Abstract
Pulmonary function measures reflect respiratory health and predict mortality, and are used in the diagnosis of chronic obstructive pulmonary disease (COPD). We tested genome-wide association with the forced expiratory volume in 1 second (FEV1) and the ratio of FEV1 to forced vital capacity (FVC) in 48,201 individuals of European ancestry, with follow-up of top associations in up to an additional 46,411 individuals. We identified new regions showing association (combined P<5×10−8) with pulmonary function, in or near MFAP2, TGFB2, HDAC4, RARB, MECOM (EVI1), SPATA9, ARMC2, NCR3, ZKSCAN3, CDC123, C10orf11, LRP1, CCDC38, MMP15, CFDP1, and KCNE2. Identification of these 16 new loci may provide insight into the molecular mechanisms regulating pulmonary function and into molecular targets for future therapy to alleviate reduced lung function.
Introduction
Pulmonary function, reliably measurable by spirometry, is a heritable trait reflecting the physiological state of the airways and lungs1. Pulmonary function measures are important predictors of population morbidity and mortality2-4, and are used in the diagnosis of chronic obstructive pulmonary disease (COPD), which ranks among the leading causes of death in developed and developing countries5,6. A reduced ratio of forced expiratory volume in 1 second (FEV1) to forced vital capacity (FVC) is used to define airway obstruction, and a reduced FEV1 is used to grade the severity of airway obstruction7.
Recently, two large genome-wide association studies (GWAS), each comprising discovery sets of more than 20,000 individuals of European ancestry identified novel loci for lung function8,9. Recognizing the need for larger datasets to increase the power to detect loci of individually modest effect size, we conducted a meta-analysis of 23 lung function GWAS comprising a total of 48,201 individuals of European ancestry (Stage 1) and followed-up potentially novel loci in 17 further studies comprising up to 46,411 individuals (Stage 2). We identified 16 additional novel loci for lung function, and provided evidence corroborating association of loci previously associated with lung function8-11. Our findings implicate a number of different mechanisms underlying regulation of lung function and highlight loci shared with complex traits and diseases, including height, lung cancer, and myocardial infarction.
Results
Genome-wide analysis (stage 1)
Meta-analyses for cross-sectional lung function measures were undertaken for approximately 2.5 million genotyped or imputed SNPs across 23 studies with a combined sample size of 48,201 adult individuals of European ancestry. Characteristics of the cohort participants and the genotyping are shown in Supplementary Tables 1A and 1B. FEV1 and FEV1/FVC were adjusted for ancestry principal components, age, age2, sex, and height as covariates. Association testing of the inverse-normal transformed residuals for FEV1 and FEV1/FVC assumed an additive genetic model and was stratified by ever-smoking (versus never-smoking) status. Meta-analyses of the smoking strata within study, and the study-specific results, were undertaken using inverse variance weighting (the inverse of the standard error squared was used as the weight). We applied genomic control twice at study level (to each smoking stratum separately and to the study level pooled estimates) and also at meta-analysis level to avoid inflation of test statistics due to cryptic population structure or relatedness (see Supplementary Table 1A for study level estimates). Our application of genomic control at the three stages is likely to be overly conservative because it has recently been shown that in large meta-analyses, test statistics are expected to be elevated under polygenic inheritance even when there is no population structure12. Test statistic inflation (λGC) prior to applying genomic control at meta-analysis level was 1.12 for FEV1 and 1.09 for FEV1/FVC. Genomic inflation estimates increase with sample size, as has been shown for other traits13-15; standardised estimates to a sample of 1000 individuals (λGC_1000) were 1.002 for FEV1 and 1.002 for FEV1/FVC. Plots of meta-analysis P-values for FEV1 and FEV1/FVC against a uniform distribution of P values expected under the null hypothesis showed deviations which were attenuated, but persisted, after removal of SNPs in loci reported previously, consistent with additional loci being associated with lung function (Supplementary Figure 1A).
Follow-up analysis (stage 2)
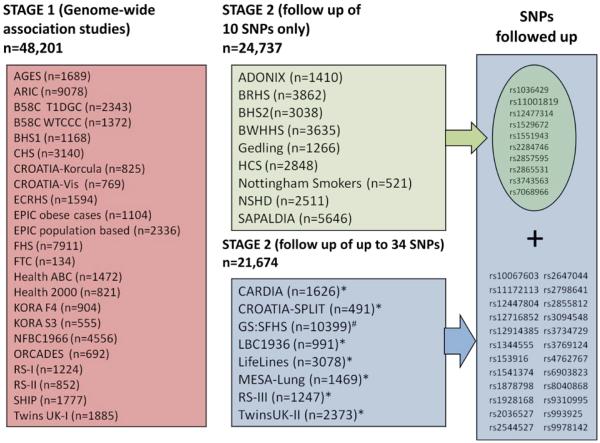
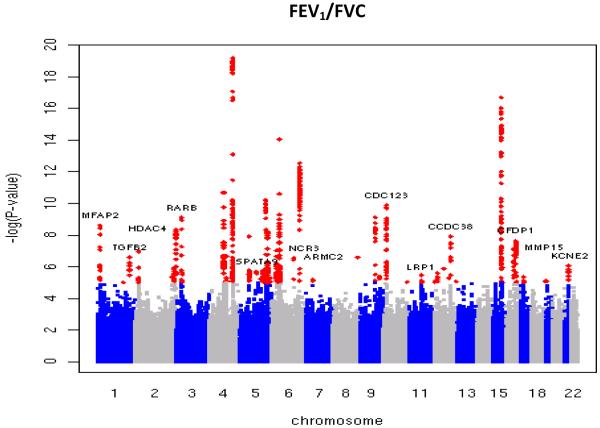
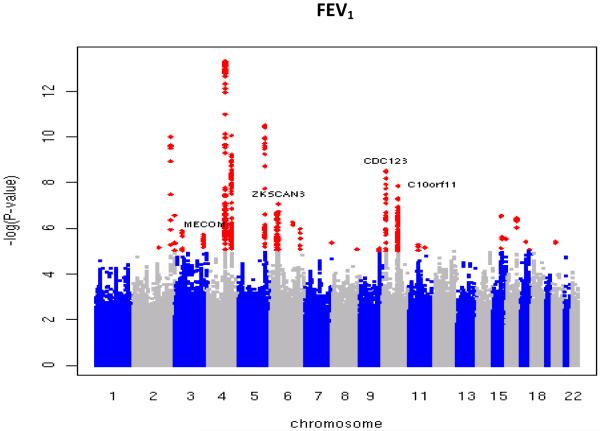
Twenty-nine new loci showing evidence of association with lung function (P<3×10−6) in Stage 1 were followed up in Stage 2 by utilizing in silico data from seven studies, and by undertaking additional genotyping in 10 studies for the 10 highest-ranked SNPs (Figure 1). Full details of the SNP selection are given in the Online Methods. Inverse variance weighting meta-analysis was performed across Stages 1 and 2, and two-sided p-values were obtained for the pooled estimates. Sixteen new loci reached genome-wide significance (P<5×10−8) and showed consistent direction of effects in both stages, comprising 12 new loci for FEV1/FVC, 3 new loci for FEV1, and one new locus reaching genome-wide significance for both traits (Figure 2, Table 1). To assess the heterogeneity across studies included in Stage 1 and Stage 2, Chi-square tests were undertaken for all 16 SNPs; and none of them was statistically significant after applying a Bonferroni correction for 16 tests. The sentinel SNPs at these loci were in or near the genes MFAP2 (1p36.13), TGFB2/LYPLAL1 (1q41), HDAC4/FLJ43879 (2q37.3), RARB (3p24.2), MECOM (EVI1) (3q26.2), SPATA9/RHOBTB3 (5q15), ARMC2 (6q21), NCR3/AIF1 (6p21.33), ZKSCAN3 (6p22.1), CDC123 (10p13), C10orf11 (10q22.3), LRP1 (12q13.3), CCDC38 (12q22), MMP15 (16q13), CFDP1 (16q23.1) and KCNE2/C21orf82 (21q22.11) (Supplementary Figures 1B and 1C). The strongest signals in AGER (rs2070600)8,9 and two of the novel signals (rs6903823 in ZKSCAN3 and rs2857595, upstream of NCR3) lie within a~3.8Mb interval at 6p21.32-22.1 which is characterised by long-range linkage disequilibrium. Nevertheless, the leading SNPs in these regions which are within the major histocompatibility complex (MHC) were statistically independent (Supplementary Note).
Figure 1.
Study design. A total of 34 SNPs showing novel evidence of association (P<3×10−6) with FEV1 and/or FEV1/FVC in a meta-analysis of the Stage 1 studies were followed up in Stage 2. Studies with a combined total of 24,737 individuals undertook genotyping and association testing of the top 10 SNPs. Seven studies (*) with a combined total of 11,275 individuals had genome-wide association data and provided results for up to 34 SNPs. #GS:SFHS undertook genotyping on a 32-SNP multiplex genotyping platform and so included the 32 top ranking SNPs (including proxies and both SNPs from regions which showed association with both FEV1 and FEV1/FVC). This assay failed for one SNP (rs3769124) which was subsequently replaced with the 33rd SNP (rs4762767). SNP rs2284746 was excluded due to poor clustering. Although rs3743563 was chosen as proxy for rs12447804 which had N effective <80% in the Stage 1 meta-analysis, BHS2 were unable to genotype rs3743563 and so undertook genotyping for rs12447804 instead. See Table 1 for definitions of abbreviations.
Figure 2.
Manhattan plots of association results for (a) FEV1/FVC and (b) FEV1. Manhattan plots ordered by chromosome position. SNPs for which −log10P>5 are indicated in red. Novel regions which reached genome-wide significance after Stage 1 + Stage 2 are labelled.
Table 1.
Loci associated with lung function. Shown are FEV1 and FEV1/FVC results for the leading SNPs, ordered by chromosome and position, for each independent locus associated (P <5×10−8) with FEV1 or FEV1/FVC in a joint analysis of up to 94,612 individuals of European ancestry from the SpiroMeta-CHARGE GWAS (Stage 1) and follow-up (Stage 2). Two-sided p-values are given for Stage 1, Stage 2 and the joint meta-analysis of all stages. P-values reaching genome-wide significance (P<5×10−8) in the joint meta-analysis of all stages are indicated in bold. SNPs reaching independent replication in Stage 2 (P=0.05/34 = 1.47×10−3) are indicated with their Stage 2 p-value in bold. The sample sizes (N) shown are the effective sample sizes. Effective sample size within each study is the product of sample size and imputation quality metric. Joint meta-analysis includes data from Stage 1 and Stage 2. Beta values reflect effect-size estimates on an inverse-normal transformed scale after adjustments for age, age2, sex, height and ancestry principal components. The estimated proportion of the variance explained by each SNP can be found in Supplementary Table 6.
| Chr. | SNP_ID(NCBI36 position), function | Coded allele freq. |
Measure | Stage 1 | Stage 2 | Joint meta-analysis of all stages |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta (s.e.m.) | P | Coded allele freq. |
N | Beta (s.e.m.) | P | Coded allele freq. |
N | Beta (s.e.m.) | P | ||||
| 1 | rs2284746(17179262), MFAP2(intron) | G | FEV1/FVC | −0.042 (0.007) | 2.47E-09 | 0.516 | 45944 | −0.038 (0.007) | 2.64E-07 | 0.522 | 35371 | −0.04 (0.005) | 7.50E-16 |
| FEV1 | 0.008 (0.007) | 2.78E-01 | 0.006 (0.007) | 3.70E-01 | 0.007 (0.005) | 1.48E-01 | |||||||
| 1 | rs993925(216926691), TGFB2(downstream) | T | FEV1/FVC | 0.04 (0.007) | 2.54E-07 | 0.308 | 42402 | 0.023 (0.01) | 1.76E-02 | 0.348 | 21414 | 0.034 (0.006) | 1.16E-08 |
| FEV1 | 0.025 (0.007) | 1.51E-03 | 0.003 (0.007) | 7.29E-01 | 0.014 (0.005) | 8.71E-03 | |||||||
| 2 | rs12477314(239542085), HDAC4(downstream) | T | FEV1/FVC | 0.052 (0.008) | 4.48E-09 | 0.202 | 45585 | 0.031 (0.008) | 8.41E-05 | 0.206 | 45821 | 0.041 (0.006) | 1.68E-12 |
| FEV1 | 0.032 (0.008) | 2.77E-04 | 0.025 (0.007) | 1.82E-04 | 0.028 (0.005) | 1.02E-07 | |||||||
| 3 | rs1529672(25495586), RARB(intron) | C | FEV1/FVC | −0.06 (0.009) | 7.75E-10 | 0.829 | 40624 | −0.038 (0.009) | 1.16E-05 | 0.831 | 45466 | −0.048 (0.006) | 3.97E-14 |
| FEV1 | −0.037 (0.009) | 1.78E-04 | −0.011 (0.007) | 9.33E-02 | −0.02 (0.006) | 2.16E-04 | |||||||
| 3 | rs1344555(170782913), MECOM(intron) | T | FEV1/FVC | −0.019 (0.008) | 2.61E-02 | 0.205 | 46067 | −0.017 (0.012) | 1.55E-01 | 0.209 | 21313 | −0.018 (0.007) | 6.65E-03 |
| FEV1 | −0.042 (0.008) | 1.91E-06 | −0.025 (0.009) | 6.44E-03 | −0.034 (0.006) | 2.65E-08 | |||||||
| 5 | rs153916(95062456), SPATA9(upstream) | T | FEV1/FVC | −0.033 (0.007) | 2.06E-06 | 0.552 | 47530 | −0.025 (0.009) | 6.67E-03 | 0.535 | 21647 | −0.031 (0.005) | 2.12E-08 |
| FEV1 | −0.001 (0.007) | 8.91E-01 | 0.004 (0.007) | 6.22E-01 | 0.001 (0.005) | 8.20E-01 | |||||||
| 6 | rs6903823(28430275), ZKSCAN3(intron)/ZNF323(intron) | G | FEV1/FVC | −0.027 (0.008) | 2.28E-03 | 0.209 | 47057 | −0.013 (0.011) | 2.34E-01 | 0.246 | 21489 | −0.021 (0.007) | 1.19E-03 |
| FEV1 | −0.046 (0.008) | 2.00E-07 | −0.029 (0.008) | 4.75E-04 | −0.037 (0.006) | 2.18E-10 | |||||||
| 6 | rs2857595(31676448), NCR3(upstream) | G | FEV1/FVC | 0.049 (0.009) | 7.86E-08 | 0.809 | 45540 | 0.028 (0.008) | 5.36E-04 | 0.796 | 46107 | 0.037 (0.006) | 2.28E-10 |
| FEV1 | 0.04 (0.009) | 1.46E-05 | 0.017 (0.007) | 9.41E-03 | 0.025 (0.005) | 1.30E-06 | |||||||
| 6 | rs2798641(109374743), ARMC2(intron) | T | FEV1/FVC | −0.047 (0.009) | 2.81E-07 | 0.183 | 46369 | −0.03 (0.012) | 1.57E-02 | 0.179 | 21173 | −0.041 (0.007) | 8.35E-09 |
| FEV1 | −0.046 (0.009) | 5.39E-07 | −0.009 (0.01) | 3.35E-01 | −0.03 (0.006) | 4.69E-06 | |||||||
| 10 | rs7068966(12317998), CDC123(intron) | T | FEV1/FVC | 0.045 (0.007) | 1.28E-10 | 0.519 | 47085 | 0.023 (0.006) | 3.86E-04 | 0.518 | 46067 | 0.033 (0.005) | 6.13E-13 |
| FEV1 | 0.04 (0.007) | 1.19E-08 | 0.022 (0.005) | 3.56E-05 | 0.029 (0.004) | 2.82E-12 | |||||||
| 10 | rs11001819(77985230), C10orf11(intron) | G | FEV1/FVC | −0.019 (0.007) | 6.50E-03 | 0.522 | 45546 | −0.006 (0.006) | 3.17E-01 | 0.506 | 45932 | −0.012 (0.005) | 7.58E-03 |
| FEV1 | −0.041 (0.007) | 1.42E-08 | −0.022 (0.005) | 3.10E-05 | −0.029 (0.004) | 2.98E-12 | |||||||
| 12 | rs11172113(55813550), LRP1(intron) | T | FEV1/FVC | −0.035 (0.007) | 1.36E-06 | 0.607 | 45387 | −0.026 (0.01) | 5.83E-03 | 0.59 | 20509 | −0.032 (0.006) | 1.24E-08 |
| FEV1 | −0.021 (0.007) | 3.55E-03 | −0.003 (0.007) | 6.94E-01 | −0.013 (0.005) | 1.19E-02 | |||||||
| 12 | rs1036429(94795559), CCDC38(intron) | T | FEV1/FVC | 0.049 (0.008) | 1.24E-08 | 0.2 | 47814 | 0.028 (0.008) | 3.35E-04 | 0.214 | 46311 | 0.038 (0.006) | 2.30E-11 |
| FEV1 | 0.01 (0.008) | 2.67E-01 | 0.004 (0.006) | 5.38E-01 | 0.006 (0.005) | 2.26E-01 | |||||||
| 16 | rs12447804(56632783), MMP15(intron) | T | FEV1/FVC | −0.053 (0.009) | 7.12E-08 | 0.208 | 35123 | −0.021 (0.01) | 4.20E-02 | 0.222 | 24398 | −0.038 (0.007) | 3.59E-08 |
| FEV1 | −0.017 (0.009) | 8.02E-02 | 0.004 (0.007) | 5.71E-01 | −0.004 (0.006) | 4.73E-01 | |||||||
| 16 | rs2865531(73947817), CFDP1(intron) | T | FEV1/FVC | 0.039 (0.007) | 2.30E-08 | 0.418 | 47594 | 0.024 (0.006) | 1.94E-04 | 0.409 | 46304 | 0.031 (0.005) | 1.77E-11 |
| FEV1 | 0.024 (0.007) | 6.30E-04 | 0.011 (0.005) | 3.89E-02 | 0.016 (0.004) | 1.09E-04 | |||||||
| 21 | rs9978142(34574109), KCNE2(upstream) | T | FEV1/FVC | −0.048 (0.009) | 8.23E-07 | 0.156 | 44577 | −0.031 (0.013) | 1.75E-02 | 0.149 | 20944 | −0.043 (0.008) | 2.65E-08 |
| FEV1 | −0.012 (0.009) | 2.47E-01 | −0.015 (0.01) | 1.35E-01 | −0.013 (0.007) | 5.57E-02 | |||||||
Gene expression
We investigated mRNA expression of the nearest gene for each of the 16 novel loci in human lung tissue and in a range of human primary cells including lung, brain, airway smooth muscle cells and bronchial epithelial cells. Transcripts were detected for all selected genes in lung tissue except CCDC38 and transcripts for most genes were also detected in airway smooth muscle cells and in bronchial epithelial cells (Table 2). As we were unable to detect expression of CCDC38 in any tissue, we also examined expression of SNPRF, which is the adjacent gene (Table 2), and found expression in all four cell types. TGFB2, MFAP2, EVI1 and MMP15 were expressed in one or more lung cell types but not in peripheral blood mononuclear cells providing evidence that these genes may exhibit tissue-specific expression.
Table 2.
Expression profiling of candidate genes in the lung and periphery. (+) indicates the gene is expressed in the cell type used and (−) indicates the gene expression at mRNA level is not detected following 40 cycles of PCR. PCR profiling of gene transcripts in the human lung demonstrates expression of all candidates except CCDC38, for which two sets of primers were designed and tested under different optimization conditions. None of these assays detected expression of CCDC38 in the cell types analysed. The SNRPF gene neighbouring CCDC38 and harbouring SNPs with strong LD with CCDC38’s sentinel SNP was assayed instead. All PCR products were sequence verified. Glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH) was used as a positive control for the cDNA and was expressed in all tissues. HASM: Human airway smooth muscle. HBEC: Human bronchial epithelial cells. PBMC: peripheral blood mononuclear cells.
| Sentinel SNP (relationship to gene) |
Chr. | Gene | Putative Function of encoded protein |
Tissue | |||
|---|---|---|---|---|---|---|---|
| Lung | HASM | HBEC | PBMC | ||||
| rs993925 (intron) |
1 | TGFB2 | Cytokine with roles in pro-fibrotic cytokine modulating epithelial repair mechanisms and extracellular matrix homeostasis including collagen deposition40 |
+ | + | − | − |
| rs2284746 (intron) |
1 | MFAP2 | Major antigen of elastin-associated microfibrils17 and a candidate for involvement in the etiology of inherited connective tissue diseases. |
+ | + | + | − |
| rs12477314 (downstream) |
2 | HDAC4 | Deacetylase of histone surrounding DNA thus influencing transcription factor access to the DNA possibly repressing gene transcription. |
+ | + | + | + |
| rs1344555 (intron) |
3 | EVI1 | Zinc finger transcription factor, encoded as part of MDS1/EVI1 complex locus (MECOM). |
+ | + | + | − |
| rs1529672 (intron) |
3 | RARB | Nuclear retinoic acid receptor responsive to retinoic acid, a vitamin A derivative and which also controls cell proliferation and differentiation. |
+ | + | + | + |
| rs153916 (intron) |
5 | SPATA9 | Initially identified as a mediator of spermatogenesis, other family members may have a role in pancreatic development and β-cell proliferation41 |
+ | + | + | + |
| rs2798641 (intron) |
6 | ARMC2 | Function unknown although other family members have been identified as having roles in cell signalling, protein degradation and cytoskeleton functions42 |
+ | + | + | + |
| rs2857595 (upstream) |
6 | NCR3 | Required for efficient cytotoxicity responses by natural killer cells against normal cells and tumours43 |
+ | − | − | + |
| rs6903823 (intron) |
6 | ZKSCAN3 | Transcription factor involved in cell growth/cell cycle/signal transduction |
+ | + | + | + |
| rs7068966 (intron) |
10 | CDC123 | Homologue in yeast shown to be a critical control protein modulating Eukaryotic initiation factor 2 in times of cell stress |
+ | + | + | + |
| rs11001819 (intron) |
10 | C10orf11 | Function unknown | + | + | + | + |
| rs11172113 (intron) |
12 | LRP1 | Potentially diverse roles including cell signalling and migration44 |
+ | + | + | + |
| rs1036429 (intron) |
12 | CCDC38 | Function unknown although other family members involved in a diverse array of functions skeletal and motor function45 |
− | − | − | − |
| rs1036429 (r2=0.96 with rs4762633 in SNRPF) |
12 | SNRPF | Small nuclear ribonucleoprotein F | + | + | + | + |
| rs12447804 (intron) |
16 | MMP15 | Member of a large protease family with diverse functional roles via protease activity and specificity including; tissue remodelling, wound healing, angiogenesis, and tumor invasion. |
+ | + | + | − |
| rs2865531 (intron) |
16 | CFDP1 | Craniofacial Development Protein 1 | + | + | + | + |
| rs9978142 (upstream) |
21 | KCNE2 | KCNQ1-KCNE2 K+ channels may modulate transepithelial anion secretion in Calu3 airway epithelial cells46. |
+ | − | − | + |
|
Reference gene |
12 | GAPDH | + | + | + | + | |
We assessed whether SNPs in these new regions, or their proxies (r2>0.6), were associated with gene expression using a database of expression-associated SNPs in lymphoblastoid cell lines16. Four loci showed regional (cis) effects on expression (P<1×10−7, Supplementary Note). A proxy for our sentinel SNP in CFDP1, rs2865531, coincided with the peak of the expression signal for CFDP1 and the strongest proxy for rs6903823 in ZKSCAN3 coincided with the peak of expression for ZSCAN12.
Plausible pathways for lung function involving new loci
The putative function of the genes within, or closest to, the association peaks identify a range of plausible mechanisms for impacting lung function. The most statistically significant new signal for FEV1/FVC (P=7.5×10−16) was in the gene encoding MFAP2, an antigen of elastin-associated microfibrils17, although correlated SNPs in the region potentially implicate other genes that could plausibly influence lung function, such as CROCC, which encodes rootletin, a component of cilia18. Our second strongest new signal, also for FEV1/FVC, was in the gene encoding the retinoic acid receptor beta (RARB). Rarb-null knockout mice exhibit premature alveolar septation19. The third most statistically significant new signal for FEV1/FVC, and the most statistically significant new signal for FEV1, was in CDC123. This was the only novel region to show genome-wide association with both traits. CDC123 encodes a homologue of a yeast cell division cycle protein which plays a critical role in modulating Eukaryotic initiation factor 2 in times of cell stress20. The fourth signal for FEV1/FVC is downstream of HDAC4 which encodes a histone deacetylase; reductions in the expression of other histone deacetylases (specifically HDAC2, HDAC5 and HDAC8) have been noted in COPD21. The regions we observed in the MHC are much more difficult to localize with multiple genes being tagged by the top SNP, including non-synonymous SNPs in ZKSCAN3, PGBD1, ZSCAN12, ZNF323, TCF19, LTA, C6orf15 and GPANK1 (also known as BAT4) (Supplementary Table 2). At 6p21.33, the strongest association with lung function was observed for rs2857595, which is in linkage disequilibrium (LD, r2=0.47) with a non-synonymous SNP in LTA (encoding lymphotoxin alpha) and with a SNP in the upstream promoter region of TNFA (encoding tumour necrosis factor alpha, r2=0.86), both of which are plausible candidates22,23. Our top SNP in MMP15 is in strong LD (r2=1) with a non-synonymous SNP (rs3743563, which has an association with FEV1/FVC at P=1.8×10−7) within the same gene. Plausible mechanisms implicated by the other novel signals of association with lung function reported here include TGF-beta signalling; TGFB2 expression is upregulated in bronchial epithelial cells in asthma24. The putative function of key genes (as defined by LD with the leading SNP) in each of the 16 loci, and relevant findings from animal models, are summarised in Table 2 and detailed in Supplementary Table 2.
Associations with lung function in children
Alleles representing 11 of the 16 novel loci showed directionally consistent effects on lung function in 6,281 children (7 to 9 years of age) (Supplementary Table 3A) suggesting that genetic determination of lung function in adults may in part act via effects on lung development, or alternatively, that some genetic determinants of lung growth and lung function decline are shared.
Association of lung function loci with other traits
Although we stratified for ever-smoking versus never-smoking, we did not adjust for the amount smoked. In order to investigate the possibility that the associations at any of our 16 novel regions were driven by an effect of the SNP on smoking behaviour, we evaluated in silico data for associations with smoking amount from the Ox-GSK consortium25 for the leading SNPs in these 16 regions. None of these 16 SNPs showed statistically significant association with the number of cigarettes smoked per day (Supplementary Table 3B).
In addition, in our Stage 1 and Stage 2 datasets combined, we assessed whether the estimated effect sizes of the variants on lung function phenotypes differed substantially between ever-smokers and never-smokers (Supplementary Table 4) across the 16 loci. For the most strongly associated trait at each locus, we tested the SNP interaction with ever-smoking (versus never-smoking). None of the 16 novel loci showed a significant interaction (Bonferroni corrected threshold for 16 independent SNPs P=0.003125). These analyses suggest that the genetic effects we have identified underlie lung function variability irrespective of smoking exposure.
Our lung function associations were adjusted for height, but there are some overlaps between loci associated with height and those associated with lung function. Therefore, we evaluated in silico data for height associations of our novel regions in the GIANT consortium14 dataset. The G allele of rs2284746 (MFAP2, intron), which was associated with decreased FEV1/FVC was associated with increased height (Supplementary Table 3C).
Given reported associations between lung cancer and either COPD or lung function decline, we also assessed in silico data for sentinel or proxy SNPs in these 16 regions for associations with lung cancer in the International Lung Cancer Consortium (ILCCO) GWAS meta-analysis26. Alleles associated with reduced lung function were associated with risk of lung cancer at the strongest available proxy SNP for rs2857595 (upstream of NCR3) at 6p21.33 (rs3099844, r2=0.67), and the strongest proxy SNP for rs6903823 (SNP in intron of ZKSCAN3 and ZNF323) at 6p22.1 (rs209181, r2=0.69) (lung cancer associations, P=2.2×10−7 and P=3.4×10−5, respectively, Supplementary Table 3D). No significant associations with lung cancer were seen at the other new loci (proxy SNPs were available for 15 of the 16 loci, Bonferroni corrected P<0.0033).
In addition to the effects on height, smoking and lung cancer described above, we examined the literature for evidence for associations with other traits for each of the 16 new loci (detailed in Supplementary Table 2). Genome-wide significant associations (P<5×10−8) have been reported in KCNE2 with myocardial infarction27, and at 6p21.33 near NCR3/AIF1, with neonatal lupus28 and with systemic lupus erythematosus29. Other significant complex disease associations have also been noted in the regions of CDC123 (type 2 diabetes30), CFDP1 (type 1 diabetes31) and MECOM (blood pressure32,33), but with weaker LD (r2<0.3) between the reported SNP and the sentinel SNP for lung function in the region (Supplementary Table 2).
Proportion of variance explained by loci discovered to date
Associations in 10 loci previously reported for lung function8,9 reached genome-wide significance (P<5×10−8) in our Stage 1 data, namely loci in or near TNS1, FAM13A, GSTCD/NPNT, HHIP, HTR4, ADAM19, AGER, GPR126, PTCH1, and TSHD4 (Supplementary Table 5A). Thus, a total of 26 regions showed genome-wide significant association with lung function in our study. In aggregate, variants at these 26 regions explain approximately 3.2% of the additive polygenic variance for FEV1/FVC and 1.5% for FEV1 (see Supplementary Note). Following the approach described by Park et al.34 we estimated that there is a total of 102 (95% CI 57-155) independent variants with similar effect sizes to the 26 variants we report. In combination these 102 variants, comprising 26 discovered variants and 76 putative undiscovered variants, collectively explain around 7.5% of the additive polygenic variance for FEV1/FVC and 3.4% for FEV1 (see Supplementary Table 6, Online Methods and Supplementary Note).
Discussion
In meta-analysis of 23 studies comprising 48,201 individuals of European ancestry and follow-up in 17 studies comprising up to 46,411 individuals, we report genome-wide significant associations with an additional 12 regions for FEV1/FVC, an additional three regions for FEV1 and one additional region associated with both FEV1 and FEV1/FVC. We also confirm genome-wide association with 10 regions previously associated with lung function, bringing to 26 the total number of loci associated with lung function in these data. Most of the new loci are in regions not previously suspected to have been involved in lung development, the control of pulmonary function or risk of developing COPD. Elucidating the mechanisms through which these regions influence lung function should lead to a more complete understanding of lung function regulation and the pathogenesis of COPD. Four of the new loci (MFAP2, ZKSCAN3, near NCR3 and near KCNE2) we show to be associated with lung function are also associated with other complex traits and diseases (P<5×10−8 for the other trait at a SNP with r2>0.3 with the top lung function SNP in the region). Understanding the intermediates underlying these pleiotropic effects could also reveal crucial insights into the pathophysiology of lung disease. One potential explanation is that these loci underlie control of the mechanisms regulating the development and resolution of inflammation and subsequent tissue remodelling in a range of tissues.
The effect sizes of the variants in the 26 loci associated with lung function explain a modest proportion of the additive genetic variance in FEV1/FVC and in FEV1, even after accounting for putative undetected variants with a similar distribution of effect sizes34. Our findings are consistent with those from other common complex traits, where it is thought that many as yet unidentified common and rare sequence variants, and potentially structural variants could explain the remaining heritability35. That our study more than doubled the number of loci known to be associated with lung function underlines the utility of large sample sizes to achieve the power to detect common variants associated with complex traits. Nevertheless, it is likely that additional variants with similar effect sizes remain undiscovered14. In addition, our study was not designed to detect rare variants or structural variants associated with lung function. Identification of rare variants associated with lung function could be helpful in narrowing the scope of ongoing functional work to those genes most likely to be causally related to the association signals we detected.
Our study focused on cross-sectional measures of lung function. Adult lung function at a particular time point is influenced by the peak lung function achieved by 25-35 years of age, as well as the rate of decline of lung function after that peak36. The 26 loci now confirmed to be associated with lung function could affect either pre- or post-natal lung development and growth or decline in lung function during adulthood, or both. We showed consistent directions of estimated effects on lung function between adults and children at 7-9 years of age for SNPs at 11 of the 16 new loci, and eight of 10 previously reported loci (Supplementary Table 3A). The results we show for lung function in children provide some indication that these loci affect lung function development, although studies in larger populations of children would provide greater clarity for SNPs in the new loci. Further investigations will be required in large populations with longitudinal data to delineate the influence of these variants on the rates of development of, and decline in, lung function and on the risk of developing COPD.
Of the sentinel SNPs at the 16 new loci associated with lung function, only rs2284746 (MFAP2) was associated with height in the GIANT consortium14 dataset. The G allele of rs2284746 was associated with both increased height and reduced lung function. A similar relationship between lung function and height was previously reported for the G allele of rs3817928 in GPR1268,14, which is associated with decreased height, but with increased FEV1/FVC. A further three of the 180 loci found to be associated with height14 showed association (for 180 loci, Bonferroni corrected threshold P=2.8×10−4) with either FEV1 (CLIC4 and BMP6) or FEV1/FVC (PIP4K2B) (Supplementary Table 3E). In each case, the allele associated with an increase in height was associated with a decrease in lung function. This is not the case for the association of rs1032296 near HHIP, which has shown consistent directions of effects on lung function and height14,11. However, the strongest SNP associated with height in the HHIP region lies within an intron of HHIP but shows no association with FEV1 or FEV1/FVC. Furthermore, while height is an important predictor of FEV1, this is not true for its ratio to FVC37. These observations argue against the associations with lung function at these loci being simply due to incomplete adjustment for height.
We stratified by ever- and never-smoker status in our analyses and in our investigation of amount smoked in the Ox-GSK consortium25 none of the sentinel SNPs in the 16 new regions showed association with the number of cigarettes smoked per day. Additionally, none of these regions was associated with ever-smoking in the Ox-GSK consortium data (Supplementary Table 3B). Thus the SNP associations with lung function we observed are unlikely to have arisen simply as a consequence of inadequate adjustment for smoking.
We did not observe any interactions with ever-smoking for any of the sentinel SNPs in the 16 new regions that exceeded a Bonferroni-corrected significance level (for 16 SNPs). Thus, the effects on lung function of the novel variants we identified are apparent in both ever-smokers and in never-smokers, and the effects of smoking and of these genetic variants may be independent and additive.
In other common complex diseases, follow up studies that incorporate common genetic risk variants into models to predict disease have not been shown to add substantially to existing risk models, particularly when such models already include family history38,39. The same may also prove to be true for the 26 genetic variants described in this paper, as the effect size of any individual variant is small, but further work is required in this area. The major utility of our findings will be in the knowledge they provide about previously unknown pathways underlying lung function. Elucidating the mechanisms that these genes are involved in will lead to improved understanding of the regulation of lung function and potentially to new therapeutic targets for COPD.
Supplementary Material
Acknowledgements
We thank the many colleagues who contributed to collection and phenotypic characterization of the clinical sampling, genotyping, and analysis of the data. We especially thank those who kindly agreed to participate in the studies.
Major funding for this work is from the following sources (alphabetical): Academy of Finland (project grants 104781, 120315, 129269, 1114194, Center of Excellence in Complex Disease Genetics (213506 and 129680) and SALVE); Althingi (Icelandic Parliament); Arthritis Research Campaign; Asthma UK; AstraZeneca; AXA Research Fund; Biotechnology and Biological Sciences Research Council (BBSRC) (BB/F019394/1, G20234); British Heart Foundation (PG/97012, PG/06/154/22043, FS05/125); British Lung Foundation; Canadian Institutes of Health Research (Grant ID MOP-82893); Cancer Research United Kingdom; Chief Scientist Office, Scottish Government Health Directorate (CZD/16/6); Croatian Institute for Public Health; UK Department of Health; Dutch Kidney Foundation; Erasmus Medical Center and Erasmus University, Rotterdam; Estonian Genome Center, University of Tartu, Estonia (SF0180142s08); EU funding (GABRIEL GRANT Number: 018996, ECRHS II Coordination Number: QLK4-CT-1999-01237); European Commission (DG XII, EURO-BLCS, FP-5 QLG1-CT-2000-01643, FP-6 LSHB-CT-2006-018996 (GABRIEL), FP-6 LSHG-CT-2006-018947 (EUROSPAN), FP-6 GenomEUtwin project QLG2-CT-2002-01254, FP7/2007-2013: HEALTH-F2-2008-201865, GEFOS, HEALTH-F2-2008-35627, TREAT-OA, HEALTH-F4-2007-201413 (ENGAGE)); Finnish Foundation for Cardiovascular Research; Flight Attendant Medical Research Institute (FAMRI); German Asthma and COPD Network (COSYCONET: BMBF grant 01GI0883); German Bundesministerium fuer Forschung und Technology (01 AK 803 A-H, 01 IG 07015 G); German Federal Ministry of Education and Research (BMBF) (03ZIK012, 01ZZ9603, 01ZZ0103, and 01ZZ0403): German National Genome Research Network (NGFN-2 and NGFN-plus); German Ministry of Cultural Affairs; GlaxoSmithKline; Gyllenberg Foundations; Healthway, Western Australia; Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany; Healthcare and Bioscience iNet (funded by the East Midlands Development Agency, part-financed by the European Regional Development Fund, delivered by Medilink East Midlands); Higher Education Funding Council for England (HEFCE); Hjartavernd (Icelandic Heart Association); Innsbruck Medical University; Institute for Anthropological Research in Zagreb; International Osteoporosis Foundation; Intramural Research Program of the NIH, National Institute on Aging and National Institute of Environmental Health Sciences; Jalmari and Rauha Ahokas Foundation; Juvenile Diabetes Research Foundation International (JDRF); Lifelong Health and Wellbeing Initiative (G0700704/84698); Medical Research Council UK (G1000861, G0501942, G0902313, G0000934, G0800582, G0500539, G0600705, PrevMetSyn/SALVE, G9901462); Medical Research Fund of the Tampere University Hospital; Ministry of Science, Education and Sport of the Republic of Croatia (108-1080315-0302); Medical Research Council Human Genetics Unit; Medisearch-The Leicester Medical Research Foundation; Munich Center of Health Sciences (MC Health) as part of LMUinnovativ; National Health and Medical Research Council of Australia (Grant ID 403981 and ID 003209); National Human Genome Research Institute (NHGRI) (U01-HG-004729, U01-HG-004402); National Institute for Health Research (NIHR) Comprehensive Biomedical Research Centres (Guy’s & St Thomas’ NHS Foundation Trust in partnership with King’s College London and Cambridge University Hospitals NHS Foundation Trust in partnership with the University of Cambridge); Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) (050-060-810); Netherlands Organization for the Health Research and Development (ZonMw); Netherlands Organization of Scientific Research NOW (1750102007006, 175.010.2005.011, 911-03-012); Northern Netherlands Collaboration of Provinces (SNN); Norwegian University of Science and Technology; Novo Nordisk; Ontario Institute of Cancer Research and Canadian Cancer Society Research Institute (CCSRI 020214); Republic of Croatia Ministry of Science, Education and Sports research grants (108-1080315-0302); Research Institute for Diseases in the Elderly (RIDE) (014-93-015: RIDE2); Research Into Ageing (251); Siemens Healthcare, Erlangen, Germany and the Federal State of Mecklenburg- West Pomerania; Social Ministry of the Federal State of Mecklenburg-West Pomerania; Structure Enhancing Fund (FES) of the Dutch government; Swedish Heart and Lung Foundation grant 20050561; Swedish Research Council for Worklife and Social research (FAS), grants 2001-0263, 2003-0139; Swiss National Science Foundation (grants no 4026-28099,3347CO-108796, 3247BO-104283, 3247BO-104288, 3247BO-104284, 32-65896.01,32-59302.99, 32-52720.97, 32-4253.94); The Asthma, Allergy and Inflammation Research Trust; The Great Wine Estates of the Margaret River region of Western Australia; The Netherlands’ Ministry of Economic Affairs, Ministry of Education, Culture and Science and Ministry for Health, Welfare and Sports; The Royal Society; The University of Split and Zagreb Medical Schools,; Tromsø University; U01 DK062418; UBS Wealth Foundation Grant BA29s8Q7-DZZ; UK Department of Health Policy Research Programme; University Hospital Oulu, Biocenter, University of Oulu, Finland (75617); University Medical Center Groningen; University of Bristol; University of Leicester HEFCE CIF award; University of Nottingham; US National Institute of Health (NIH) (1P50 CA70907, RO1 CA121197, U19 CA148127, CA55769, CA127219, R01HL071051, R01HL071205, R01HL071250, R01HL071251, R01HL071252, R01HL071258, R01HL071259, UL1RR025005, contracts HHSN268200625226C, HHSN268200782096C, R01-HL084099); US NIH National Cancer Institute (RO1CA111703); US NIH National Center for Research Resources (grants M01-RR00425 and 5M01 RR00997); US NIH National Eye Institute (NEI); US NIH National Heart, Lung and Blood Institute (contracts N01-HC-85079 through N01-HC-85086, N01-HC-35129, N01 HC-15103, N01 HC-55222, N01-HC-75150, N01-HC-45133, N01-HC-95095, N01-HC-48047, N01-HC-48048, N01-HC-48049, N01-HC-48050, N01-HC-45134, N01-HC-05187, N01-HC-45205, N01-HC-45204, N01 HC-25195, N01-HC-95159 through N01-HC-95169, RR-024156, N02-HL-6-4278, R01 HL-071022,R01 HL-077612, R01 HL-074104, RC1 HL100543, HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C, grants HL080295, HL087652, HL105756, R01-HL-084099, R01HL087641, R01HL59367,R01HL086694, HL088133, HL075336 5R01HL087679-02 through the STAMPEED program (1RL1MH083268-01), 1K23HL094531-01); US NIH National Institute of Allergy and Infectious Diseases (NIAID); US NIH National Institute of Child Health and Human Development (NICHD); US NIH National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) (DK063491); US NIH National Institute of Environmental Health Sciences (NIEHS) (ZO1 ES49019, ES015794); US NIH National Institute of Mental Health (NIMH) (5R01MH63706:02); US NIH National Institute of Neurological Disorders and Stroke (NINDD); US NIH National Institute on Aging (NIA) (R01 AG032098, RC1 AG035835, N01AG12100, N01AG62101, N01AG62103, N01AG62106, 1R01AG032098-01A1, AG-023269, AG-15928, AG-20098, AG-027058); Wellcome Trust (077016/Z/05/Z, GR069224, 068545/Z/02, 076113/B/04/Z, 079895).
APPENDIX
Online Methods
Study design
The study consisted of two stages. Stage 1 was a meta-analysis conducted on directly genotyped and imputed SNPs from individuals of European ancestry in 23 studies, with a total of 48,201 individuals. Supplementary Table 1A gives details of these studies. Thirty-four SNPs selected according to the results in Stage 1 were followed up in Stage 2. The ten leading SNPs were followed up in up to 46,411 individuals of European origin and the remaining 24 SNPs were followed up in a subset of up to 21,674 individuals (Figure 1).
Stage 1 samples
A total of 23 studies, 17 from the SpiroMeta consortium and six studies from the CHARGE consortium, formed Stage 1: AGES, ARIC, B58C T1DGC, B58C WTCCC, BHS1, CHS, ECRHS, EPIC (obese cases and population-based studies), the EUROSPAN studies (CROATIA-Korcula, ORCADES and CROATIA-Vis), FHS, FTC (incorporating the FinnTwin16 and Finnish Twin Study on Aging), Health 2000, Health ABC, KORA F4, KORA S3, NFBC1966, RS-I, RS-II, SHIP and TwinsUK-I (see Supplementary Table 1 for definitions of abbreviations). Measurements of spirometry for each study are described in the Supplementary Note. The genotyping platforms and quality-control criteria implemented by each study are described in Supplementary Table 1B.
Imputation
Imputation of non-genotyped SNPs was undertaken with MACH47, IMPUTE48 or BIMBAM49 with pre-imputation filters and parameters as shown in Supplementary Table 1B. SNPs were excluded if the imputation information, assessed using r2.hat (MACH), .info (IMPUTE) or OEvar (BIMBAM), was <0.3. In total, 2,706,349 SNPs were analyzed.
Transformation of data and genotype-phenotype association analysis
Linear regression of age, age2, sex, height and ancestry principal components was undertaken on FEV1 (milliliters) and FEV1/FVC (percent). The residuals were transformed to ranks and then transformed to normally distributed z-scores. These transformed residuals were then used as the phenotype for association testing under an additive genetic model, separately for ever-smokers and for never-smokers. The software used is specified in Supplementary Table 1B. Appropriate tests for association in related individuals were applied where necessary, as described in the Supplementary Note.
Meta-analysis of Stage 1 data
All Stage 1 study effect estimates, both for ever-smokers and never-smokers were corrected using genomic control50 and were oriented to the forward strand of the NCBI build 36 reference sequence of the human genome, consistently using the alphabetically higher allele as the coded allele. Study-specific lambda estimates are shown in Supplementary Table 1. For each study, effect estimates and standard errors for ever-smokers and never-smokers were meta-analysed using inverse variance weighting. Genomic control was applied again to the pooled effect-size estimates for each study. Finally effect-size estimates and standard errors were combined across studies using inverse variance weighting meta-analysis, and genomic control was applied to the pooled effect-size estimates. To describe the effect of imperfect imputation on power, for each SNP we report the effective sample size (N effective); the sum of the study-specific products of the sample size and the imputation quality metric. Meta-analysis statistics and figures were produced using R version 2.9.2.
Selection of SNPs for Stage 2
All regions selected for follow-up in Stage 2 contained a lead SNP with novel evidence of association (all with P<3×10−6) with FEV1 and/or FEV1/FVC, an N effective ≥70% of the total Stage 1 sample size and the association signals from surrounding SNPs were consistent with their correlation (linkage disequilibrium) with the leading SNP. Twenty-nine independent regions with a leading SNP meeting these criteria were assessed in Stage 2. Regions were defined as independent if the leading SNP from one region was >500kb from the leading SNP of any other region. Long range linkage disequilibrium was also investigated between leading SNPs of regions in or near the MHC on chromosome 6 (Supplementary Note). For two regions, the leading SNP had an N effective ≥70% but <80% of the Stage 1 sample size and therefore a proxy SNP (r2=1 and 0.97) was also taken forward. For three regions, there were different leading SNPs for FEV1 and FEV1/FVC and so both leading SNPs were assessed. A total of 34 SNPs were analysed in Stage 2 and are listed in Supplementary Table 5B. Previously reported regions8-11,51,52 were not followed up. We present association test statistics in Stage 1 only for relevant SNPs from previously reported regions in Supplementary Table 5A.
Stage 2 samples
The 34 SNPs were followed up in up to 11,275 individuals from seven studies with in silico data; CARDIA, CROATIA-Split, LifeLines, LBC1936, MESA-Lung, RS-III and TwinsUK-II (Supplementary Table 1). SNP rs2647044 was not available from TwinsUK-II.
The 34 SNPs were ranked by P value (for association with either FEV1 or FEV1/FVC) and the top ten leading SNPs selected for follow up by genotyping in up to 35,136 individuals from ADONIX, BHS2, BRHS, BWHHS, Gedling, GS:SFHS, HCS, Nottingham Smokers, NSHD and SAPALDIA (Supplementary Table 1). If a SNP within the top ten had an N effective <80%, only the proxy SNP was included in the top ten for follow up. For regions which showed association with both FEV1 and FEV1/FVC, only the leading SNP with the lowest P value for either trait was included if it was within the top ten SNPs. The study design is illustrated in Figure 1.
Meta-analysis of Stage 2 data
All Stage 2 studies provided effect estimates for ever-smokers and never-smokers, apart from Nottingham Smokers since they only had smokers. Studies with family data (BHS2 and GS:SFHS) analysed ever- and never-smokers together to account for the family correlation, adding the smoking status as a covariate in the model, and therefore provided smoking adjusted effect estimates. All Stage 2 study effect estimates were oriented to the forward strand of the NCBI build 36 reference sequence of the human genome, consistently using the alphabetically higher allele as the coded allele. For each study with separate results for ever- and for never-smokers, effect estimates and standard errors for ever- and never-smokers were meta-analysed using inverse variance weighting. Genomic control was applied to the pooled effect sizes of those studies with in silico data that undertook the analysis genome-wide. Effect estimates and standard errors were combined across Stage 2 studies using inverse variance weighting meta-analysis.
Combined analysis of Stage 1 and Stage 2 samples
Meta-analysis of Stage 1 and Stage 2 results was undertaken using inverse variance weighting. We described associations as genome-wide significant if P<5×10−8.
PCR expression profiling
The mRNA expression profiles of TGFB2, MFAP2, HDAC4, EVI1, RARB, SPATA9, ARMC2, NCR3, CDC123, LRP1, CCDC38, SNRPF, MMP15, CFDP1, ZKSCAN3, KCNE2 and C10orf11 were determined in human lung tissue and primary cell samples using RT-PCR, including RNA from lung (Ambion/ABI), brain, airway smooth muscle cells and human bronchial epithelial cells (Clonetics42). Primer sequences are listed in Supplementary Table 2. Full details are provided in the Supplementary Note.
Lung function associations in our data of SNPs previously associated with lung function
In order to permit comparison of findings with recent studies of relevance to the field, we present association test statistics (in Stage 1 only) for relevant SNPs from previously reported regions (Supplementary Table 5A). We included regions: (i) reported as showing genome-wide significant association (P<5×10−8) with lung function; (ii) reported as showing genome-wide significant association with COPD, providing that there was additional evidence of association with lung function and; (iii) DAAM2, which reached borderline significance in the SpiroMeta consortium9. Within each of these regions, if multiple SNPs had been reported, we included all relevant SNPs and also the SNP that showed the strongest association in our data.
Association to other traits of lung function associated SNPs
Regions associated (P<5×10−8) with lung function or COPD (and also associated with lung function) were looked up for other traits. Where multiple SNPs were reported for different traits or by different investigators, we aimed to include all relevant SNPs, except those with r2>0.9 with another SNP in the region. We also included the SNPs that showed the strongest association in our data for each region. The following related traits were assessed: (i) lung function in children (Supplementary Table 3A); (ii) smoking amount and ever-smoking versus never-smoking in the Ox-GSK consortium25 dataset (Supplementary Table 3B); (iii) height in the GIANT consortium14 dataset (Supplementary Table 3C and 3E) and (iv) lung cancer in the International Lung Cancer Consortium (ILCCO) GWAS meta-analysis26 (Supplementary Table 3D).
Estimation of the number of undiscovered variants and calculation of the proportion of variance explained
We used the approach proposed by Park et al.34 to estimate the number of independent variants associated with lung function measures that have similar effect sizes to the variants already reported, and to calculate the proportion of the variance explained by them. We excluded discovery data when estimating effect sizes to avoid winner’s curse bias, and obtained the number of undiscovered variants using the discovery power to detect the unbiased effect sizes (Supplementary Table 6 and Supplementary Note).
Additional analyses
The top SNPs from our novel loci, and proxies, were searched for correlation with known common copy number variants and expression SNPs. Analyses to identify common pathways underlying the association signals for lung function were undertaken using MAGENTA v253 and GRAIL54. Full methods and results are given in the Supplementary Note.
Footnotes
AUTHOR CONTRIBUTIONS
Author contributions are listed in alphabetical order. See Supplementary Note for definitions of study acronyms.
Project conception, design and management. Stage 1 GWAS, AGES: G.E., M.G., V.G., T.B.H., L.J.L. ARIC: S.J.L., N.F., L.R.L., D.J.C., D.B.H., B.R.J., A.C.M., K.E.N. B58C-T1DGC: D.P.S. B58C-WTCCC: D.P.S. BHS1: A.L.J., A.W.M., L.J.P., CHS: S.A.G., S.R.H., T.L., B.M.P. CROATIA-Korcula: H.C., I.G., S.J., I.R., A.F.W., L.Z. CROATIA-Vis: H.C., C.H., O.P., I.R., A.F.W. ECRHS: D.L.J., E.O., I.P, M.W. EPIC: N.J.W. FHS: J.B.W., G.T.O. FTC: J.K., K.H.P., T. Rantanen. Health ABC: M.C.A., P.A.C., T.B.H., S.B.K., Y.L., B.M. Health 2000: M.H., M.K. KORA F4: J. Heinrich. KORA S3: C.G., H.E.W. NFBC1966: P.E., A-L.H., M-R.J., A.P. ORCADES: H.C., S.H.W., J.F.W., A.F.W. RS: A. Hofman. SHIP: S.G., G.H., B.K., H.V. TwinsUK: T.D.S., G.Z. Stage 2 follow-up, ADONIX: J. Brisman., A-C.O. BHS2: J. Beilby. BRHS: R.W.M., S.G.W., P.H.W. BWHHS: G.D.S., S.E., D.A.L., P.H.W. CARDIA: A.S.CROATIA-Split: M.B., I.K., T.Z. GS:SFHS: C.M.J., S.M.K., A.D.M, D.J.P. HCS: C.C., J.W.H., A.A.S. LBC1936: I.J.D., S.E.H., J.M.S. LifeLines: H.M.B., D.S.P., J.M.V., C. W. MESA-Lung: R.G.B., J.L.H. Nottingham smokers: I.P.H. NSHD: R.H., D.K. SAPALDIA: N.P.-H., T. Rochat. Look-up studies, ALSPAC: R.G., J. Henderson. ILCCO: ILCCO data. Ox-GSK: C.F., J.M.
Phenotype collection and data management. Stage 1 GWAS, AGES: T.A. ARIC: D.J.C., N.F., L.R.L., A.C.M., K.E.N. B58C-T1DGC: A.R.R., D.P.S. B58C-WTCCC: A.R.R., D.P.S. BHS1: A.L.J., A.W.M., L.J.P. CHS: S.A.G., S.R.H., T.L., B.M.P.CROATIA-Korcula: I.G., S.J., O.P., I.R., L.Z. CROATIA-Vis: H.C., C.H., O.P., I.R., A.F.W. ECRHS: D.L.J., E.O., I.P, M.W. EPIC: N.J.W. FHS: J.B.W., G.T.O. FTC: J.K., K.H.P., T. Rantanen. Health ABC: P.A.C., B.M., W.T. Health 2000: M.H., M.K. KORA F4: S.K, H.S. KORA S3: N.P.-H. NFBC1966: P.E., A-L.H., M-R.J., A.P. ORCADES: H.C., S.H.W., J.F.W. RS: G.G.B, M.E., D.W.L., B.H.Ch.S. SHIP: S.G., B.K., H.V. TwinsUK: C.J.H., P.G.Hysi., M.M., T.D.S., G.Z. Stage 2 follow-up, ADONIX: J. Brisman., A-C.O. BHS2: J. Beilby., M.L.H. BRHS: R.W.M., S.G.W., P.H.W. BWHHS: G.D.S., S.E., D.A.L., P.H.W. CARDIA: O.D.W. CROATIA-Split: M.B., I.K., T.Z. GS:SFHS: C.M.J., A.D.M. HCS: C.C., K.A.J., A.A.S. LBC1936: I.J.D., L.M.L., J.M.S. LifeLines: D.S.P., J.M.V. MESA-Lung: R.G.B., J.L.H. Nottingham smokers: K.A.A-B., J.D.B., I.P.H., A. Henry., M.O., I. Sayers. NSHD: R.H., D.K. SAPALDIA: N.P-H. Look up studies, ALSPAC: R.G., J. Henderson. ILCCO: ILCCO. Raine: W.Q.A., P.G. Holt., C.E.P., P.D.S
Genotyping. Stage 1 GWAS, B58C-T1DGC: W.L.M. B58C-WTCCC: W.L.M. BHS1: A.L.J., A.W.M., L.J.P. CHS: S.R.H., B.M.P., J.I.R. CROATIA-Vis: C.H., I.R., A.F.W. ECRHS: M.W. EPIC: I.B., R.J.F.L., J.H.Z. FTC: J.K. Health ABC: Y.L., K.L. Health 2000: S.R., I. Surakka. KORA F4: N.K. KORA S3: C.G. NFBC1966: P.E., A-L.H., M-R.J., A.P., A.R. ORCADES: H.C., J.F.W. RS: F.R., A.G.U. SHIP: G.H. TwinsUK: C.J.H., S-Y.S. Stage 2 follow-up, ADONIX: S.D., F.N., A-C.O. BHS2: J. Beilby, G.C., J.H. BRHS: A.D.H., R.W.M. BWHHS: S.E., D.A.L. CARDIA: M.F., X.G. CROATIA-Split: V.B., T.Z. Gedling: J.R.B., T.M. GS:SFHS: C.M.J., S.M.K., D.J.P. HCS: J.W.H. LBC1936: I.J.D., S.E.H., L.M.L., J.M.S. LifeLines: C.W. MESA-Lung: S.S.R. NSHD: D.K., A.W. SAPALDIA: M.I., F.K. Look up studies, ALSPAC: S.M.R., W.L.M. ILCCO: ILCCO. Raine: W.Q.A., C.E.P.
Data analysis. Stage 1 GWAS, AGES: G.K.G., A.V.S. ARIC: N.F., D.B.H., L.R.L. B58C-T1DGC: A.R.R., D.P.S., B58C-WTCCC: A.R.R., D.P.S. BHS1: N.M.W. CHS: K.D.M., J.I.R. CROATIA-Korcula: C.H., J.E.H, V.V. CROATIA-Vis: C.H, V.V. ECRHS: D.L.J., A.R. EPIC: J.H.Z. FHS: J.B.W. FTC: I. Surakka. Health ABC: P.A.C., Y.L., K.L., W.T. Health 2000: M.K., S.R., I. Surakka. KORA S3: E.A. NFBC1966: A.R. ORCADES: C.H., V.V. RS: M.E., D.W.L. SHIP: S.G., G.H., B.K, H.V. TwinsUK: M.M., G.Z. Stage 2 follow-up studies, ADONIX: S.D., F.N. BHS2: G.C. BRHS: R.W.M. BWHHS: D.A.L. CARDIA: M.F., X.G. HCS: J.W.H., K.A.J. LBC1936: L.M.L. LifeLines: H.M.B. MESA-Lung: A.M., S.S.R. Nottingham smokers: I. Sayers., A. Henry. NSHD: D.G., R.H. SAPALDIA: I.C., M.I. Look up studies, ALSPAC: D.M.E. ILCCO: ILCCO. Ox-GSK: J.Z.L. Raine: W.Q.A.
Analysis group: SpiroMeta consortium, I.P.H., T.J., M.S.A., M.D.T., L.V.W. CHARGE consortium, N.F., S.J.L., D.W.L., K.D.M., A.V.S., W.T., J.B.W.
Expression profiling and bioinformatics group: SpiroMeta consortium, I.P.H., M.O., I.Sayers, M.S.A., M.D.T., L.V.W. CHARGE consortium, S.A.G., D.W.L.
Writing group: SpiroMeta consortium, P.E., I.P.H., M.O., M.S.A., D.P.S., M.D.T., L.V.W. CHARGE consortium, S.J.L., D.W.L., S.A.G., G.T.O., V.G., B.H.Ch.S., W.T.
COMPETING FINANCIAL INTERESTS:
Inês Barroso and spouse own stock in Incyte Ltd and GlaxoSmithKline. Fredrik Nyberg is employed by AstraZeneca R&D, 431 83 Mölndal, Sweden. Professor Postma has received unrestricted research grants from and has been consultant to AstraZeneca, Boehringer Ingelheim, Chiesi, GlaxoSmithKline, Nycomed, TEVA. Clyde Francks is a full-time employee of the company GlaxoSmithKline (GSK) and GSK also funded several aspects of the study as detailed in the ACKNOWLEDGMENTS section for Ox-GSK.
References
- 1.Wilk JB, et al. Evidence for major genes influencing pulmonary function in the NHLBI family heart study. Genet Epidemiol. 2000;19:81–94. doi: 10.1002/1098-2272(200007)19:1<81::AID-GEPI6>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 2.Hole DJ, et al. Impaired lung function and mortality risk in men and women: findings from the Renfrew and Paisley prospective population study. BMJ. 1996;313:711–5. doi: 10.1136/bmj.313.7059.711. discussion 715-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strachan DP. Ventilatory function, height, and mortality among lifelong non-smokers. J Epidemiol Community Health. 1992;46:66–70. doi: 10.1136/jech.46.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young RP, Hopkins R, Eaton TE. Forced expiratory volume in one second: not just a lung function test but a marker of premature death from all causes. Eur Respir J. 2007;30:616–22. doi: 10.1183/09031936.00021707. [DOI] [PubMed] [Google Scholar]
- 5.Lopez AD, et al. Chronic obstructive pulmonary disease: current burden and future projections. Eur Respir J. 2006;27:397–412. doi: 10.1183/09031936.06.00025805. [DOI] [PubMed] [Google Scholar]
- 6.Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3:e442. doi: 10.1371/journal.pmed.0030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabe KF, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176:532–55. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- 8.Hancock DB, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Repapi E, et al. Genome-wide association study identifies five loci associated with lung function. Nat Genet. 2010;42:36–44. doi: 10.1038/ng.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pillai SG, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5:e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilk JB, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5:e1000429. doi: 10.1371/journal.pgen.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J, et al. Genomic inflation factors under polygenic inheritance. Eur J Hum Genet. 2011 doi: 10.1038/ejhg.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elks CE, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet. 2010;42:1077–85. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–8. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindgren CM, et al. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet. 2009;5:e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dixon AL, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–7. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 17.Gibson MA, Hughes JL, Fanning JC, Cleary EG. The major antigen of elastin-associated microfibrils is a 31-kDa glycoprotein. The Journal of biological chemistry. 1986;261:11429–36. [PubMed] [Google Scholar]
- 18.Yang J, et al. Rootletin, a novel coiled-coil protein, is a structural component of the ciliary rootlet. J Cell Biol. 2002;159:431–40. doi: 10.1083/jcb.200207153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massaro GD, et al. Retinoic acid receptor-beta: an endogenous inhibitor of the perinatal formation of pulmonary alveoli. Physiol Genomics. 2000;4:51–7. doi: 10.1152/physiolgenomics.2000.4.1.51. [DOI] [PubMed] [Google Scholar]
- 20.Bieganowski P, Shilinski K, Tsichlis PN, Brenner C. Cdc123 and checkpoint forkhead associated with RING proteins control the cell cycle by controlling eIF2gamma abundance. The Journal of biological chemistry. 2004;279:44656–66. doi: 10.1074/jbc.M406151200. [DOI] [PubMed] [Google Scholar]
- 21.Ito K, et al. Decreased histone deacetylase activity in chronic obstructive pulmonary disease. The New England journal of medicine. 2005;352:1967–76. doi: 10.1056/NEJMoa041892. [DOI] [PubMed] [Google Scholar]
- 22.Wu H, et al. Parental smoking modifies the relation between genetic variation in tumor necrosis factor-alpha (TNF) and childhood asthma. Environ Health Perspect. 2007;115:616–22. doi: 10.1289/ehp.9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruse CE, et al. Tumour necrosis factor gene complex polymorphisms in chronic obstructive pulmonary disease. Respir Med. 2007;101:340–4. doi: 10.1016/j.rmed.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 24.Chu HW, et al. Transforming growth factor-beta2 induces bronchial epithelial mucin expression in asthma. The American journal of pathology. 2004;165:1097–106. doi: 10.1016/s0002-9440(10)63371-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu JZ, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42:436–40. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landi MT, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–91. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kathiresan S, et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–41. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clancy RM, et al. Identification of candidate loci at 6p21 and 21q22 in a genome-wide association study of cardiac manifestations of neonatal lupus. Arthritis and rheumatism. 2010;62:3415–24. doi: 10.1002/art.27658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harley JB, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–10. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nature genetics. 2008;40:638–45. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrett JC, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–7. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levy D, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–87. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newton-Cheh C, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–76. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park JH, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010;42:570–5. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat Genet. 2011;43:519–25. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohansal R, et al. The natural history of chronic airflow obstruction revisited: an analysis of the framingham offspring cohort. Am J Respir Crit Care Med. 2009;180:3–10. doi: 10.1164/rccm.200901-0047OC. [DOI] [PubMed] [Google Scholar]
- 37.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med. 1999;159:179–87. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- 38.Talmud PJ, et al. Utility of genetic and non-genetic risk factors in prediction of type 2 diabetes: Whitehall II prospective cohort study. BMJ. 340:b4838. doi: 10.1136/bmj.b4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wacholder S, et al. Performance of common genetic variants in breast-cancer risk models. N Engl J Med. 362:986–93. doi: 10.1056/NEJMoa0907727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson HGR, Mih JD, Krasieva TB, Tromberg BJ, George SC. Epithelial-derived TGF-beta2 modulates basal and wound-healing subepithelial matrix homeostasis. American journal of physiology Lung cellular and molecular physiology. 2006;291:L1277–85. doi: 10.1152/ajplung.00057.2006. [DOI] [PubMed] [Google Scholar]
- 41.Maran C, Tassone E, Masola V, Onisto M. The Story of SPATA2 (Spermatogenesis-Associated Protein 2): From Sertoli Cells to Pancreatic Beta-Cells. Current genomics. 2009;10:361–3. doi: 10.2174/138920209788920976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tewari R, Bailes E, Bunting KA, Coates JC. Armadillo-repeat protein functions: questions for little creatures. Trends in cell biology. 2010;20:470–81. doi: 10.1016/j.tcb.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 43.Pende D, et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. The Journal of experimental medicine. 1999;190:1505–16. doi: 10.1084/jem.190.10.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lillis AP, Mikhailenko I, Strickland DK. Beyond endocytosis: LRP function in cell migration, proliferation and vascular permeability. Journal of thrombosis and haemostasis: JTH. 2005;3:1884–93. doi: 10.1111/j.1538-7836.2005.01371.x. [DOI] [PubMed] [Google Scholar]
- 45.Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends in cell biology. 2001;11:82–8. doi: 10.1016/s0962-8924(00)01898-5. [DOI] [PubMed] [Google Scholar]
- 46.Cowley EA, Linsdell P. Characterization of basolateral K+ channels underlying anion secretion in the human airway cell line Calu-3. The Journal of physiology. 2002;538:747–57. doi: 10.1113/jphysiol.2001.013300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, Abecasis GR. Mach 1.0: Rapid haplotype construction and missing genotype inference. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- 48.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 49.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 51.Cho MH, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42:200–2. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pillai SG, et al. Loci identified by genome-wide association studies influence different disease-related phenotypes in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182:1498–505. doi: 10.1164/rccm.201002-0151OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segre AV, Groop L, Mootha VK, Daly MJ, Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010:6. doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raychaudhuri S, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.