [13,14] called into question the monophyly of Papilionoidea, although both studies suffered from minimal taxon sampling of butterflies and close relatives. We increased taxon sampling of butterflies and skippers and inferred that Papilionoidea as currently understood is a paraphyletic entity, with Papilionidae being sister to skippers, hedylids and all other butterflies. In addition, we found strong support for a sister-group relationship between Hedylidae and Hesperiidae. In this study, the identical shape of the third axillary sclerite of the forewing (51 : 1 and 52 : 0; electronic supplementary material, figure S2a), formerly considered an apomorphy of Hesperiidae, and the flat ridges on the mesophragma (38 : 1; electronic supplementary material, figure S2b) are morphological characters that could support the sister-group relationship between Hedylidae and Hesperiidae.
These results differ radically from a similar previous study [9], although we note that the Bayesian analysis of three genes in the previous study finds Papilionidae to be sister to the rest, including Hedylidae and Hesperiidae (figure 1b in the study of Wahlberg et al. [9]). That hypothesis was discounted as it changed with the addition of new data [9]. Our current study adds new data, both morphological and molecular, and evidently the new data show a stronger phylogenetic signal, as the positions of Papilionidae, Hedylidae and Hesperiidae appear robust to various perturbations of the data (figure 1 and electronic supplementary material, figure S1).
One of the major uncertainties in butterfly phylogeny has been the position of Pieridae [7]. Two hypotheses have dominated: Pieridae and Papilionidae as sister-groups [37–39] or Pieridae sister to Lycaenidae (s.l.) + Nymphalidae [9,40,41]. The MP analysis of the morphological data alone suggests that Pieridae is the sister-group of Papilionidae, in accordance with one hypothesis [37–39]. However, this relationship is only supported by five homoplastic characters (electronic supplementary material, figure S2c) and no molecular studies have suggested the sister relationship of Pieridae and Papilionidae. Most other analyses in our study place Pieridae with Nymphalidae and Riodinidae + Lycaenidae.
The difficulty in resolving the relationships between the families of butterflies and skippers is evidently owing to the rapid divergences of the lineages leading to Papilionidae, Hedylidae + Hesperiidae, Pieridae, Nymphalidae and Riodinidae + Lycaenidae (figure 2). It appears that these divergences, happening within a ca 8 Myr period in the Early Cretaceous (110–102 Myr ago), may not have left any synapomorphies behind which we could have detected to be used to unite lineages. Given the error inherent in estimating divergences that have happened over 100 Ma, such a time span is essentially not different from a hard polytomy, despite the visual representation of dichotomously branching lineages (figure 2).
Our results suggest that the division of butterflies, skippers and butterfly moths into three different superfamilies is not tenable. We suggest that the families Papilionidae, Hedylidae, Hesperiidae, Pieridae, Nymphalidae, Riodinidae and Lycaenidae be placed in a single superfamily Papilionoidea. This clade appears to be stable within Lepidoptera, and the constituent families are strongly supported clades that are well-defined morphologically [12,14]. Any changes of the relationships of the families based on increased data would not affect the classification as suggested here.
(b). Rates of diversification and the evolutionary history of butterflies
The diversification analysis found the net rate of diversification of butterflies and skippers to be low (r = 0.0506 lineages per Myr). It is notable that the species-rich skipper family Hesperiidae (ca 4000 species [42]) has a slower rate of diversification (r = 0.0327) than the overall tempo of diversification across our phylogenetic tree. Our results indicate that the lineage leading to extant Hesperiidae diverged early in the evolution of butterflies (around 100 Ma) and had a relatively long delay before it began to diversify after 65 Myr. The lineage thus survived the mass extinction event at the K–Pg boundary. It appears that despite having a slower net rate of diversification, the species richness of skippers is relatively high owing to a steady rise in the number of lineages over a long span of time. Whether the skippers were diverse in the Cretaceous is not known, thus we are unable to say whether there was a long period of time during which the skipper lineage survived with low diversity, or whether simply one lineage survived the K–Pg event. A previous study on the subfamily Satyrinae (Nymphalidae) suggests that a radical change in the environment allowed the butterflies to exploit the new habitat that became available with the drying up and cooling down of the Earth in the Oligocene [5]. Clearly, a more detailed study of the evolutionary history of Hesperiidae is needed to investigate the reasons behind the ‘long fuse’ before the diversification of the skippers.
A previous study proposed that 10 or 12 lineages of nymphalids survived the K–Pg event and radiated with elevated rates of diversification during the Tertiary diversification [3]. In this paper, we find support for our previous findings, as our Nymphalidae + (Riodinidae + Lycaenidae) clade has a higher net diversification rate (r = 0.0814) than the background tempo for all the butterflies and skippers (r = 0.0506; figure 2). A more detailed diversification analysis including more terminals for the species-rich Nymphalidae + (Riodinidae + Lycaenidae) clades would shed light on the different rates of diversification for specific clades, as there are many lineages with very few species, such as the riodinid Stygini, that might have suffered a deceleration of their net diversification rate. Similar findings may be found by studying Satyrinae at a finer scale, which is thought to have undergone a rapid radiation [5].
The rapid diversification of the lineages leading to the families, and the apparent deceleration of diversification until after the K–Pg boundary can be explained in several ways. It may be that the butterflies and skippers were more diverse in the Cretaceous, and suffered from extinctions during the K–Pg event, as has been suggested for Nymphalidae [3]. An alternative scenario would suggest that the ecological conditions for diversification of day-flying Lepidoptera only became favourable in the Palaeogene, e.g. in conjunction with the diversification of angiosperm plants [43], on which the majority of butterflies and skippers are specialized as larvae [44].
It is interesting to note that our findings of the tempo of diversification of butterflies and skippers are comparable with the rates found in the evolutionary history of vertebrates [19], which have high net rates of diversification for species-rich lineages that began diversifying simultaneously with the butterflies, around 100 Ma, and have comparable numbers of extant species. For example, Percomorpha fishes, which include around 20 000 extant species, have a net diversification rate of 0.082 lineages per million years. This is comparable with our estimated rate of 0.0814 lineages per million years for the clade Nymphalidae + (Riodinidae + Lycaenidae) that include ca 12 000 butterfly species.
In summary, we found Papilionoidea to be paraphyletic with regard to Hedyloidea and Hesperioidea, as well as strong support for a sister relationship between the butterfly–moths (Hedylidae) and the skippers (Hesperiidae). Therefore, as currently understood, Papilionoidea is a paraphyletic entity, and we suggest it should include Hedylidae and Hesperiidae. As in previous molecular studies, our study recovered the higher level relationships among the rest of butterflies to be (Pieridae + (Nymphalidae + (Riodinidae + Lycaenidae))). The butterfly families probably diverged from each other in the Early Cretaceous, between 110 and 100 Ma, but extant lineages began diversifying only after the K–Pg event at 65 Ma. We found that the pattern of the diversification of butterflies is characterized by three changes in the rates of speciation along their history. The species Baronia brevicornis should be regarded as a ‘living fossil’, as it is a descendant of an old lineage with an extremely low rate of diversification (r = 1.528 × 10e−16). The skippers had a period of stasis following their divergence from their sister-group Hedylidae, while an acceleration in the rate of diversification occurred in the surviving lineages of the K–Pg event that led to the clade Nymphalidae + (Riodinidae + Lycaenidae) (r = 0.0814).
Acknowledgements
We thank the following institutions and persons for providing material for the morphological study: Finnish Museum of Natural History (MZH), Australian National Insect Collection (ANIC), Texas A&M University, McGuire Centre for Lepidotera and Biodiversity (Florida Museum of Natural History, FLMNH), Coll. Kimmo Silvonen, Nico Elfferich; Kimmo Silvonen for the photos of the pierid, lycaenid and nymphalid pupae and of the riodinid larva, and Dan Janzen and Winnie Hallwachs for the photo of the hedylid larva (voucher code 03-SRNP-13034.1; for more information see http://janzen.sas.upenn.edu/caterpillars/database.lasso) all in the electronic supplementary material, figure S1. We also thank Marjatta Mikkonen, Ville Heimala and Pekka Malinen of MZH for their help. We are grateful to Dan Janzen and Winnie Hallwachs (through US NSF award nos DEB0072730 and DEB0515699), Felix Sperling, Chris Müller and Heikki Roininen for providing tissue samples crucial for this study. We thank Fabien Condamine, Thomas Simonsen and two anonymous referees for constructive comments on the manuscript. This study was funded by the Academy of Finland (projects 110906 to L.K. and 118369 to N.W.).
References
- 1.Nieukerken E. J. V., et al. In press. Order Lepidoptera. In Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness (ed. Zhang Z.-Q.), Zootaxa: [DOI] [PubMed] [Google Scholar]
- 2.Wahlberg N. 2006. That awkward age for butterflies: insights from the age of the butterfly subfamily Nymphalinae. Syst. Biol. 55, 703–714 10.1080/10635150600913235 (doi:10.1080/10635150600913235) [DOI] [PubMed] [Google Scholar]
- 3.Wahlberg N., Leneveu J., Kodandaramaiah U., Peña C., Nylin S., Freitas A. V. L., Brower A. V. Z. 2009. Nymphalid butterflies diversify following near demise at the Cretaceous/Tertiary boundary. Proc. R. Soc. B 276, 4295–4302 10.1098/rspb.2009.1303 (doi:10.1098/rspb.2009.1303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wheat C. W., Vogel H., Wittstock U., Braby M. F., Underwood D., Mitchell-Olds T. 2007. The genetic basis of a plant-insect coevolutionary key innovation. Proc. Natl Acad. Sci. USA 104, 20 427–20 431 10.1073/pnas.0706229104 (doi:10.1073/pnas.0706229104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peña C., Wahlberg N. 2008. Prehistorical climate change increased diversification of a group of butterflies. Biol. Lett. 4, 274–278 10.1098/rsbl.2008.0062 (doi:10.1098/rsbl.2008.0062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simonsen T. J., Zakharov E. V., Djernaes M., Cotton A. M., Vane-Wright R. I., Sperling F. A. H. 2011. Phylogenetics and divergence times of Papilioninae (Lepidoptera) with special reference to the enigmatic genera Teinopalpus and Meandrusa. Cladistics 27, 113–137 10.1111/j.1096-0031.2010.00326.x (doi:10.1111/j.1096-0031.2010.00326.x) [DOI] [PubMed] [Google Scholar]
- 7.Ackery P. R., de Jong R., Vane-Wright R. I. 1999. The butterflies: Hedyloidea, Hesperoidea and Papilionoidea. In Lepidoptera, moths and butterflies. I. Evolution, systematics and biogeography. Handbook of zoology, vol. 4, part 35: Lepidoptera (ed. Kristensen N. P.), pp. 263–300 Berlin, Germany: de Gruyter [Google Scholar]
- 8.Campbell D. L., Brower A. V. Z., Pierce N. E. 2000. Molecular evolution of the wingless gene and its implications for the phylogenetic placement of the butterfly family Riodinidae (Lepidoptera: Papilionoidea). Mol. Biol. Evol. 17, 684–696 [DOI] [PubMed] [Google Scholar]
- 9.Wahlberg N., et al. 2005. Synergistic effects of combining morphological and molecular data in resolving the phylogeny of butterflies and skippers. Proc. R. Soc. B 272, 1577–1586 10.1098/rspb.2005.3124 (doi:10.1098/rspb.2005.3124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weller S. J., Pashley D. P. 1995. In search of butterfly origins. Mol. Phylogenet. Evol. 4, 235–246 10.1006/mpev.1995.1022 (doi:10.1006/mpev.1995.1022) [DOI] [PubMed] [Google Scholar]
- 11.Scoble M. J. 1986. The structure and affinities of the Hedyloidea: a new concept of butterflies. Bull. Br. Mus. Nat. His. Entomol. 53, 251–286 [Google Scholar]
- 12.de Jong R., Vane-Wright R. I., Ackery P. R. 1996. The higher classification of butterflies (Lepidoptera): problems and prospects. Entomol. Scand. 27, 65–101 [Google Scholar]
- 13.Regier J. C., et al. 2009. Toward reconstructing the evolution of advanced moths and butterflies (Lepidoptera: Ditrysia): an initial molecular study. BMC Evol. Biol. 9, 280. 10.1186/1471-2148-9-280 (doi:10.1186/1471-2148-9-280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mutanen M., Wahlberg N., Kaila L. 2010. Comprehensive gene and taxon coverage elucidates radiation patterns in moths and butterflies. Proc. R. Soc. B 277, 2839–2848 10.1098/rspb.2010.0392 (doi:10.1098/rspb.2010.0392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim M. I., Wan X., Kim M. J., Jeong H. C., Ahn N.-H., Kim K.-G., Han Y. S., Kim I. 2010. Phylogenetics relationships of true butterflies (Lepidoptera: Papilionoidea) inferred from COI, 16S rRNA and EF-1α sequences. Mol. Cells 30, 409–425 10.1007/s10059-010-0141-9 (doi:10.1007/s10059-010-0141-9) [DOI] [PubMed] [Google Scholar]
- 16.Rokas A., Carroll S. B. 2006. Bushes in the tree of life. PLoS Biol. 4, e352. 10.1371/journal.pbio.0040352 (doi:10.1371/journal.pbio.0040352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitfield J. B., Lockhart P. J. 2007. Deciphering ancient rapid radiations. Trends Ecol. Evol. 22, 258–265 10.1016/j.tree.2007.01.012 (doi:10.1016/j.tree.2007.01.012) [DOI] [PubMed] [Google Scholar]
- 18.Whitfield J. B., Kjer K. M. 2008. Ancient rapid radiations of insects: challenges for phylogenetic analysis. Annu. Rev. Entomol. 53, 449–472 10.1146/annurev.ento.53.103106.093304 (doi:10.1146/annurev.ento.53.103106.093304) [DOI] [PubMed] [Google Scholar]
- 19.Alfaro M. E., Santini F., Brock C., Alamillo H., Dornburg A., Rabosky D. L., Carnevale G., Harmon L. J. 2009. Nine exceptional radiations plus high turnover explain species diversity of jawed vertebrates. Proc. Natl Acad. Sci. USA 106, 13 410–13 414 10.1073/pnas.0811087106 (doi:10.1073/pnas.0811087106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warren A. D., Ogawa J. R., Brower A. V. Z. 2009. Revised classification of the family Hesperiidae (Lepidoptera: Hesperioidea) based on combined molecular and morphological data. Syst. Entomol. 34, 467–523 10.1111/j.1365-3113.2008.00463.x (doi:10.1111/j.1365-3113.2008.00463.x) [DOI] [Google Scholar]
- 21.Wahlberg N., Wheat C. W. 2008. Genomic outposts serve the phylogenomic pioneers: designing novel nuclear markers for genomic DNA extractions of Lepidoptera. Syst. Biol. 57, 231–242 10.1080/10635150802033006 (doi:10.1080/10635150802033006) [DOI] [PubMed] [Google Scholar]
- 22.Goloboff P. A., Farris J. S., Nixon K. C. 2008. TNT, a free program for phylogenetic analysis. Cladistics 24, 774–786 10.1111/j.1096-0031.2008.00217.x (doi:10.1111/j.1096-0031.2008.00217.x) [DOI] [Google Scholar]
- 23.Fitch W. M. 1971. Toward defing the course of evolution: minimum change for a specified tree topology. Syst. Zool. 20, 406–416 10.2307/2412116 (doi:10.2307/2412116) [DOI] [Google Scholar]
- 24.Peña C., Wahlberg N., Weingartner E., Kodandaramaiah U., Nylin S., Freitas A. V. L., Brower A. V. Z. 2006. Higher level phylogeny of Satyrinae butterflies (Lepidoptera: Nymphalidae) based on DNA sequence data. Mol. Phylogenet. Evol. 40, 29–49 10.1016/j.ympev.2006.02.007 (doi:10.1016/j.ympev.2006.02.007) [DOI] [PubMed] [Google Scholar]
- 25.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 10.1093/bioinformatics/btl446 (doi:10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 26.Tao N., Richardson R., Bruno W., Kuiken C. 2010. FindModel. Los Alamos, NM: Los Alamos National Security. See http://www.hiv.lanl.gov/content/sequence/findmodel/findmodel.html.
- 27.Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 10.1093/bioinformatics/btg180 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 28.Lewis P. O. 2001. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst. Biol. 50, 913–925 10.1080/106351501753462876 (doi:10.1080/106351501753462876) [DOI] [PubMed] [Google Scholar]
- 29.Drummond A. J., Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214. 10.1186/1471-2148-7-214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drummond A. J., Ho S. Y. W., Phillips M. J., Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4, e88. 10.1371/journal.pbio.0040088 (doi:10.1371/journal.pbio.0040088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emmel T. C., Minno M. C., Drummond B. A. 1992. Florissant butterflies: a guide to the fossil and present-day species of Central Colorado. Stanford, CA: Stanford University Press [Google Scholar]
- 32.Braby M. F., Vila R., Pierce N. E. 2006. Molecular phylogeny and systematics of the Pieridae (Lepidoptera: Papilionoidea): higher classification and biogeography. Zool. J. Linn. Soc. 147, 239–275 10.1111/j.1096-3642.2006.00218.x (doi:10.1111/j.1096-3642.2006.00218.x) [DOI] [Google Scholar]
- 33.R Development Core Team 2010. A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 34.Harmon L. J., Weir J. T., Brock C. D., Glor R. E., Challenger W. 2008. GEIGER: investigating evolutionary radiations. Bioinformatics 24, 129–131 10.1093/bioinformatics/btm538 (doi:10.1093/bioinformatics/btm538) [DOI] [PubMed] [Google Scholar]
- 35.Lamas G. 2004. Atlas of neotropical Lepidoptera. Checklist: part 4A, Hesperioidea–Papilionoidea. Gainsville, FL: Scientific Publishers [Google Scholar]
- 36.Hurvich C. M., Tsai L. 1989. Regression and time series model selection in small samples. Biometrika 76, 297. 10.1093/biomet/76.2.297 (doi:10.1093/biomet/76.2.297) [DOI] [Google Scholar]
- 37.Ehrlich P. R. 1958. The comparative morphology, phylogeny and higher classification of the butterflies (Lepidoptera: Papilionoidea). Univ. Kansas Sci. Bull. 39, 305–370 [Google Scholar]
- 38.Scott J. A. 1985. The phylogeny of butterflies (Papilionoidea and Hesperoidea). J. Res. Lepid. 23, 241–281 [Google Scholar]
- 39.Scott J. A., Wright D. M. 1990. Butterfly phylogeny and fossils. In Butterflies of Europe, vol. 2 (ed. Kudrna O.), pp. 152–208 Wiesbaden, Germany: Aula-Verlag [Google Scholar]
- 40.Kristensen N. P. 1976. Remarks on the family-level phylogeny of butterflies (Insecta, Lepidoptera, Rhopalocera). Z. Zool. Syst. Evol. 14, 25–33 10.1111/j.1439-0469.1976.tb00515.x (doi:10.1111/j.1439-0469.1976.tb00515.x) [DOI] [Google Scholar]
- 41.Weller S. J., Pashley D. P., Martin J. A. 1996. Reassessment of butterfly family relationships using independent genes and morphology. Ann. Entomol. Soc. Am. 89, 184–192 [Google Scholar]
- 42.Warren A. D., Ogawa J. R., Brower A. V. Z. 2008. Phylogenetic relationships of subfamilies and circumscriptions of tribes in the family Hesperiidae (Lepidoptera: Hesperioidea). Cladistics 24, 642–676 10.1111/j.1096-0031.2008.00218.x (doi:10.1111/j.1096-0031.2008.00218.x) [DOI] [Google Scholar]
- 43.Bell C. D., Soltis D. E., Soltis P. S. 2010. The age and diversification of the angiosperms re-revisited. Am. J. Bot. 97, 1296–1303 10.3732/ajb.0900346 (doi:10.3732/ajb.0900346) [DOI] [PubMed] [Google Scholar]
- 44.Janz N., Nylin S. 1998. Butterflies and plants: a phylogenetic study. Evolution 52, 486–502 10.2307/2411084 (doi:10.2307/2411084) [DOI] [PubMed] [Google Scholar]