Abstract
Several vertebrates choose their mate according to genetic heterozygosity and relatedness, and use odour cues to assess their conspecifics' genetic make-up. In birds, although several species (including the black-legged kittiwake) exhibit non-random mating according to genetic traits, the cues used to assess genetic characteristics remain unknown. The importance of olfaction in birds' social behaviour is gaining attention among researchers, and it has been suggested that, as in other vertebrates, bird body scent may convey information about genetic traits. Here, we combined gas chromatography data and genetic analyses at microsatellite loci to test whether semiochemical messages in preen secretion of kittiwakes carried information about genetic heterozygosity and relatedness. Semiochemical profile was correlated with heterozygosity in males and females, while semiochemical distance was correlated with genetic distance only in male–male dyads. Our study is the first to demonstrate a link between odour and genetics in birds, which sets the stage for the existence of sophisticated odour-based mechanisms of mate choice also in birds.
Keywords: genetic compatibility, heterozygosity, olfactory communication, birds, black-legged kittiwake
1. Introduction
Many species exhibit non-random mating, and numerous traits affect mate choice. Although most studies have focused on morphological and behavioural traits [1], evidence is accruing for multiple genetic criteria for mate choice [2,3]. Females may choose males with ‘good genes’, such as those with alleles or allele combinations increasing fitness [4,5] or with high heterozygosity [6–8]. Alternatively, but not exclusively, females may choose males with ‘compatible genes’ [3,9] to enhance offspring heterozygosity, thus positively affecting offspring viability through increased resistance to infectious diseases and inbreeding avoidance [9–11].
As genes are not directly assessable, they need to be expressed in the phenotype to influence mate choice. Good genes may be assessed visually, acoustically or olfactorily, through ornamentation, colour, dominance, song or body scent [12–16]. Studies on compatible gene assessment are relatively scarce. However, genetic relatedness has been demonstrated to be advertised by olfactory cues in a growing number of species. For instance, women prefer the sweat odour of genetically dissimilar men [17]. Female bank voles Myodes glareolus [18], house mice Mus musculus [19], agile antechinuses Antechinus agilis [20] and sand lizards Lacerta agilis [11] prefer spending time near the odour of genetically dissimilar males. Ring-tailed lemurs Lemur catta can detect their relatedness to conspecifics using the odour of glandular secretions [13]. Finally, in Arctic charrs Salvelinus alpinus, genetic distance between individuals is related to olfactory cues [21].
Most of these studies have shown a link between genetic relatedness at major histocompatibility complex (MHC) loci and body scent [11,17,19,21,22]. The MHC is an extremely polymorphic complex of genes that plays a critical role in immune response and disease resistance. MHC heterozygotes have an advantage over homozygotes because they have better immune capacities and resist a wider range of pathogens [23–25]. MHC gene products are found in bodily secretions [26], and MHC-associated odour-mediated mating preferences have been documented in various vertebrates. However, in addition to MHC, ‘background’ genes or other highly polymorphic markers, such as the major urinary proteins in mice, can influence body scent [27,28]. Furthermore, if scent reflects genetic characteristics as suggested by these studies, scent signals have been suggested to systematically reflect more genome-wide variation than MHC variation only [13]. Accordingly, several studies have shown a link between body scent and genome-wide relatedness measured through neutral loci or pedigree analyses [20,29–32].
Several bird species preferentially mate with heterozygous or unrelated individuals [10,33–39], or adjust their reproductive effort in response to the genetic similarity of their partners [40,41]. Sexually selected traits such as colour, song and body size have been shown to correlate with inbreeding or heterozygosity in several bird species [36,42–44]. However, the phenotypic cues used in the assessment of genetic relatedness in birds have not yet been determined. Most birds do not patently scent-investigate their conspecifics, and the use of chemical communication in bird social behaviour was doubted for a long time. However, bird body scent may convey information about species, sex, hormonal state or individuality [45–49], and recent studies have shown that birds used olfaction in various social contexts [50–55]. As in other vertebrates, birds' body scent may thus convey information about genetic make-up [51,56].
Black-legged kittiwakes Rissa tridactyla preferentially mate with genetically dissimilar individuals [33] and do not seem to use vocal cues to achieve such discrimination [57]. Furthermore, second-hatched chicks that are heterozygous at neutral loci grow faster and survive longer than homozygous chicks [33]. It is therefore likely that, as in many other species [58–60], heterozygous adults have higher fitness than homozygous adults and are preferred mates. Furthermore, kittiwakes can smell [61], and their preen secretion odour conveys information about individuality [49]. Kittiwakes have thus been suggested to use chemical cues to assess the genetic make-up of their conspecifics. Here, we combine gas chromatography-mass spectrometry (GCMS) data and microsatellite analyses to test whether semiochemical compounds in preen secretion of kittiwakes carry information about genome-wide heterozygosity and relatedness.
2. Methods
(a). Study site
Samples were collected in the pre-laying period, between 9 May and 3 June 2008 (mean laying date was 1 June 2008), in a population of black-legged kittiwakes nesting on an abandoned US Air Force radar tower on Middleton Island (59°26′ N, 146°20′ W), Gulf of Alaska (mean temperature in May 2008: 5.9 ± 0.3°C, mean humidity in May 2008: 82 ± 1%) [62]. Odourant and genetic samples were collected from 15 females and 19 males. Adult sexing was based on molecular sexing (n = 19 birds), and on copulation and courtship feeding during the pre-laying period (n = 15 birds) [63].
(b). Odourant sample collection and analyses
We collected preen oil as in an earlier study [49], by gently pressing the base of the gland and collecting the exudates with a glass capillary. We stored samples at −20°C in 2 ml polytetrafluoroethylene-faced septum vials, until extraction of organic compounds. Extraction and analyses followed our protocol described earlier [49]. To further identify the acid and alcohol portions of ester compounds, we transesterified representative samples. Three millilitres of methanol with 5 per cent sodium methoxylate were added to the secretion sample. The mixture was left for 1 h at ambient temperature. GCMS analyses of transesterified products of the secretions were carried out on a GCMS Shimadzu QP2010 plus apparatus equipped with splitless injection and Supelco SLB-5 ms capillary column. The scan range of the mass spectrometer was 38–600 m/z. The oven temperature programme was as follows: 7°C min–1 from 50°C to 200°C, and then 3°C min–1 to 290°C. Identification of compounds was based on mass-spectral fragmentation pattern using various libraries and a chromatographic retention index (NIST 2005).
In all, 224 compounds were detected in preen secretions. As we could not control for the amount of oil collected, we did not rely on the absolute abundance of chromatogram peaks in our statistical analyses (as described in earlier studies [29,45,46,64,65]). Rather, each peak was quantified as the relative proportion of the peak size to the overall total area of the chromatogram. For statistical analyses, we retained peaks that comprised at least 0.1 per cent of the overall area of the chromatogram (n = 94 peaks). These compounds represented on average 98.3 ± 0.1% of the overall chromatogram area and were present in all individuals. To describe the semiochemistry of kittiwakes, we used four indices. First, we calculated Simpson and Shannon indices, which are commonly used by community ecologists to describe diversity [66,67]. Both of these indices account for abundance and evenness of detected compounds, but whereas the Shannon index is most strongly influenced by the relatively common compounds that are of intermediate abundance, the Simpson index is more sensitive to compounds that show the greatest relative abundance [66]. Genetic characteristics such as heterozygosity may be encoded by a set of particular compounds rather than by the overall diversity in compounds [68]. We thus generated a principal component analysis (PCA) on relative abundances of compounds and used the first two principal components (PC1 and PC2). This allowed us to summarize individual chemical information into synthetic variables, each mainly reflecting a particular set of compounds, and therefore to determine the compounds that encode the genetic heterozygosity. To describe the dissimilarity between semiochemical profiles, we used two indices: the relative Euclidean distance, and the difference in PC1 (DPC1) and PC2 (DPC2) for each dyad of individuals. The relative Euclidean distance reflects the overall distance between two profiles and considers each compound equally. However, many chemical compounds are probably silent in kin recognition [68]. Using differences in PC allowed us to identify the chemicals that show the greatest correlation with relatedness. This has been suggested to be an attractive way forward in the understanding of the relationship between body scent and genetic relatedness [68].
(c). Genetic analyses
At capture, blood was taken from the alar vein using a 1 ml syringe and a 25 gauge needle, and kept in a preservative solution (Longmire buffer [69]). Genomic DNA was extracted from each blood sample using the DNeasy Blood and Tissue Kit (Qiagen Group) following the supplier's guidelines. We used diversity at microsatellite loci as a proxy of genome-wide diversity (but see [70]). Samples were genotyped at 10 microsatellite loci [33], distributed in two multiplexes (electronic supplementary material, table A1). We amplified 1 µl extracted DNA in 10 µl reactions using 0.75X Multiplex PCR Master Mix (Qiagen Group), and 0.05–0.45 µM of each primer (electronic supplementary material, table A1). For the multiplex 1, the PCR thermal profile consisted of denaturation at 95°C for 15 min, followed by 35 cycles of 95°C denaturation for 30 s, 57°C annealing for 1 min 30 s, 72°C elongation for 1 min and a final elongation at 60°C for 30 min. For multiplex 2, the thermal profile was the same except that annealing was done at 60°C. PCR product (1 µl) was then mixed with 8.7 µl of highly deionized formamide and 0.3 µl of Genescan 600 LIZ size standard (Applied Biosystems). The mixtures were denatured at 95°C for 5 min before separation with a 48-capillary 3730 DNA Analyzer (Applied Biosystems) using POP-7 polymer and the manufacturer's default electrophoresis run settings. Data analysis and genotyping were performed with GeneMapper software (Applied Biosystems).
Mean heterozygosity (H; proportion of heterozygous loci in a given individual) and internal relatedness (IR; IR = (2Hz − ∑ fi)/(2N − ∑ fi), where Hz is the number of loci that are homozygous, N is the number of loci and fi is the frequency of the ith allele contained in the genotype) [71] were used as the estimate of genetic quality and were calculated using the Nausicaa software [33]. We found a wide range of variation in H and IR (range: 0.20 to 0.90 and −0.18 to 0.68, respectively; mean ± s.e.: 0.68 ± 0.02 and 0.07 ± 0.03, respectively). H and IR were highly correlated (r2 = 0.95; p < 0.0001). Two estimates of genetic relatedness between each dyad of individuals were calculated using IDENTIX software [72]: identity (RID) [73] and Queller & Goodnight (RQG) [74]. These estimates were transformed into estimates of genetic distances using the formula, D = 1 – R. To obtain more accurate estimates of the relatedness between our focal subjects, we calculated these estimates using a larger dataset (n = 371 individuals). Five individuals included in this study were only genotyped using the method described by Mulard et al. [33]. The correspondence between the two methods was tested by genotyping 30 individuals of the population using the two methods. Correspondence was found not to match for loci K32 and RBG20. The genetic distance between those five individuals and the 371 other individuals was thus calculated without those two loci.
(d). Statistical analyses
The effect of heterozygosity (H and IR) on chemical profile (as described by PC1, PC2, Simpson and Shannon indices) was examined using General Linear Models. Kittiwakes' semiochemical compounds differ between males and females, and vary according to the breeding stage [49]. Sex and breeding stage (i.e. number of days between sampling and laying) were thus included as covariates in the model.
The relationship between chemical distances (DPC1, DPC2 and relative Euclidean distances) and genetic distances (DID and DQG) in female–female (FF) and male–male (MM) dyads was analysed with partial Mantel tests and 5000 data randomizations (VEGAN package in R). Difference in breeding stage between the members of the dyad was included as a covariate in these tests. The male–female (MF) matrix was not square and thus we could not use the Mantel test. Instead, we used a permutation test to compare the empirical F-value against a distribution of F-values generated by resampling. We calculated the empirical F-value of a model with chemical distance as the dependent variable and genetic distance and difference in breeding stage as fixed effects, using the raw data. Then, we shuffled the empirical genetic distances and recalculated the simulated F-values. This shuffling and scoring procedure was repeated 1000 times to generate a distribution of simulated F-values. To generate a p-value, we counted how many times the value of the simulated correlation coefficient exceeded the value of the empirical correlation coefficient, and then divided this number by 1000 (as in the study of Boulet et al. [31]). All statistical tests were performed with R statistical software [75].
3. Results
(a). Semiochemical diversity
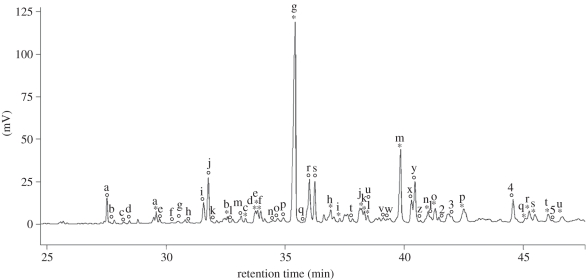
PC1 represented 29 per cent of the variance and was strongly positively correlated with several minor compounds (r > 0.6, p < 0.0001; figure 1 and table 1a). Considering only the major compounds, PC1 was not correlated with the two main compounds (hexadecyl octanoate and octadecyl octanoate; r = −0.13, p = 0.43 and r = −0.16, p = 0.31, respectively) but was positively correlated with the next eight most abundant compounds (tridecyl 2-methyloctanoate: r = 0.65; tridecyl 2-methyldecanoate: r = 0.73; pentadecyl 2-methyloctanoate: r = 0.77; pentadecyl 2-methyldecanoate: r = 0.82; heptadecyl 2-methyloctanoate: r = 0.78; pentadecyl 2-methyldodecanoate: r = 0.82; heptadecyl 2-methyldecanoate: r = 0.79; heptadecyl 2-methyldodecanoate: r = 0.77; all p < 0.0001; figure 1 and table 1a).
Figure 1.
Chromatogram of preen secretion in kittiwakes. Circles indicate compounds that are correlated with PC1 (p < 0.0001), whereas asterisks indicate compounds that are correlated with PC2 (p < 0.0001). Letters and numbers refer to compounds listed in table 1.
Table 1.
Names of the compounds that are correlated with (a) PC1 and (b) PC2 (see references in figure 1).
| (a) compounds correlated with PC1 | letter/no. (figure 1) | correlation coefficient (r) |
|---|---|---|
| tridecyl 2-methyloctanoate | a | 0.65 |
| tetradecyl 2-methylheptanoate | b | −0.84 |
| unidentified | c | −0.76 |
| 2-methyltridecyl 2-methyloctanoate | d | −0.80 |
| pentadecyl 2-methylheptanoate | e | −0.64 |
| 2-methyltetradecyl 2-methyloctanoate | f | −0.77 |
| 2-methyldodecyl 2-methyldecanoate | g | −0.88 |
| dodecyl decanoate | h | −0.77 |
| tridecyl 2-methyldecanoate | i | 0.73 |
| pentadecyl 2-methyloctanoate | j | 0.77 |
| unidentified | k | −0.70 |
| 2-methyltetradecyl 2-methylnonanoate | l | −0.93 |
| tridecyl decanoate | m | −0.66 |
| unidentified | n | −0.80 |
| 2-methylpentadecyl 2-methylnonanoate | o | −0.89 |
| unidentified | p | −0.83 |
| unidentified | q | −0.84 |
| pentadecyl 2-methyldecanoate | r | 0.82 |
| heptadecyl 2-methyloctanoate | s | 0.78 |
| tetradecyl nonanoate | t | −0.72 |
| octadecyl 2-methyloctanoate | u | −0.60 |
| 2-methylhexadecyl 2-methyldecanoate | v | −0.85 |
| unidentified | w | −0.89 |
| pentadecyl 2-methyldodecanoate | x | 0.82 |
| heptadecyl 2-methyldecanoate | y | 0.79 |
| nonadecyl 2-methyloctanoate | z | 0.61 |
| unidentified | 1 | −0.62 |
| unidentified | 2 | −0.78 |
| hexadecyl undecanoate | 3 | −0.71 |
| heptadecyl 2-methyldodecanoate | 4 | 0.77 |
| unidentified | 5 | −0.75 |
| (b) compounds correlated with PC2 | letter/no. (figure 1) | correlation coefficient (r) |
| tetradecyl 2-methyloctanoate | a | 0.62 |
| unidentified | b | 0.60 |
| unidentified | c | 0.71 |
| tetradecyl 2-methyldecanoate | d | 0.61 |
| pentadecyl 2-methylnonanoate | e | 0.68 |
| hexadecyl 2-methyloctanoate | f | 0.63 |
| hexadecyl octanoate | g | −0.91 |
| 2-methylpentadecyl 2-methyldecanoate | h | 0.59 |
| 2-methylheptadecyl nonanoate | i | 0.64 |
| pentadecyl 2-methylundecanoate | j | 0.59 |
| heptadecyl 2-methylnonanoate | k | 0.73 |
| octadecyl 2-methyloctanoate | l | 0.60 |
| octadecyl octanoate | m | −0.74 |
| pentadecyl 2-methyltridecanoate | n | 0.72 |
| 2-methylheptadecyl 2-methyldecanoate | o | 0.84 |
| heptadecyl 2-methylundecanoate | p | 0.61 |
| unidentified | q | 0.60 |
| unidentified | r | 0.70 |
| unidentified | s | 0.63 |
| unidentified | t | 0.69 |
| unidentified | u | 0.65 |
PC2 represented 18 per cent of the variance and was negatively correlated with the two major compounds (hexadecyl octanoate and octadecyl octanoate; r = −0.91, p < 0.0001 and r = −0.74, p < 0.0001, respectively; figure 1 and table 1b), and positively with moderately abundant compounds (figure 1 and table 1b).
(b). Semiochemical diversity and heterozygosity
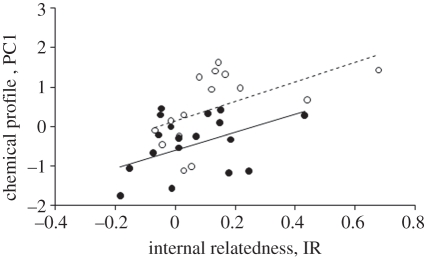
PC1 was significantly correlated with IR (F1,30 = 9.34, p = 0.0047, figure 2 and see electronic supplementary material, figure A2) and H (F1,30 = 7.07, p = 0.013). PC1 was lower in males than in females (F1,30 = 14.28, p = 0.0007; figure 2), a finding consistent with the study of Leclaire et al. [49]. PC1 increased as laying date approached similarly in both males and females (F1,30 = 11.75, p = 0.0018; sex × time before laying: F1,29 = 0.78, p = 0.38).
Figure 2.
Relationship between genetic heterozygosity (internal relatedness) and semiochemical profile (as described by PC1) in males (solid line and filled circles) and females (dashed line and open circles). A greater internal relatedness indicates decreased heterozygosity.
PC2, Simpson and Shannon indices did not relate to IR, H or sex (all p > 0.35). Shannon index and PC2 decreased, and Simpson index tended to decrease as laying date approached similarly in both males and females (F1,33 = 4.26, p = 0.047; F1,33 = 6.07, p = 0.019; and F1,33 = 3.48, p = 0.071, respectively; sex × time-before-laying: all p > 0.18).
(c). Semiochemical compounds and genetic distances
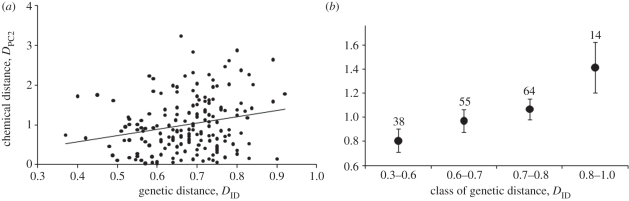
In MM dyads, chemical distance as described by the difference in PC2 (DPC2) increased significantly with genetic distance (DID: r = 0.23, p = 0.004; figure 3; DQG: r = 0.21, p = 0.009). Distance in PC1 (DPC1) did not correlate with genetic distance (DID: r = −0.17, p = 0.97; DQG: r = −0.17, p = 0.98). In the same way, relative Euclidean distance did not correlate with DID or DQG (DID: r = 0.03, p = 0.40; DQG: r = 0.05, p = 0.30).
Figure 3.
(a) Relationship between genetic distances (DID) and chemical distances (as described by differences in PC2) in MM dyads. (b) For ease of representation, we also illustrate the relationship using mean chemical distance per class of genetic distance.
In FF dyads, neither relative Euclidean distances nor DPC2 appeared related to DID and DQG (all p > 0.80). However, DPC1 tended to increase with DID and DQG (r = 0.15, p = 0.099 and r = 0.14, p = 0.10, respectively).
In MF dyads, neither relative Euclidean distances nor DPC1 and DPC2 appeared related to DID or DQG (all p > 0.10).
4. Discussion
To the best of our knowledge, we provide the first evidence that birds' semiochemical compounds can provide information about genetic make-up. Several studies have shown persistent differences in preen gland secretion between individuals or populations, suggesting a genetic basis to preen oil chemistry [45–49]. However, no previous studies have demonstrated a correlative link between preen oil chemicals and genetic heterozygosity or relatedness in birds.
(a). Semiochemical compounds and heterozygosity
The chemical profile of preen secretion was found to reflect genetic heterozygosity in kittiwakes (figures 2 and 3). In mice, ring-tailed lemurs Lemur catta and humans, heterozygosity is also encoded in body scent, and females prefer the scent of heterozygous over homozygous males [13,14,29,76]. It is unclear how olfactory signals come to represent an individual's heterozygosity [26,77]. One mediating mechanism may implicate genetic polymorphism in the enzymes involved in the biosynthesis of chemicals or in the proteins that bind fatty acids [78,79]. As a result, heterozygous individuals are suggested to have more complex chemical profiles than homozygous individuals. Consistently, lemur males that express more compounds in scrotal secretion are more heterozygous [29]. In kittiwakes, heterozygosity is not related to diversity indices in preen secretion compounds, but rather to a particular set of compounds. Preen secretion of heterozygous individuals contains several minor compounds in higher abundance, but several major compounds (i.e. odd carbon-numbered alcohols esterified with even carbon-numbered 2-methyl fatty acids: tridecyl 2-methyloctanoate, tridecyl 2-methyldecanoate, pentadecyl 2-methyloctanoate, pentadecyl 2-methyldecanoate, heptadecyl 2-methyloctanoate, pentadecyl 2-methyldodecanoate, heptadecyl 2-methyldecanoate and heptadecyl 2-methyldodecanoate) in lower abundance than preen secretion of homozygous individuals (figures 1 and 2; table 1a; electronic supplementary material, figure A2). This may suggest a more even composition of compound abundances in heterozygous than in homozygous kittiwakes (electronic supplementary material, figure A2).
The compounds encoding heterozygosity were also found to encode sex. Homozygous males have more female-like preen secretions, whereas heterozygous females have more male-like secretions (figure 2) [49]. Similarly, in dark-eyed juncos Junco hyemalis, lower-quality males (as expressed by shorter wing length) have more female-like preen odour [80].
In many species, heterozygosity is associated with traits of individual quality, such as growth, parasite load, fecundity or survival [58–60]. In kittiwakes, heterozygosity is related to chick growth rate and chick survival [33], and it probably also affects adult fitness. Kittiwakes are thus expected to choose their mate according to heterozygosity, as found in other birds such as wire-tailed manakins Pipra filicauda [36], blue tits Cyanistes caeruleus [37] and house sparrows Passer domesticus [34]. In kittiwakes, integument coloration reflects heterozygosity [42]. Odour may thus be one of the cues used by birds to assess the genetic quality of their potential partners.
(b). Semiochemical compounds and genetic relatedness
Distances in chemical profiles were found to be positively correlated with genetic distances in MM dyads (figure 3). The compounds encoding this relation were mainly hexadecyl octanoate and octadecyl octanoate, which are the two major compounds in kittiwakes' preen secretion, and on preen down feathers and neck feathers [49] (Sarah Leclaire 2010, unpublished data; figure 1 and table 1b). In humans, mandrills Mandrillus sphinx, ring-tailed lemurs, giant pandas Ailuropoda melanoleuca, beavers Castor canadensis, mice, sand lizards Lacerta agilis and Arctic charrs Salvelinus alpinus, genetic relatedness is also encoded in semiochemicals, and in some of these species, females have been shown to prefer the scent of unrelated males [11,17,19,21,22,30–32].
Kittiwakes do not display much social behaviour except within the pair. Therefore, they are not expected to be nepotistic towards their kin. Kittiwakes preferentially mate with unrelated individuals [33]. The assessment of MF relatedness should thus be more strongly selected than the assessment of MM or FF relatedness. We did not, however, find any evidence for a relationship between genetic relatedness and semiochemical distance in MF dyads. This may be owing to our relatively small sample size. Similarly, in giant pandas, information about kinship was only found in the urine of males [32]. Genetic relatedness may be assessed by self-referent or known-kin matching, with individuals avoiding breeding with individuals that have scent signatures to their own or their known kin [81]. In birds, sexual imprinting, by which mate preferences are affected by learning at a very young age, usually using a parent as the model, is very common [82,83]. Our results suggest that females may use a matching mechanism with a known kin male, such as their father, to assess their relatedness to potential mates. Similarly, in house mice Mus musculus domesticus, cross-fostering experiments have shown that females negatively imprint on familiar MHC-determined odour [84].
Most studies on body scent as a cue in the assessment of genetic relatedness have shown a link between body scent and relatedness at MHC loci [22]. The mechanism by which MHC genes influence odour is still unclear, but the mounting evidence for genetically unrelated mate choice might be explained by widespread MHC odour types in vertebrates including birds. CD1 genes, which are found in birds and mammals, are evolutionary predecessors of the MHC genes [85,86]. These genes code for proteins that present antigens such as fatty acids and glycolipids to T-cells, rather than the more typical presentation of peptides [87]. These genes might also play a role in regulating similar fatty acid compositions as found in preen gland secretions [87]. Additionally, preen esters may be metabolic by-products of preen gland bacterial flora, the composition of which may partly depend on MHC [26]. Finally, background genes can also influence odour profiles [27]. Polymorphism in the gene products that govern the biosynthesis of wax esters could mediate the relationship between odour and genetic make-up [78,79,88]. Studies are needed to determine whether odours are influenced more by MHC than by overall genetic relatedness in kittiwakes.
Our study is the first to demonstrate a link between odour and genetics in birds, which sets the stage for the existence of sophisticated odour-based mechanisms of mate choice also in birds. Our study thus contributes to an increasing body of evidence suggesting that odour cues play a greater role in communication in birds than previously assumed. However, whether birds use odour cues to choose their mates still needs to be demonstrated.
Acknowledgements
We are grateful to E. Moëc, B. Planade and C. Bello Marín for their help in the field. We thank C. Veyssière for her help with genetic analyses, and G. Giacinti for her help with chemical analyses. We acknowledge the Genomic Platform of Genopole Toulouse Midi Pyrénées, where genotyping was performed. This work was supported by the French Polar Institute Paul-Emile Victor (IPEV Programme 1162 SexCoMonArc), and by an interdisciplinary programme of the CNRS (PIR Maladies infectieuses et environnement numéro 74164). Experiments were carried out in accordance with United States laws and with due permits from the US Fish and Wildlife Service and State of Alaska. Any use of trade names is for descriptive purposes only and does not imply endorsement of the US Government.
References
- 1.Andersson M. 1994. Sexual selection. Princeton, NJ: Princeton University Press [Google Scholar]
- 2.Mays H. L., Hill G. E. 2004. Choosing mates: good genes versus genes that are a good fit. Trends Ecol. Evol. 19, 554–559 10.1016/j.tree.2004.07.018 (doi:10.1016/j.tree.2004.07.018) [DOI] [PubMed] [Google Scholar]
- 3.Neff B. D., Pitcher T. E. 2005. Genetic quality and sexual selection: an integrated framework for good genes and compatible genes. Mol. Ecol. 14, 19–38 10.1111/j.1365-294X.2004.02395.x (doi:10.1111/j.1365-294X.2004.02395.x) [DOI] [PubMed] [Google Scholar]
- 4.Ekblom R., Saether S. A., Grahn M., Fiske P., Kalas J. A., Hoglund J. 2004. Major histocompatibility complex variation and mate choice in a lekking bird, the great snipe (Gallinago media). Mol. Ecol. 13, 3821–3828 10.1111/j.1365-294X.2004.02361.x (doi:10.1111/j.1365-294X.2004.02361.x) [DOI] [PubMed] [Google Scholar]
- 5.Eizaguirre C., Yeates S. E., Lenz T. L., Kalbe M., Milinski M. 2009. MHC-based mate choice combines good genes and maintenance of MHC polymorphism. Mol. Ecol. 18, 3316–3329 10.1111/j.1365-294X.2009.04243.x (doi:10.1111/j.1365-294X.2009.04243.x) [DOI] [PubMed] [Google Scholar]
- 6.Schwensow N., Fietz J., Dausmann K., Sommer S. 2008. MHC-associated mating strategies and the importance of overall genetic diversity in an obligate pair-living primate. Evol. Ecol. 22, 617–636 10.1007/s10682-007-9186-4 (doi:10.1007/s10682-007-9186-4) [DOI] [Google Scholar]
- 7.Husseneder C., Simms D. M. 2008. Size and heterozygosity influence partner selection in the Formosan subterranean termite. Behav. Ecol. 19, 764–773 10.1093/beheco/arn041 (doi:10.1093/beheco/arn041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts S. C., Gosling L. M. 2003. Genetic similarity and quality interact in mate choice decisions by female mice. Nat. Genet. 35, 103–106 10.1038/ng1231 (doi:10.1038/ng1231) [DOI] [PubMed] [Google Scholar]
- 9.Tregenza T., Wedell N. 2000. Genetic compatibility, mate choice and patterns of parentage: invited review. Mol. Ecol. 9, 1013–1027 10.1046/j.1365-294x.2000.00964.x (doi:10.1046/j.1365-294x.2000.00964.x) [DOI] [PubMed] [Google Scholar]
- 10.Freeman-Gallant C. R., Meguerdichian M., Wheelwright N. T., Sollecito S. V. 2003. Social pairing and female mating fidelity predicted by restriction fragment length polymorphism similarity at the major histocompatibility complex in a songbird. Mol. Ecol. 12, 3077–3083 10.1046/j.1365-294X.2003.01968.x (doi:10.1046/j.1365-294X.2003.01968.x) [DOI] [PubMed] [Google Scholar]
- 11.Olsson M., Madsen T., Nordby J., Wapstra E., Ujvari B., Wittsell H. 2003. Major histocompatibility complex and mate choice in sand lizards. Proc. R. Soc. Lond. B 270, S254–S256 10.1098/rsbl.2003.0079 (doi:10.1098/rsbl.2003.0079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ditchkoff S. S., Lochmiller R. L., Masters R. E., Hoofer S. R., Van Den Bussche R. A. 2001. Major-histocompatibility-complex-associated variation in secondary sexual traits of white-tailed deer (Odocoileus virginianus): evidence for good-genes advertisement. Evolution 55, 616–625 10.1554/0014-3820(2001)055[0616:MHCAVI]2.0.CO;2 (doi:10.1554/0014-3820(2001)055[0616:MHCAVI]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 13.Charpentier M. J. E., Crawford J. C., Boulet M., Drea C. M. 2010. Message ‘scent’: lemurs detect the genetic relatedness and quality of conspecifics via olfactory cues. Anim. Behav. 80, 101–108 10.1016/j.anbehav.2010.04.005 (doi:10.1016/j.anbehav.2010.04.005) [DOI] [Google Scholar]
- 14.Thom M. D., Stockley P., Jury F., Ollier W. E. R., Beynon R. J., Hurst J. L. 2008. The direct assessment of genetic heterozygosity through scent in the mouse. Curr. Biol. 18, 619–623 10.1016/j.cub.2008.03.056 (doi:10.1016/j.cub.2008.03.056) [DOI] [PubMed] [Google Scholar]
- 15.Hoikkala A., Aspi J., Suvanto L. 1998. Male courtship song frequency as an indicator of male genetic quality in an insect species, Drosophila montana. Proc. R. Soc. Lond. B 265, 503–508 10.1098/rspb.1998.0323 (doi:10.1098/rspb.1998.0323) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Oosterhout C., Trigg R. E., Carvalho G. R., Magurran A. E., Hauser L., Shaw P. W. 2003. Inbreeding depression and genetic load of sexually selected traits: how the guppy lost its spots. J. Evol. Biol. 16, 273–281 10.1046/j.1420-9101.2003.00511.x (doi:10.1046/j.1420-9101.2003.00511.x) [DOI] [PubMed] [Google Scholar]
- 17.Wedekind C., Seebeck T., Bettens F., Paepke A. J. 1995. MHC-dependent mate preferences in humans. Proc. R. Soc. Lond. B 260, 245–249 10.1098/rspb.1995.0087 (doi:10.1098/rspb.1995.0087) [DOI] [PubMed] [Google Scholar]
- 18.Radwan J., Tkacz A., Kloch A. 2008. MHC and preferences for male odour in the bank vole. Ethology 114, 827–833 10.1111/j.1439-0310.2008.01528.x (doi:10.1111/j.1439-0310.2008.01528.x) [DOI] [Google Scholar]
- 19.Yamazaki K., Yamaguchi M., Baranoski L., Bard J., Boyse E. A., Thomas L. 1979. Recognition among mice: evidence from the use of a Y-maze differentially scented by congenic mice of different major histocompatibility types. J. Exp. Med. 150, 755–760 10.1084/jem.150.4.755 (doi:10.1084/jem.150.4.755) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parrott M. L., Ward S. J., Temple-Smith P. D. 2007. Olfactory cues, genetic relatedness and female mate choice in the agile antechinus (Antechinus agilis). Behav. Ecol. Sociobiol. 61, 1075–1079 10.1007/s00265-006-0340-8 (doi:10.1007/s00265-006-0340-8) [DOI] [Google Scholar]
- 21.Olsen K. H., Grahn M., Lohm J., Langefors A. 1998. MHC and kin discrimination in juvenile Arctic charr, Salvelinus alpinus (L.). Anim. Behav. 56, 319–327 10.1006/anbe.1998.0837 (doi:10.1006/anbe.1998.0837) [DOI] [PubMed] [Google Scholar]
- 22.Setchell J. M., Vaglio S., Abbott K. M., Moggi-Cecchi J., Boscaro F., Pieraccini G., Knapp L. A. 2011. Odour signals major histocompatibility complex genotype in an Old World monkey. Proc. R. Soc. B 278, 274–280 10.1098/rspb.2010.0571 (doi:10.1098/rspb.2010.0571) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonneaud C., Mazuc J., Chastel O., Westerdahl H., Sorci G. 2004. Terminal investment induced by immune challenge and fitness traits associated with major histocompatibility complex in the house sparrow. Evolution 58, 2823–2830 [DOI] [PubMed] [Google Scholar]
- 24.Westerdahl H., Waldenstrom J., Hansson B., Hasselquist D., von Schantz T., Bensch S. 2005. Associations between malaria and MHC genes in a migratory songbird. Proc. R. Soc. B 272, 1511–1518 10.1098/rspb.2005.3113 (doi:10.1098/rspb.2005.3113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wedekind C., Walker M., Portmann J., Cenni B., Muller R., Binz T. 2004. MHC-linked susceptibility to a bacterial infection, but no MHC-linked cryptic female choice in whitefish. J. Evol. Biol. 17, 11–18 10.1046/j.1420-9101.2004.00669.x (doi:10.1046/j.1420-9101.2004.00669.x) [DOI] [PubMed] [Google Scholar]
- 26.Penn D. J. 2002. The scent of genetic compatibility: sexual selection and the major histocompatibility complex. Ethology 108, 1–21 10.1046/j.1439-0310.2002.00768.x (doi:10.1046/j.1439-0310.2002.00768.x) [DOI] [Google Scholar]
- 27.Willse A., Kwak J., Yamazaki K., Preti G., Wahl J. H., Beauchamp G. K. 2006. Individual odortypes: interaction of MHC and background genes. Immunogenetics 58, 967–982 10.1007/s00251-006-0162-x (doi:10.1007/s00251-006-0162-x) [DOI] [PubMed] [Google Scholar]
- 28.Cheetham S. A., Thom M. D., Jury F., Ollier W. E. R., Beynon R. J., Hurst J. L. 2007. The genetic basis of individual-recognition signals in the mouse. Curr. Biol. 17, 1771–1777 10.1016/j.cub.2007.10.007 (doi:10.1016/j.cub.2007.10.007) [DOI] [PubMed] [Google Scholar]
- 29.Charpentier M. J. E., Boulet M., Drea C. M. 2008. Smelling right: the scent of male lemurs advertises genetic quality and relatedness. Mol. Ecol. 17, 3225–3233 10.1111/j.1365-294X.2008.03831.x (doi:10.1111/j.1365-294X.2008.03831.x) [DOI] [PubMed] [Google Scholar]
- 30.Sun L. X., Muller-Schwarze D. 1998. Anal gland secretion codes for relatedness in the beaver, Castor canadensis. Ethology 104, 917–927 10.1111/j.1439-0310.1998.tb00041.x (doi:10.1111/j.1439-0310.1998.tb00041.x) [DOI] [Google Scholar]
- 31.Boulet M., Charpentier M. J. E., Drea C. M. 2009. Decoding an olfactory mechanism of kin recognition and inbreeding avoidance in a primate. BMC Evol. Biol. 9, 281. 10.1186/1471-2148-9-281 (doi:10.1186/1471-2148-9-281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu D. Z., Wei R. P., Zhang G. Q., Yuan H., Wang Z. P., Sun L. X., Zhang J. X., Zhang H. M. 2008. Male panda (Ailuropoda melanoleuca) urine contains kinship information. Chin. Sci. Bull. 53, 2793–2800 10.1007/s11434-008-0373-7 (doi:10.1007/s11434-008-0373-7) [DOI] [Google Scholar]
- 33.Mulard H., Danchin E., Talbot S. L., Ramey A. M., Hatch S. A., White J. F., Helfenstein F., Wagner R. H. 2009. Evidence that pairing with genetically similar mates is maladaptive in a monogamous bird. BMC Evol. Biol. 9, 147. 10.1186/1471-2148-9-147 (doi:10.1186/1471-2148-9-147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonneaud C., Chastel O., Federici P., Westerdahl H., Sorci G. 2006. Complex MHC-based mate choice in a wild passerine. Proc. R. Soc. B 273, 1111–1116 10.1098/rspb.2005.3325 (doi:10.1098/rspb.2005.3325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foerster K., Delhey K., Johnsen A., Lifjeld J. T., Kempenaers B. 2003. Females increase offspring heterozygosity and fitness through extra-pair matings. Nature 425, 714–717 10.1038/nature01969 (doi:10.1038/nature01969) [DOI] [PubMed] [Google Scholar]
- 36.Ryder T. B., Tori W. P., Blake J. G., Loiselle B. A., Parker P. G. 2010. Mate choice for genetic quality: a test of the heterozygosity and compatibility hypotheses in a lek-breeding bird. Behav. Ecol. 21, 203–210 10.1093/beheco/arp176 (doi:10.1093/beheco/arp176) [DOI] [Google Scholar]
- 37.Garcia-Navas V., Ortego J., Sanz J. J. 2009. Heterozygosity-based assortative mating in blue tits (Cyanistes caeruleus): implications for the evolution of mate choice. Proc. R. Soc. B 276, 2931–2940 10.1098/rspb.2009.0417 (doi:10.1098/rspb.2009.0417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dreiss A. N., Silva N., Richard M., Moyen F., Thery M., Moller A. P., Danchin E. 2008. Condition-dependent genetic benefits of extrapair fertilization in female blue tits Cyanistes caeruleus. J. Evol. Biol. 21, 1814–1822 10.1111/j.1420-9101.2008.01578.x (doi:10.1111/j.1420-9101.2008.01578.x) [DOI] [PubMed] [Google Scholar]
- 39.Griggio M., Biard C., Penn D. J., Hoi H. 2011. Female house sparrows ‘count on’ male genes: experimental evidence for MHC-dependent mate preference in birds. BMC Evol. Biol. 11, (doi:10.1186/1471-2148-11-44) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arct A., Rutkowska J., Martyka R., Drobniak S. M., Cichon M. 2010. Kin recognition and adjustment of reproductive effort in zebra finches. Biol. Lett. 6, 762–764 10.1098/rsbl.2010.0417 (doi:10.1098/rsbl.2010.0417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Potvin D. A., MacDougall-Shackleton E. A. 2009. Parental investment amplifies effects of genetic complementarity on growth rates in song sparrows, Melospiza melodia. Anim. Behav. 78, 943–948 10.1016/j.anbehav.2009.07.023 (doi:10.1016/j.anbehav.2009.07.023) [DOI] [Google Scholar]
- 42.Leclaire S., White J., Arnoux E., Faivre B., Vetter N., Hatch S. A., Danchin E. 2011. Integument coloration signals reproductive success, heterozygosity and antioxidant levels in chick-rearing black-legged kittiwakes. Naturwissenschaften 98, 773–782 10.1007/s00114-011-0827-7 (doi:10.1007/s00114-011-0827-7) [DOI] [PubMed] [Google Scholar]
- 43.Bolund E., Martin K., Kempenaers B., Forstmeier W. 2010. Inbreeding depression of sexually selected traits and attractiveness in the zebra finch. Anim. Behav. 79, 947–955 10.1016/j.anbehav.2010.01.014 (doi:10.1016/j.anbehav.2010.01.014) [DOI] [Google Scholar]
- 44.Marshall R. C., Buchanan K. L., Catchpole C. K. 2003. Sexual selection and individual genetic diversity in a songbird. Proc. R. Soc. Lond. B 270, S248–S250 10.1098/rsbl.2003.0081 (doi:10.1098/rsbl.2003.0081) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mardon J., Saunders S. M., Anderson M. J., Couchoux C., Bonadonna F. 2010. Species, gender, and identity: cracking petrels' sociochemical code. Chem. Senses 35, 309–321 10.1093/chemse/bjq021 (doi:10.1093/chemse/bjq021) [DOI] [PubMed] [Google Scholar]
- 46.Bonadonna F., Miguel E., Grosbois V., Jouventin P., Bessière J. M. 2007. Individual odor recognition in birds: an endogenous olfactory signature on petrels' feathers? J. Chem. Ecol. 33, 1819–1829 10.1007/s10886-007-9345-7 (doi:10.1007/s10886-007-9345-7) [DOI] [PubMed] [Google Scholar]
- 47.Whittaker D. J., Soini H. A., Atwell J. W., Hollars C., Novotny M. V., Ketterson E. D. 2010. Songbird chemosignals: volatile compounds in preen gland secretions vary among individuals, sexes, and populations. Behav. Ecol. 21, 608–614 10.1093/beheco/arq033 (doi:10.1093/beheco/arq033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karlsson A. C., Jensen P., Elgland M., Laur K., Fyrner T., Konradsson P., Laska M. 2010. Red junglefowl have individual body odors. J. Exp. Biol. 213, 1619–1624 10.1242/jeb.040279 (doi:10.1242/jeb.040279) [DOI] [PubMed] [Google Scholar]
- 49.Leclaire S., Merkling T., Raynaud C., Giacinti G., Bessière J. M., Hatch S. A., Danchin E. 2011. An individual and a sex odor signature in kittiwakes? Study of the semiochemical composition of preen secretion and preen down feathers. Naturwissenschaften 98, 615–624 10.1016/j.yhbeh.2008.07.002 (doi:10.1016/j.yhbeh.2008.07.002) [DOI] [PubMed] [Google Scholar]
- 50.Balthazart J., Taziaux M. 2009. The underestimated role of olfaction in avian reproduction? Behav. Brain Res. 200, 248–259 10.1016/j.bbr.2008.08.036 (doi:10.1016/j.bbr.2008.08.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hagelin J. C., Jones I. L. 2007. Bird odors and other chemical substances: a defense mechanism or overlooked mode of intraspecific communication? Auk 124, 741–761 10.1642/0004-8038(2007)124[741:BOAOCS]2.0.CO;2 (doi:10.1642/0004-8038(2007)124[741:BOAOCS]2.0.CO;2) [DOI] [Google Scholar]
- 52.Balthazart J., Schoffeniels E. 1979. Pheromones are involved in the control of sexual behavior in birds. Naturwissenschaften 66, 55–56 10.1007/BF00369365 (doi:10.1007/BF00369365) [DOI] [PubMed] [Google Scholar]
- 53.Hirao A., Aoyama M., Sugita S. 2009. The role of uropygial gland on sexual behavior in domestic chicken Gallus gallus domesticus. Behav. Process. 80, 115–120 10.1016/j.beproc.2008.10.006 (doi:10.1016/j.beproc.2008.10.006) [DOI] [PubMed] [Google Scholar]
- 54.Bonadonna F., Nevitt G. A. 2004. Partner-specific odor recognition in an Antarctic seabird. Science 306, 835. 10.1126/science.1103001 (doi:10.1126/science.1103001) [DOI] [PubMed] [Google Scholar]
- 55.Whittaker D. J., Reichard D. G., Dapper A. L., Ketterson E. D. 2009. Behavioral responses of nesting female dark-eyed juncos Junco hyemalis to hetero- and conspecific passerine preen oils. J. Avian Biol. 40, 579–583 10.1111/j.1600-048X.2009.04813.x (doi:10.1111/j.1600-048X.2009.04813.x) [DOI] [Google Scholar]
- 56.Soini H. A., Schrock S. E., Bruce K. E., Wiesler D., Ketterson E. D., Novotny M. V. 2007. Seasonal variation in volatile compound profiles of preen gland secretions of the dark-eyed junco ( Junco hyemalis). J. Chem. Ecol. 33, 183–198 10.1007/s10886-006-9210-0 (doi:10.1007/s10886-006-9210-0) [DOI] [PubMed] [Google Scholar]
- 57.Mulard H. 2007. Behavioural implications of strict monogamy: individual recognition and genetic bases of mate choice in the black-legged kittiwake, Rissa tridactyla. PhD thesis, Université Pierre et Marie Curie, Paris, France [Google Scholar]
- 58.Acevedo-Whitehouse K., Gulland F., Greig D., Amos W. 2003. Disease susceptibility in California sea lions. Nature 422, 35. 10.1038/422035a (doi:10.1038/422035a) [DOI] [PubMed] [Google Scholar]
- 59.Keller L. F., Waller D. M. 2002. Inbreeding effects in wild populations. Trends Ecol. Evol. 17, 230–241 10.1016/S0169-5347(02)02489-8 (doi:10.1016/S0169-5347(02)02489-8) [DOI] [Google Scholar]
- 60.Roldan E. R. S., Cassinello J., Abaigar T., Gomendio M. 1998. Inbreeding, fluctuating asymmetry, and ejaculate quality in an endangered ungulate. Proc. R. Soc. Lond. B 265, 243–248 10.1098/rspb.1998.0288 (doi:10.1098/rspb.1998.0288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leclaire S., Mulard H., Wagner R. H., Hatch S. A., Danchin E. 2009. Can Kittiwakes smell? Experimental evidence in a Larid species. Ibis 151, 584–587 10.1111/j.1474-919X.2009.00935.x (doi:10.1111/j.1474-919X.2009.00935.x) [DOI] [Google Scholar]
- 62.Gill V. A., Hatch S. A. 2002. Components of productivity in black-legged kittiwakes Rissa tridactyla: response to supplemental feeding. J. Avian Biol. 33, 113–126 10.1034/j.1600-048X.2002.330201.x (doi:10.1034/j.1600-048X.2002.330201.x) [DOI] [Google Scholar]
- 63.Jodice P. G. R., Lanctot R. B., Gill V. A., Roby D. D., Hatch S. A. 2000. Sexing adult black-legged kittiwakes by DNA, behavior, and morphology. Waterbirds 23, 405–415 [Google Scholar]
- 64.Jordan N. R., Mwanguhya F., Kyabulima S., Ruedi P., Cant M. A. 2010. Scent marking within and between groups of wild banded mongooses. J. Zool. 280, 72–83 10.1111/j.1469-7998.2009.00646.x (doi:10.1111/j.1469-7998.2009.00646.x) [DOI] [Google Scholar]
- 65.Lawson R. E., Putman R. J., Fielding A. H. 2001. Chemical communication in Eurasian deer (Cervidae): do individual odours also code for attributes? J. Zool. 253, 91–99 10.1017/S0952836901000085 (doi:10.1017/S0952836901000085) [DOI] [Google Scholar]
- 66.McCune B., Grace J. B., Urban D. L. 2002. Analysis of ecological communities. Gleneden Beach, OR: MjM Software Design [Google Scholar]
- 67.Legendre P., Legendre L. 1998. Numerical ecology. Amsterdam, The Netherlands: Elsevier Science BV [Google Scholar]
- 68.Hurst J. L., Beynon R. J. 2010. Making progress in genetic kin recognition among vertebrates. J. Biol. 9, 13. 10.1186/jbiol221 (doi:10.1186/jbiol221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Longmire J., et al. 1988. Isolation and molecular characterization of a highly polymorphic centromeric tandem repeat in the family Falconidae. Genomics 2, 14–24 10.1016/0888-7543(88)90104-8 (doi:10.1016/0888-7543(88)90104-8) [DOI] [PubMed] [Google Scholar]
- 70.Vali U., Einarsson A., Waits L., Ellegren H. 2008. To what extent do microsatellite markers reflect genome-wide genetic diversity in natural populations? Mol. Ecol. 17, 3808–3817 10.1111/j.1365-294X.2008.03876.x (doi:10.1111/j.1365-294X.2008.03876.x) [DOI] [PubMed] [Google Scholar]
- 71.Amos W., Worthington Wilmer J., Fullard K., Burg T. M., Croxall J. P., Bloch D., Coulson T. 2001. The influence of parental relatedness on reproductive success. Proc. R. Soc. Lond. B 268, 2021–2027 10.1098/rspb.2001.1751 (doi:10.1098/rspb.2001.1751) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Belkhir K., Castric V., Bonhomme F. 2002. IDENTIX, a software to test for relatedness in a population using permutation methods. Mol. Ecol. Notes 2, 611–614 10.1046/j.1471-8286.2002.00273.x (doi:10.1046/j.1471-8286.2002.00273.x) [DOI] [Google Scholar]
- 73.Mathieu E., Autem M., Roux M., Bonhomme F. 1990. Épreuves de la validation dans l'analyse de structures génétiques multivariées: comment tester l'équilibre panmictique? Revue de statistiques appliquées 38, 47–66 [Google Scholar]
- 74.Queller D. C., Goodnight K. F. 1989. Estimating relatedness using genetic markers. Evolution 43, 258–275 10.2307/2409206 (doi:10.2307/2409206) [DOI] [PubMed] [Google Scholar]
- 75.R Development Core Team. 2008. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 76.Ilmonen P., Stundner G., Thoss M., Penn D. J. 2009. Females prefer the scent of outbred males: good-genes-as-heterozygosity? BMC Evol. Biol. 9, (doi:10.1186/1471-2148-9-104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boehm T., Zufall F. 2006. MHC peptides and the sensory evaluation of genotype. Trends Neurosci. 29, 100–107 10.1016/j.tins.2005.11.006 (doi:10.1016/j.tins.2005.11.006) [DOI] [PubMed] [Google Scholar]
- 78.Takahashi A., Tsaur S.-C., Coyne J., Wu I. 2001. The nucleotide changes governing cuticular hydrocarbon variation and their evolution in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 98, 3920–3925 10.1073/pnas.061465098 (doi:10.1073/pnas.061465098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roelofs W., Rooney A. 2003. Molecular genetics and evolution of pheromone biosynthesis in Lepidoptera. Proc. Natl Acad. Sci. USA 100, 9179–9184 10.1073/pnas.1233767100 (doi:10.1073/pnas.1233767100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Whittaker D. J., Richmond K. M., Miller A. K., Kiley R., Burns C. B., Atwell J. W., Ketterson E. D. In press Intraspecific preen oil odor preferences in dark-eyed juncos ( Junco hyemalis). Behav. Ecol. (doi:10.1093/beheco/arr122) [Google Scholar]
- 81.Mateo J. M. 2003. Kin recognition in ground squirrels and other rodents. J. Mammal. 84, 1163–1181 10.1644/BLe-011 (doi:10.1644/BLe-011) [DOI] [Google Scholar]
- 82.Hauber M. E., Sherman P. W. 2001. Self-referent phenotype matching: theoretical considerations and empirical evidence. Trends Neurosci. 24, 609–616 10.1016/S0166-2236(00)01916-0 (doi:10.1016/S0166-2236(00)01916-0) [DOI] [PubMed] [Google Scholar]
- 83.Bateson P. P. G. 1966. Characteristics and context of imprinting. Biol. Rev. Camb. Phil. Soc. 41, 177–217 10.1111/j.1469-185X.1966.tb01489.x (doi:10.1111/j.1469-185X.1966.tb01489.x) [DOI] [PubMed] [Google Scholar]
- 84.Penn D., Potts W. 1998. MHC-disassortative mating preferences reversed by cross-fostering. Proc. R. Soc. Lond. B 265, 1299–1306 10.1098/rspb.1998.0433 (doi:10.1098/rspb.1998.0433) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miller M. M., et al. 2005. Characterization of two avian MHC-like genes reveals an ancient origin of the CD1 family. Proc. Natl Acad. Sci. USA 102, 8674–8679 10.1073/pnas.0500105102 (doi:10.1073/pnas.0500105102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Salomonsen J., et al. 2005. Two CD1 genes map to the chicken MHC, indicating that CD1 genes are ancient and likely to have been present in the primordial MHC. Proc. Natl Acad. Sci. USA 102, 8668–8673 10.1073/pnas.0409213102 (doi:10.1073/pnas.0409213102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Porcelli S. A., Modlin R. L. 1999. The CD1 system: antigen-presenting molecules for T cell recognition of lipids and glycolipids. Annu. Rev. Immunol. 17, 297–329 10.1146/annurev.immunol.17.1.297 (doi:10.1146/annurev.immunol.17.1.297) [DOI] [PubMed] [Google Scholar]
- 88.Turkish A. R., Sturley S. L. 2009. The genetics of neutral lipid biosynthesis: an evolutionary perspective. Am. J. Physiol. Endocrinol. Metab. 297, E19–E27 10.1152/ajpendo.90898.2008 (doi:10.1152/ajpendo.90898.2008) [DOI] [PMC free article] [PubMed] [Google Scholar]