Abstract
Measles is one of the most contagious viral diseases, and remains a major cause of childhood morbidity and mortality worldwide. The measles virus (MV), a member of the family Paramyxoviridae, enters cells through a cellular receptor, the signaling lymphocyte activation molecule (SLAM), CD46 or nectin-4. Entry is mediated by two MV envelope glycoproteins, the hemagglutinin (H) and the fusion (F) protein. The H protein mediates receptor attachment, while the F protein causes membrane fusion. The interaction between the H and F proteins is essential to initiate the cell entry process. Recently determined crystal structures of the MV-H protein unbound and bound to SLAM or CD46 have provided insights into paramyxovirus entry and the effectiveness of measles vaccine.
Keywords: measles virus, hemagglutinin, structure, fusion, entry, measles vaccine, glycoprotein, receptor
Introduction
Measles virus (MV), the agent that causes measles, is an enveloped, non-segmented, negative-strand RNA virus, a member of the genus Morbillivirus, the family Paramyxoviridae (Griffin, 2007). Paramyxoviruses also include, among others, mumps virus, Newcastle disease virus (NDV), human parainfluenza viruses (hPIV), and emerging Hendra virus (HeV) and Nipah virus (NiV), members of the genus Henipavirus (Lamb and Parks, 2007). MV enters cells through membrane fusion in a pH-independent manner, like other paramyxoviruses. MV possesses two glycoproteins on its envelope, an attachment protein hemagglutinin (H) (MV-H) and a fusion (F) protein (MV-F) (Griffin, 2007). MV-H and MV-F form hetero-oligomer, which is required to induce membrane fusion. Upon receptor binding, MV-H is thought to undergo a conformational change, which in turn would trigger a structural rearrangement of MV-F from the metastable pre-fusion form to the intermediate and post-fusion forms (Plemper et al., 2011). This conformational change of MV-F would drive the fusion between the viral envelope and the host cell membrane. Currently, the detailed structural organization of MV-H and MV-F remains elusive.
The paramyxovirus attachment proteins are classified into three groups based on their functions, the hemagglutinin–neuraminidase (HN) protein, the H protein, and the G protein. Although these attachment proteins may have a common assembly process, their interaction with the F protein may be viral species-dependent, and different paramyxoviruses may use different strategies for fusion activation (Lamb and Parks, 2007; Iorio and Mahon, 2008; Connolly et al., 2009; Smith et al., 2009; Plemper et al., 2011). Among paramyxoviruses, only those belonging to the genera Morbillivirus and Henipavirus recognize protein receptors expressed on the target cell surface. This is in marked contrast to most other members of the family Paramyxoviridae, which contain the neuraminidase domain in their HN proteins and recognize sialic acid-containing receptors.
Three protein molecules have been identified as MV receptors, the signaling lymphocyte activation molecule (SLAM), CD46, and nectin-4. On the other hand, NiV and HeV utilize as receptors ephrin (Eph) B2 and EphB3, which belong to the highly conserved Eph family of tyrosine kinase receptors (Bonaparte et al., 2005; Negrete et al., 2005, 2006). Although both MV and henipaviruses recognize protein receptors, there is a large structural difference between MV-H and henipavirus G proteins (Bowden et al., 2010). The G proteins exhibit a structural-fold property closer to that of HN proteins. This suggests that MV-H and henipavirus G proteins have independently evolved the ability to recognize protein receptors.
One clinical isolate obtained from a child in 1954 known as the Edmonston strain gave rise to the majority of current MV vaccine strains (Katz, 2009). After 50 years, its progeny (Edmonston lineage) are still effective as live attenuated vaccines. It is estimated that more than 4.5 million measles deaths have been prevented annually through implementation of the vaccination strategies developed by WHO and UNISEF (Bellini and Rota, 2011).
Here, we review recent advances in our understanding of MV entry, based on crystal structures of MV-H–receptor complexes as well as functional studies. We compare the attachment protein-receptor complex of MV with those of other paramyxoviruses, and provide a general principle of paramyxovirus entry. We also discuss why MV has only one major serotype and why our current MV vaccine prepared more than five decades ago from a single strain is still effective.
MV Receptors
CD46
Currently, three receptors are identified, which facilitate MV entry. CD46 (also called membrane cofactor protein) is the first MV receptor discovered by two groups independently in 1993 (Dorig et al., 1993; Naniche et al., 1993). CD46 is a type I membrane protein that belongs to the regulators of complement activation (RCA) family. CD46 comprises an N-terminal signal sequence, four short consensus repeats (SCR1–SCR4), a transmembrane region, and a C-terminal cytoplasmic tail. Each SCR module contains ∼60 amino acids that fold into a compact β-barrel structure (Persson et al., 2010). N-terminal domains SCR1 and SCR2 participate in the interaction with MV-H (Santiago et al., 2010).
CD46 is expressed on all nucleated human cells, and acts as a cofactor in the proteolytic inactivation of C3b and C4b by factor I. Monkey, but not human, erythrocytes express CD46, allowing hemagglutination by MV (Cole et al., 1985; Kemper and Atkinson, 2009). Interestingly, only vaccine and laboratory-adapted strains of MV can utilize CD46 as a receptor, while clinically isolated MV strains employ different receptors for entry.
Signaling lymphocyte activation molecule
Measles viruses were found to be efficiently isolated from clinical specimens by using the marmoset B cell line B95a or other human B cell lines (Kobune et al., 1990; Schneider-Schaulies et al., 1995). Further, MVs obtained using these cell lines were unable to downregulate CD46 or induce cell-to-cell fusion in CD46-positive cell lines (Lecouturier et al., 1996; Bartz et al., 1998; Tanaka et al., 1998). These observations suggested the presence of MV receptor(s) other than CD46. In 2000, SLAM (also known as CD150) was identified as a principal cellular receptor for all MV strains (Tatsuo et al., 2000).
Signaling lymphocyte activation molecule, a member of the immunoglobulin (Ig) superfamily, is a type I membrane protein that possesses an N-terminal signal sequence, two Ig-like domains (V-set and C2-set), a transmembrane region, and a cytoplasmic tail (Cocks et al., 1995). Each Ig-like domain has ∼110 amino acid residues. Only the membrane-distal V-set domain is necessary and sufficient for MV-H binding. The V domain is comprised of the BED and AGFCC′C″ β-sheet, which utilize a Cys32–Cys132 disulfide bond to stabilize the A–G interstrand interaction (Hashiguchi et al., 2011).
In humans, SLAM is expressed on thymocytes, activated lymphocyte, mature dendritic cells, macrophages, and platelets. However, it is absent from monocytes, natural killer cells, and granulocytes (Schwartzberg et al., 2009). The distribution of SLAM nicely explains the tropism and immunosuppressive nature of MV (Yanagi et al., 2009).
Nectin-4
Although epithelial cells do not express SLAM, MV antigens, and syncytia have been observed in the epithelia of various organs from measles patients and experimentally infected monkeys. It was found that a human lung adenocarcinoma cell line NCI-H358 supports SLAM- and CD46-independent MV infection (Takeda et al., 2007). Furthermore, several polarized epithelial cell lines have been reported to be susceptible to MV (Takeda et al., 2007; Leonard et al., 2008; Tahara et al., 2008). Together, these data hinted at the presence of a third MV receptor.
Recent studies have identified nectin-4 as an epithelial cell receptor (Muhlebach et al., 2011; Noyce et al., 2011). Human nectin-4 is expressed mainly in placenta cells, and to a lesser degree in trachea cells (Reymond et al., 2001). Nectin-4 is a member of the poliovirus receptor-like proteins (PVRLs), which are adhesion receptors of the Ig superfamily. Similar to SLAM, this type I membrane protein contains an N-terminal signal sequence, three Ig-like domains (V-set and two C2-set), a transmembrane region, and a cytoplasmic tail. Each Ig-like domain is predicted to be ∼105 amino acid residues. Both wild type and the Edmonston lineage MV strains can use nectin-4 as a receptor.
MV-H Protein
The MV-H is a type II membrane glycoprotein comprised of an N-terminal cytoplasmic tail, a transmembrane region, a stalk, and a C-terminal receptor-binding head domain. The head domain of MV-H exhibits a six-bladed β-propeller fold, a feature conserved among all head domains of paramyxovirus attachment proteins thus far crystallized (Colf et al., 2007; Hashiguchi et al., 2007). Without a neuraminidase domain, MV-H targets protein receptors, SLAM, CD46, and nectin-4. The difference in receptor preference between H and HN proteins reflects the difference in their head domain structures (see below).
Crystal structure of the MV-H head domain reveals a top pocket reminiscent of the sialic acid binding cavity found in both HN proteins and the neuraminidase (NA) of influenza virus (Flu). However, the pocket in MV-H is enlarged and lacks several key residues that contribute to sialic acid binding. In addition, the N-linked sugar at position 215 of MV-H (located at the rim of the pocket) renders the top pocket inaccessible to sialic acid, antibodies or other molecules. The MV-H head domain exhibits a cubic shape, and forms a homodimer (Hashiguchi et al., 2007). Two neighboring N-linked sugars at position 200 (one each from both monomers), located at the homodimer interface, likely block access to the region, which in HN of NDV, has been proposed to contain a second sialic acid binding site. Similarly, HNs of hPIV3 and PIV5 do not contain a second sialic acid binding site due to N-linked sugar at Asn523 and Asn497, respectively.
Interaction between MV-H and Receptors
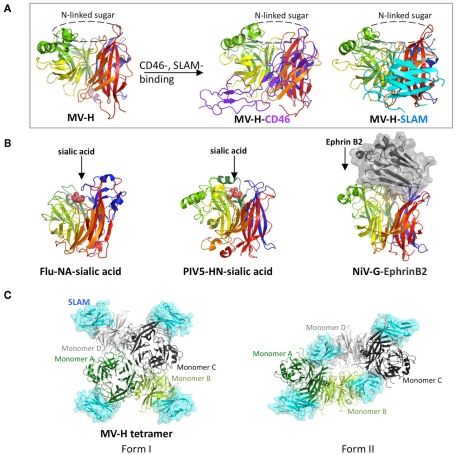
Mutagenesis studies have suggested that receptor-binding sites on MV-H are located on the side of the β-propeller, which was subsequently confirmed by the crystal structures of MV-H complexed with SCR1 and SCR2 of CD46 or with the V domain of SLAM (Figure 1A; Santiago et al., 2010; Hashiguchi et al., 2011). By contrast, attachment proteins of NDV, hPIV3, and PIV5, NA of Flu and G proteins of NiV and HeV utilize the top pocket of the β-propeller structure for binding to their corresponding receptors or ligand (Figure 1B; Burmeister et al., 1992; Crennell et al., 2000; Lawrence et al., 2004; Yuan et al., 2005; Bowden et al., 2008; Xu et al., 2008).
Figure 1.
Structures of viral glycoproteins exhibiting the β-propeller fold unbound or bound to their receptors or ligand. (A) Side view of the MV-H monomer (rainbow colors) unbound (left) or bound to CD46 (middle) or SLAM (right). CD46 (purple) and SLAM (cyan) are indicated in a ribbon diagram. The top pocket of the MV-H monomer is masked by N-linked sugar. (B) Side views of Flu-NA bound to sialic acid (left), PIV5-HN bound to sialic acid (middle), and NiV-G bound to ephrin B2 (right). Sialic acid (white and red) is indicated as sphere models, whereas ephrin B2 (black) is shown in surface presentation. (C) Top views of two forms of MV-H tetramer with each monomer bound to SLAM (cyan). MV-H monomers A (green) and B (light green) form one dimer, whereas monomers C (black) and D (gray) form another.
Crystal structure of MV-H bound to CD46 reveals that β-sheet 4 (β4) and β5 of the β-propeller structure interact with SCR1 and SCR2 of CD46 (Figure 1A; Santiago et al., 2010). Key residues Tyr481 and Gly546 on MV-H make hydrogen bonds with the main chain oxygen group of Cys65 and the main chain amino group of Glu63 on CD46, respectively. Furthermore, a serine-to-glycine mutation at position 546 on MV-H (found in some CD46-using MV strains) likely increases flexibility of the β5s3–β5s4 region, which is favorable for CD46 binding. Pro38 and Pro39 on CD46 are sandwiched between β4 residues Leu464 and Leu500 and β5 residues Tyr541 and Tyr543 on MV-H, while Tyr67 on CD46 makes a hydrophobic interaction with Val451 on MV-H. The interaction between MV-H and CD46 is relatively week (Kd, 2.2 μM; Hashiguchi et al., 2007).
Crystal structure of MV-H in complex with SLAM shows that β4–6 and loop regions on the lateral surface of the MV-H β-propeller structure interact with GFCC′C″ region of the SLAM-V domain (Figure 1A; Hashiguchi et al., 2011). This interaction is also confirmed by functional assays, surface plasmon resonance and infectious assay in vitro. Salt bridges formed by residues Asp530 and Arg533 on MV-H and Glu123 on SLAM play a key role in stabilizing the MV-H–SLAM complex. An intermolecular β-sheet, comprised of residues Pro191-Arg195 (β6) of MV-H and residues Ser127-Phe131 (G sheet) of SLAM-V, further stabilizes the MV-H–SLAM complex. Mouse SLAM does not contain the MV-H-interacting key residues, including His61 and Arg130, and therefore does not act as a MV receptor (Ono et al., 2001). Although SLAM interacts with another SLAM through its V domain, the affinity of SLAM–SLAM interaction (Kd of ∼200 μM) is over 400-fold lower than that of the MV-H–SLAM interaction (Kd of 0.29 ∼ 0.43 μM; Hashiguchi et al., 2007). Therefore, upon MV infection, SLAM preferentially interacts with MV-H rather than adjacent SLAM.
Mutagenesis studies have mapped nectin-4 binding sites in a region partly shared by SLAM-binding sites (Leonard et al., 2008; Tahara et al., 2008). Amino acids in this region are highly conserved among morbilliviruses, whereas CD46 binding sites are not. Thus, it is likely that many morbilliviruses utilize this region of the H protein to infect immune and epithelial cells.
Like other paramyxovirus attachment proteins, MV-H forms a homodimer (or tetramer, see below). Interestingly, two monomers forming the MV-H dimer are highly tilted from each other, in contrast to other paramyxovirus attachment proteins (Hashiguchi et al., 2007). As a result, the receptor-binding sites on MV-H (located at the side of the β-propeller) are oriented upward in such a way that they are readily accessible to cellular receptors. This structural property and the inaccessibility of the top pocket due to N-linked sugar at position 215 account for the difference in receptor-binding sites between MV-H and other paramyxovirus attachment proteins.
MV-H Tetramer and Membrane Fusion Triggering
The mechanism by which receptor-binding leads to F protein-mediated membrane fusion is not well understood. However, crystal structures of MV-H–CD46 and MV-H–SLAM have also provided clues for this intriguing process (Santiago et al., 2010; Hashiguchi et al., 2011). No large structural change is observed in the MV-H monomer between receptor-free and receptor-bound forms. Furthermore, the relative orientation of two monomers in the MV-H dimer also remains essentially identical before and after receptor binding (r.m.s. deviation of 1.95 Å for 751 Ca atoms in MV-H–SLAM; r.m.s. deviation of 1.33 Å for 791 Ca atoms in MV-H–CD46).
Interestingly, crystal structures of the MV-H–SLAM complex reveal two different tetrameric configurations (dimer of dimers), form I and form II (Figure 1C; Hashiguchi et al., 2011). The tetrameric formation of MV-H is also detected by native PAGE and immunoblotting when the full-length MV-H is transiently expressed in cells. Recently reported crystal structures of NDV and PIV5 also reveal a four-helix bundle stalk (Bose et al., 2011; Yuan et al., 2011). MV-H monomers A and B form one dimer, while monomers C and D form another, and SLAM is bound to each MV-H monomer. Form I exhibits a conformation similar to previously reported head domains of NDV-HN, hPIV3-HN, PIV5-HN, and Flu-NA, although the relative position of monomers in the dimer units varies depending on the viral protein. In form II, a shift in the dimer of dimers occurs, enabling SLAM to bridge two neighboring monomers. Monomers A and C form the dimer–dimer interface in form I (contact area of 1.312 Å), whereas monomers B and D mainly form the dimer–dimer interface in form II (contact area of 2.099 Å). Based on these structural features, we propose the following fusion triggering mechanism. Upon the virus–cell interaction, the MV-H–SLAM complex in form I is formed. This structure renders the orientation of SLAM parallel rather than perpendicular in relation to the host membrane, and substantially reduces the distance between viral envelope and host membrane. This may prepare the environment suitable for subsequent conformational changes of MV-H and MV-F. The ensuing conformational shift of MV-H from form I to form II reorganizes the stalk region, allowing MV-F to refold and interact with the target cell membrane (Hashiguchi et al., 2011).
In a recent study, disulfide bonds were introduced at the protein interface to covalently anchor the two head domains in the MV-H dimer, leading to the block of its fusion-support activity (Navaratnarajah et al., 2011). The authors proposed that the two head domains in an MV-H dimer twist relative to each other upon receptor binding, which triggers membrane fusion. However, twisting of the head domains is not consistent with crystal structures of MV-H bound and unbound to receptors (Hashiguchi et al., 2007, 2011; Santiago et al., 2010). These introduced disulfide bonds might somehow prevent the proper formation of MV-H tetramer or its conformational shift upon receptor binding (Saphire and Oldstone, 2011). The similar experiment with the NDV-HN protein did not affect its fusion-support activity (Mahon et al., 2008).
Two models have been proposed for fusion triggering in paramyxoviruses. In one model, the attachment protein undergoes a conformational change upon receptor binding. This conformational change directly affects the F protein, causing its refolding which in turn drives membrane fusion. This model is consistent with the data for viruses using sialic acid as a receptor. In cells infected with hPIV3 and PIV5, the HN protein is associated with the F protein on the cell surface, but not in the endoplasmic reticulum (ER) (Paterson et al., 1997). Furthermore, the increased strength of the HN-F interaction enhances fusion activity. These results suggest that upon receptor binding, HN actively acts on the F protein and facilitates its refolding (Connolly et al., 2009). In the second model, the attachment protein serves as a clamp that stabilizes the F protein in its pre-fusion state. Receptor binding of the attachment protein releases the F protein from the clamp to facilitate its spontaneous conformational change. MV entry is consistent with this model. MV-H is already associated with MV-F within the ER (Plemper et al., 2001), and the conformational shift of the tetramer is likely to facilitate the release of MV-F from the heteromeric H–F oligomers. This release model is also supported by the data that a weaker interaction between MV-H and MV-F results in increased fusogenicity (Plemper et al., 2002; Corey and Iorio, 2009). A similar release model has also been proposed for NiV and HeV (Aguilar et al., 2006, 2007; Bishop et al., 2007). Thus, different paramyxoviruses may use different mechanisms of fusion triggering, although the overall cell entry mechanism may be similar among them.
Structural Insight into MV Serotype and Vaccine
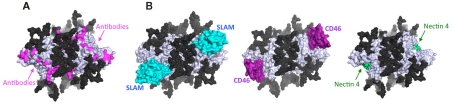
Crystal structure of MV-H sheds light on why measles vaccine has been successful for a long time (Hashiguchi et al., 2007, 2011; Ruigrok and Gerlier, 2007). The structure suggests that N-linked sugars (at positions 168, 187, 200, and 215) on MV-H cover a considerable portion of its surface, only exposing a small area for receptor and antibody bindings (Figures 2A,B). Indeed, crystal structures of MV-H–SLAM and MV-H–CD46 complexes indicate that both receptors target this exposed area. Mutagenesis studies have also revealed that nectin-4 binds to this region as well. Furthermore, a majority of MV-H monoclonal antibodies are mapped onto this exposed receptor-binding area, indicating that this area acts as the epitope “hot spot.” This overlap of binding sites for receptors and neutralizing antibodies explains why measles vaccine, derived from a single strain, remains effective against all 23 distinct MV genotypes. A functional importance (receptor binding) likely exerts a restrain that renders this region highly unfavorable to mutation. As a result, MV still occurs in only a single serotype (Hashiguchi et al., 2011). Interestingly, this receptor-binding area is highly conserved not only among MV strains but also among morbilliviruses, suggesting that this region could serve as a template for a universal vaccine targeting all morbilliviruses (Hashiguchi et al., 2007). The human immunodeficiency virus glycoprotein lacks the similarly conserved region to be targeted by antibodies, which has hampered the vaccine development.
Figure 2.
Structural basis of the effectiveness of measles vaccine. (A) Epitopes of anti-MV-H monoclonal antibodies (red) on the MV-H homodimer (blue white) with potential N-linked sugars (mesh, black) as viewed downwards from the top. (B) Receptor-binding sites on MV-H. Left, crystal structure of MV-H–SLAM (SLAM shown in cyan). Middle, crystal structure of MV-H–CD46 (CD46 in purple). Right, putative nectin-4 binding sites (green), based on site-directed mutagenesis of MV-H.
Perspective
Recent advances in our understanding of MV-H structures, combined with functional studies, have provided new insights into the mechanism of MV entry. Although the receptor-binding mode of MV-H is different from that of other paramyxovirus HN and G proteins, the overall entry process may be common among paramyxoviruses. MV-H forms tetramer (dimer of dimers), like other HN and G proteins, and we propose a model for fusion triggering, based on the presence of two forms of MV-H tetramer. However, further structural studies of MV-H and MV-F are required to completely understand the whole process of MV entry.
Crystal structures have also illuminated how a sugar-uncovered, exposed area of MV-H is targeted by receptors as well as by neutralizing antibodies. This nicely explains why only a single serotype occurs for MV and why measles vaccine, derived from one strain, has been effective for a long time. It is hoped that better understanding of the MV entry process will not only contribute to our measles eradication effort worldwide, but also allow us to develop effective vaccines for other challenging pathogens.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank A. P. P. Zhang for discussion and reviewing the manuscript. Our work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology and the Ministry of Health, Labor and Welfare of Japan. Takao Hashiguchi is supported by the Japan Society for the Promotion of Science Research Fellowship for Young Scientists.
References
- Aguilar H. C., Matreyek K. A., Choi D. Y., Filone C. M., Young S., Lee B. (2007). Polybasic KKR motif in the cytoplasmic tail of Nipah virus fusion protein modulates membrane fusion by inside-out signaling. J. Virol. 81, 4520–4532 10.1128/JVI.02205-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar H. C., Matreyek K. A., Filone C. M., Hashimi S. T., Levroney E. L., Negrete O. A., Bertolotti-Ciarlet A., Choi D. Y., Mchardy I., Fulcher J. A., Su S. V., Wolf M. C., Kohatsu L., Baum L. G., Lee B. (2006). N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J. Virol. 80, 4878–4889 10.1128/JVI.80.10.4878-4889.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz R., Firsching R., Rima B., Ter Meulen V., Schneider-Schaulies J. (1998). Differential receptor usage by measles virus strains. J. Gen. Virol. 79(Pt 5), 1015–1025 [DOI] [PubMed] [Google Scholar]
- Bellini W. J., Rota P. A. (2011). Biological feasibility of measles eradication. Virus Res. 162, 72–79 10.1016/j.virusres.2011.09.023 [DOI] [PubMed] [Google Scholar]
- Bishop K. A., Stantchev T. S., Hickey A. C., Khetawat D., Bossart K. N., Krasnoperov V., Gill P., Feng Y. R., Wang L., Eaton B. T., Wang L. F., Broder C. C. (2007). Identification of Hendra virus G glycoprotein residues that are critical for receptor binding. J. Virol. 81, 5893–5901 10.1128/JVI.02022-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaparte M. I., Dimitrov A. S., Bossart K. N., Crameri G., Mungall B. A., Bishop K. A., Choudhry V., Dimitrov D. S., Wang L. F., Eaton B. T., Broder C. C. (2005). Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. U.S.A. 102, 10652–10657 10.1073/pnas.0504887102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose S., Welch B. D., Kors C. A., Yuan P., Jardetzky T. S., Lamb R. A. (2011). Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J. Virol. 85, 12855–12866 10.1128/JVI.06350-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden T. A., Aricescu A. R., Gilbert R. J., Grimes J. M., Jones E. Y., Stuart D. I. (2008). Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat. Struct. Mol. Biol. 15, 567–572 10.1038/nsmb.1435 [DOI] [PubMed] [Google Scholar]
- Bowden T. A., Crispin M., Jones E. Y., Stuart D. I. (2010). Shared paramyxoviral glycoprotein architecture is adapted for diverse attachment strategies. Biochem. Soc. Trans. 38, 1349–1355 10.1042/BST0381349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister W. P., Ruigrok R. W., Cusack S. (1992). The 2.2 A resolution crystal structure of influenza B neuraminidase and its complex with sialic acid. EMBO J. 11, 49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocks B. G., Chang C.-C. J., Carballido J. M., Yssel H., De Vries J. E., Aversa G. (1995). A novel receptor involved in T-cell activation. Nature 376, 260–263 10.1038/376260a0 [DOI] [PubMed] [Google Scholar]
- Cole J. L., Housley G. A., Jr., Dykman T. R., Macdermott R. P., Atkinson J. P. (1985). Identification of an additional class of C3-binding membrane proteins of human peripheral blood leukocytes and cell lines. Proc. Natl. Acad. Sci. U.S.A. 82, 859–863 10.1073/pnas.82.3.859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colf L. A., Juo Z. S., Garcia K. C. (2007). Structure of the measles virus hemagglutinin. Nat. Struct. Mol. Biol. 14, 1227–1228 10.1038/nsmb1342 [DOI] [PubMed] [Google Scholar]
- Connolly S. A., Leser G. P., Jardetzky T. S., Lamb R. A. (2009). Bimolecular complementation of paramyxovirus fusion and hemagglutinin-neuraminidase proteins enhances fusion: implications for the mechanism of fusion triggering. J. Virol. 83, 10857–10868 10.1128/JVI.01191-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey E. A., Iorio R. M. (2009). Measles virus attachment proteins with impaired ability to bind CD46 interact more efficiently with the homologous fusion protein. Virology 383, 1–5 10.1016/j.virol.2008.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crennell S., Takimoto T., Portner A., Taylor G. (2000). Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 7, 1068–1074 10.1038/81002 [DOI] [PubMed] [Google Scholar]
- Dorig R. E., Marcil A., Chopra A., Richardson C. D. (1993). The human CD46 molecule is a receptor for measles virus (Edmonston strain). Cell 75, 295–305 10.1016/0092-8674(93)80071-L [DOI] [PubMed] [Google Scholar]
- Griffin D. E. (2007). “Measles virus,” in Fields Virology, 5th Edn, eds Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E. (Philadelphia: Lippincott Williams & Wilkins; ), 1551–1585 [Google Scholar]
- Hashiguchi T., Kajikawa M., Maita N., Takeda M., Kuroki K., Sasaki K., Kohda D., Yanagi Y., Maenaka K. (2007). Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc. Natl. Acad. Sci. U.S.A. 104, 19535–19540 10.1073/pnas.0707830104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi T., Ose T., Kubota M., Maita N., Kamishikiryo J., Maenaka K., Yanagi Y. (2011). Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat. Struct. Mol. Biol. 18, 135–141 10.1038/nsmb.1969 [DOI] [PubMed] [Google Scholar]
- Iorio R. M., Mahon P. J. (2008). Paramyxoviruses: different receptors – different mechanisms of fusion. Trends Microbiol. 16, 135–137 10.1016/j.tim.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz S. L. (2009). John F. Enders and measles virus vaccine – a reminiscence. Curr. Top. Microbiol. Immunol. 329, 3–11 10.1007/978-3-540-70523-9_1 [DOI] [PubMed] [Google Scholar]
- Kemper C., Atkinson J. P. (2009). Measles virus and CD46. Curr. Top. Microbiol. Immunol. 329, 31–57 10.1007/978-3-540-70523-9_3 [DOI] [PubMed] [Google Scholar]
- Kobune F., Sakata H., Sugiura A. (1990). Marmoset lymphoblastoid cells as a sensitive host for isolation of measles virus. J. Virol. 64, 700–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R. A., Parks G. D. (2007). “Paramyxoviridae: the viruses and their replication,” in Fields Virology, 5th Edn, eds Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E. (Philadelphia: Lippincott Williams & Wilkins; ), 1449–1496 [Google Scholar]
- Lawrence M. C., Borg N. A., Streltsov V. A., Pilling P. A., Epa V. C., Varghese J. N., Mckimm-Breschkin J. L., Colman P. M. (2004). Structure of the haemagglutinin-neuraminidase from human parainfluenza virus type III. J. Mol. Biol. 335, 1343–1357 10.1016/j.jmb.2003.11.032 [DOI] [PubMed] [Google Scholar]
- Lecouturier V., Fayolle J., Caballero M., Carabana J., Celma M. L., Fernandez-Munoz R., Wild T. F., Buckland R. (1996). Identification of two amino acids in the hemagglutinin glycoprotein of measles virus (MV) that govern hemadsorption, HeLa cell fusion, and CD46 downregulation: phenotypic markers that differentiate vaccine and wild-type MV strains. J. Virol. 70, 4200–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard V. H., Sinn P. L., Hodge G., Miest T., Devaux P., Oezguen N., Braun W., Mccray P. B., Jr., Mcchesney M. B., Cattaneo R. (2008). Measles virus blind to its epithelial cell receptor remains virulent in rhesus monkeys but cannot cross the airway epithelium and is not shed. J. Clin. Invest. 118, 2448–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon P. J., Mirza A. M., Musich T. A., Iorio R. M. (2008). Engineered intermonomeric disulfide bonds in the globular domain of Newcastle disease virus hemagglutinin-neuraminidase protein: implications for the mechanism of fusion promotion. J. Virol. 82, 10386–10396 10.1128/JVI.00581-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlebach M. D., Mateo M., Sinn P. L., Prufer S., Uhlig K. M., Leonard V. H., Navaratnarajah C. K., Frenzke M., Wong X. X., Sawatsky B., Ramachandran S., Mccray P. B., Cichutek K., Von Messling V., Lopez M., Cattaneo R. (2011). Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature. 10.1038/nature10639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naniche D., Varior-Krishnan G., Cervoni F., Wild T. F., Rossi B., Rabourdin-Combe C., Gerlier D. (1993). Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J. Virol. 67, 6025–6032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaratnarajah C. K., Oezguen N., Rupp L., Kay L., Leonard V. H., Braun W., Cattaneo R. (2011). The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nat. Struct. Mol. Biol. 18, 128–134 10.1038/nsmb.1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrete O. A., Levroney E. L., Aguilar H. C., Bertolotti-Ciarlet A., Nazarian R., Tajyar S., Lee B. (2005). EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 436, 401–405 [DOI] [PubMed] [Google Scholar]
- Negrete O. A., Wolf M. C., Aguilar H. C., Enterlein S., Wang W., Muhlberger E., Su S. V., Bertolotti-Ciarlet A., Flick R., Lee B. (2006). Two key residues in ephrinB3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog. 2, e7. 10.1371/journal.ppat.0020007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyce R. S., Bondre D. G., Ha M. N., Lin L. T., Sisson G., Tsao M. S., Richardson C. D. (2011). Tumor cell marker PVRL4 (Nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog. 7, e1002240. 10.1371/journal.ppat.1002240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono N., Tatsuo H., Tanaka K., Minagawa H., Yanagi Y. (2001). V domain of human SLAM (CDw150) is essential for its function as a measles virus receptor. J. Virol. 75, 1594–1600 10.1128/JVI.75.4.1594-1600.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson R. G., Johnson M. L., Lamb R. A. (1997). Paramyxovirus fusion (F) protein and hemagglutinin-neuraminidase (HN) protein interactions: intracellular retention of F and HN does not affect transport of the homotypic HN or F protein. Virology 237, 1–9 10.1006/viro.1997.8759 [DOI] [PubMed] [Google Scholar]
- Persson B. D., Schmitz N. B., Santiago C., Zocher G., Larvie M., Scheu U., Casasnovas J. M., Stehle T. (2010). Structure of the extracellular portion of CD46 provides insights into its interactions with complement proteins and pathogens. PLoS Pathog. 6, e1001122. 10.1371/journal.ppat.1001122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper R. K., Brindley M. A., Iorio R. M. (2011). Structural and mechanistic studies of measles virus illuminate paramyxovirus entry. PLoS Pathog. 7, e1002058. 10.1371/journal.ppat.1002058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper R. K., Hammond A. L., Cattaneo R. (2001). Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J. Biol. Chem. 276, 44239–44246 10.1074/jbc.M105967200 [DOI] [PubMed] [Google Scholar]
- Plemper R. K., Hammond A. L., Gerlier D., Fielding A. K., Cattaneo R. (2002). Strength of envelope protein interaction modulates cytopathicity of measles virus. J. Virol. 76, 5051–5061 10.1128/JVI.76.10.5051-5061.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond N., Fabre S., Lecocq E., Adelaide J., Dubreuil P., Lopez M. (2001). Nectin4/PRR4, a new afadin-associated member of the nectin family that trans-interacts with nectin1/PRR1 through V domain interaction. J. Biol. Chem. 276, 43205–43215 10.1074/jbc.M103810200 [DOI] [PubMed] [Google Scholar]
- Ruigrok R. W., Gerlier D. (2007). Structure of the measles virus H glycoprotein sheds light on an efficient vaccine. Proc. Natl. Acad. Sci. U.S.A. 104, 20639–20640 10.1073/pnas.0709995105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago C., Celma M. L., Stehle T., Casasnovas J. M. (2010). Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat. Struct. Mol. Biol. 17, 124–129 10.1038/nsmb.1726 [DOI] [PubMed] [Google Scholar]
- Saphire E. O., Oldstone M. B. (2011). Measles virus fusion shifts into gear. Nat. Struct. Mol. Biol. 18, 115–116 10.1038/nsmb0211-115 [DOI] [PubMed] [Google Scholar]
- Schneider-Schaulies J., Schnorr J.-J., Brinckmann U., Dunster L. M., Baczko K., Liebert U. G., Schneider-Schaulies S., Ter Meulen V. (1995). Receptor usage and differential downregulation of CD46 by measles virus wild-type and vaccine strains. Proc. Natl. Acad. Sci. U.S.A. 92, 3943–3947 10.1073/pnas.92.9.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzberg P. L., Mueller K. L., Qi H., Cannons J. L. (2009). SLAM receptors and SAP influence lymphocyte interactions, development and function. Nat. Rev. Immunol. 9, 39–46 10.1038/nri2456 [DOI] [PubMed] [Google Scholar]
- Smith E. C., Popa A., Chang A., Masante C., Dutch R. E. (2009). Viral entry mechanisms: the increasing diversity of paramyxovirus entry. FEBS J. 276, 7217–7227 10.1111/j.1742-4658.2009.07401.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara M., Takeda M., Shirogane Y., Hashiguchi T., Ohno S., Yanagi Y. (2008). Measles virus infects both polarized epithelial and immune cells by using distinctive receptor-binding sites on its hemagglutinin. J. Virol. 82, 4630–4637 10.1128/JVI.02691-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M., Tahara M., Hashiguchi T., Sato T. A., Jinnouchi F., Ueki S., Ohno S., Yanagi Y. (2007). A human lung carcinoma cell line supports efficient measles virus growth and syncytium formation via a SLAM- and CD46-independent mechanism. J. Virol. 81, 12091–12096 10.1128/JVI.01264-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Xie M., Yanagi Y. (1998). The hemagglutinin of recent measles virus isolates induces cell fusion in a marmoset cell line, but not in other CD46-positive human and monkey cell lines, when expressed together with the F protein. Arch. Virol. 143, 213–225 10.1007/s007050050281 [DOI] [PubMed] [Google Scholar]
- Tatsuo H., Ono N., Tanaka K., Yanagi Y. (2000). SLAM (CDw150) is a cellular receptor for measles virus. Nature 406, 893–897 10.1038/35022579 [DOI] [PubMed] [Google Scholar]
- Xu K., Rajashankar K. R., Chan Y. P., Himanen J. P., Broder C. C., Nikolov D. B. (2008). Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc. Natl. Acad. Sci. U.S.A. 105, 9953–9958 10.1073/pnas.0707082105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi Y., Takeda M., Ohno S., Hashiguchi T. (2009). Measles virus receptors. Curr. Top. Microbiol. Immunol. 329, 13–30 10.1007/978-3-540-70523-9_2 [DOI] [PubMed] [Google Scholar]
- Yuan P., Swanson K. A., Leser G. P., Paterson R. G., Lamb R. A., Jardetzky T. S. (2011). Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc. Natl. Acad. Sci. U.S.A. 108, 14920–14925 10.1073/pnas.1018669108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P., Thompson T. B., Wurzburg B. A., Paterson R. G., Lamb R. A., Jardetzky T. S. (2005). Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 13, 803–815 10.1016/j.str.2005.02.019 [DOI] [PubMed] [Google Scholar]