Abstract
T cell activation leads to a dramatic demand for energy and biosynthetic precursors that is met by increased glucose and glutamine metabolism. In this issue of Immunity, Wang et al. (2011) show that Myc is essential in T cell metabolic reprogramming.
Adaptive immunity requires that lymphocytes clonally expand to combat invading pathogens. Cell division, however, must be preceded by cell growth. This fundamental necessity for growth has propelled efforts to understand basic mechanisms to generate essential lipids, nucleic acids, and proteins. One field that has approached this challenge head-on is cancer biology, in which it has been known for nearly a century that cellular transformation leads to increased glucose consumption and lactate secretion, a metabolic program termed aerobic glycolysis. The key to why cancer would promote an energy inefficient glycolytic metabolism has been the realization that it's not just the ATP that matters. Rather, aerobic glycolysis is an ideal metabolic program to provide biosynthetic substrates. In this issue of Immunity, Wang et al. (2011) address the role of the oncogenic transcription factor Myc in T cell activation to show it plays an essential role in the metabolic switch that supports T cell growth.
If proliferation leads to a sharp increase in metabolic demands, stimulated T cells are as demanding as any mammalian cell. After an initial lag, activated T cells can cycle extremely quickly, with a doubling of mass and cell division as rapidly as every 4 to 6 hr. To support this robust growth, activated T cells quickly increase glucose uptake and glycolysis as well as glutamine metabolism and become, in metabolic respects, very similar to cancer cells. This metabolic switch is essential for T cell proliferation and function, given that inhibition of glucose or glutamine metabolism can prevent growth and division as well as selectively impair some cytokine production of activated T cells (Jacobs et al., 2008; Wang et al., 2011). Conversely, increased glucose uptake by transgenic expression of the glucose transporter Glut1 can strengthen T cell responses (Jacobs et al., 2008). These findings suggest that targeting metabolism may be a new strategy to modulate immunity. A key question that remains is how are these metabolic programs regulated?
Evidence is now emerging that the very same mechanisms that drive aerobic glycolysis in cancer are responsible for the metabolic reprogramming of activated T cells. This is perhaps not surprising because many of the same signaling pathways that drive oncogenesis and the metabolic phenotype of tumors are activated and essential upon lymphocyte stimulation. The best example is the post-translational regulation of aerobic glycolysis by the Akt and mTOR pathway. This pathway is a promising target to suppress cancer cell growth and is also critical for T cell activation and effector function. The transcriptional regulation of T cell metabolism, however, has been largely unclear. Many proteins could fill this role, but Myc has been a prime candidate to transcriptionally promote T cell glycolysis given its clear role in the metabolic program of cancer cells (Dang et al., 2009) as well as T cell development and function (Douglas et al., 2001).
Starting with the expression of a variety of genes involved in glucose and glutamine metabolism, Wang et al. undertook a computational approach to identify transcription factors that are likely to drive metabolic reprogramming in T cell activation. Although a number of interesting transcription factors were suggested, two of the top candidates were Myc and hypoxia inducible factor-1α (HIF-1α). Using Myc- and HIF-1α-deficient T cells, the authors then showed by measurement of gene expression, metabolite levels, and flux through metabolic pathways that acute loss of Myc, but surprisingly not HIF-1α, dramatically suppressed metabolic reprogramming in the initial day after T cell activation that leads to the onset of rapid cell divisions.
Myc has a large number of potential gene targets and it could be argued that failure to upregulate expression of metabolic genes may not be entirely due to failure of specific Myc association and regulation of these genes. Rather, the critical role for Myc in another process, such as cell cycle regulation or ribosome biogenesis, may lead to a feedback to control the expression of metabolic genes. Although direct binding of Myc to metabolic targets in T cells remains to be formally established, Myc is well known to directly bind and regulate many of these same metabolic genes in other settings. A further consideration is that T cells do not enter the cell cycle until long after initial stimulation. Changes in T cell metabolism were observed, however, within 3 to 10 hr of stimulation. Thus, the metabolic effects of Myc, direct or otherwise, were rapid and appear to be cell cycle independent.
The metabolic pathways in particular that stood out as Myc dependent were glycolysis, glutamine oxidation, and polyamine synthesis (Figure 1). Glucose and glutamine metabolism would be anticipated on the basis of the role of Myc in these pathways in cancer cells (Dang et al., 2009). The Myc-dependent regulation of polyamine synthesis, however, was more unexpected and may indicate a previously unappreciated regulation of these important metabolites in T cells. Indeed, ornithine decarboxylase, which can promote polyamine synthesis, has been shown to be a Myc target and is essential for lymphomagenesis (Nilsson et al., 2005). Polyamine metabolism and molecular function are not well understood, and this may be a case in which T cell metabolism research may inform the cancer cell metabolism field.
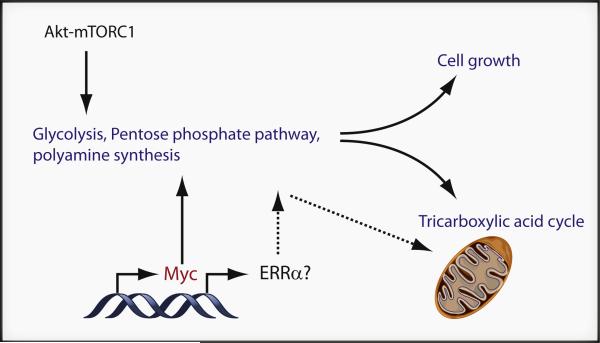
Figure 1. Regulatory Mechanisms that Control T Cell Metabolic Reprogramming in Activation.
T cell activation leads to increased glucose, glutamine, and mitochondrial oxidative metabolism that are essential for cell growth and proliferation. Data presented by Wang et al. (2011) shows Myc plays a key role in upregulation of glucose, glutamine, and polyamine metabolism. In addition, the Akt-mTOR pathway promotes posttranslational events in aerobic glycolysis, and ERRα may play a role in regulating mitochondrial metabolism in T cell activation.
Equally intriguing as the pathways that were Myc dependent were the pathways that were neither Myc nor HIF-1α dependent. In particular, mitochondrial pathways were not strongly affected by Myc deficiency, given that lipid oxidation decreased and oxygen consumption increased in T cell activation regardless of Myc or HIF-1α status. Clearly, T cell metabolism is regulated in a somewhat piecemeal fashion and additional factors also contribute to regulate specific arms of the metabolic program. For example, the nuclear hormone receptor estrogen-related receptor-α (ERRα) can also play a role in T cell metabolism and may mediate the mitochondrial arm of T cell metabolic reprogramming (Michalek et al., 2011b). HIF-1α may also be involved, albeit at later times and in specific T cell subsets, such as Th17 cells (Shi et al., 2011). The cumulative action of these multiple transcription factors, together with posttranslational regulation, provides flexibility to coordinate T cell metabolic reprogramming in distinct settings (Figure 1).
Observations that T cell metabolic programming occurs through a set of regulatory events open important new doors to understand fundamental T cell biology. A challenge, however, is to extend beyond correlation of cellular metabolic phenotype and functional outcome. As this field grows, therefore, it is important to directly manipulate the specific metabolic pathways in question to determine their role and sufficiency to impact T cell activity. This is a daunting prospect, however, because a coordinated action of multiple metabolic pathways is likely to be essential and replacement of a single metabolite or metabolic gene may simply be insufficient. Nevertheless, Wang et al. do address this challenge and show that addition of polyamines can partially rescue the limited cell division of T cells upon glutamine deficiency. Specific metabolic pathways that are Myc dependent, therefore, are key components of T cell growth and proliferation.
One additional unresolved issue in this study and in the field of T cell metabolism in general is how metabolic changes occur and regulate T cell function in vivo. Wang et al. (2011) show that the inhibition of Myc clearly prevented T cell proliferation in vivo. Likewise, inhibition of glucose or glutamine metabolism by whole-animal treatment with glycolysis or glutaminolysis inhibitors also prevented T cell proliferation in vivo. Given that antigen-presenting and other cells also utilize these pathways, however, it is unclear to what extent T cell metabolism itself is limiting and what pathways may be most important in vivo. T cells isolated during an acute graft-versus-host reaction, for example, appear to be highly sensitive to inhibition of mitochondrial rather than glucose metabolism (Gatza et al., 2011). The in vitro environment has been highly optimized for maximal cell growth and the different array of nutrients and stimuli in vivo may have a critical effect on T cell metabolism and, therefore, how T cells respond to inhibition of metabolic pathways.
In the end, much as the field of cancer metabolism has a goal of targeting aerobic glycolysis to control or eliminate cancer, targeting the metabolic phenotype of activated T cells may provide a new approach to immunosuppression. A key consideration in these efforts that goes beyond investigation of the initial steps of T cell activation described by Wang et al. is the differentiation of T cells into specific effector, regulatory, or memory cell subsets. Importantly, each CD4 T cell effector subset appears to have a distinct metabolic character from regulatory T cells (Michalek et al., 2011a; Shi et al., 2011) and memory CD8 T cells (Pearce et al., 2009), given that the latter primarily utilize lipids rather than glucose as a metabolic fuel. Thus, targeting the aerobic glycolysis of T cells has the potential to selectively impede effector, but not regulatory or memory T cells. Understanding the regulation of metabolism in T cell activa tion has great potential to modulate immunity and the clear identification of Myc in this process is a strong step forward.
REFERENCES
- Dang CV, Le A, Gao P. Clin. Cancer Res. 2009;15:6479–6483. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas NC, Jacobs H, Bothwell AL, Hayday AC. Nat. Immunol. 2001;2:307–315. doi: 10.1038/86308. [DOI] [PubMed] [Google Scholar]
- Gatza E, Wahl DR, Opipari AW, Sundberg TB, Reddy P, Liu C, Glick GD, Ferrara JL. Sci. Transl. Med. 2011;3:67ra68. doi: 10.1126/scitranslmed.3001975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SR, Herman CE, Maciver NJ, Wofford JA, Wieman HL, Hammen JJ, Rathmell JC. J. Immunol. 2008;180:4476–4486. doi: 10.4049/jimmunol.180.7.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, Sullivan SA, Nichols AG, Rathmell JC. J. Immunol. 2011a;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek RD, Gerriets VA, Nichols AG, Inoue M, Kazmin D, Chang CY, Dwyer MA, Nelson ER, Pollizzi KN, Ilkayeva O, et al. Proc. Natl. Acad. Sci. USA. 2011b;108:18348–18353. doi: 10.1073/pnas.1108856108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson JA, Keller UB, Baudino TA, Yang C, Norton S, Old JA, Nilsson LM, Neale G, Kramer DL, Porter CW, Cleveland JL. Cancer Cell. 2005;7:433–444. doi: 10.1016/j.ccr.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LZ, Wang R, Huang G, Vogel P, Neale G, Green DR, Chi H. J. Exp. Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, Green DR. Immunity. 2011;35(this issue):871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]