Abstract
The fractional guanine + cytosine (GC) contents of sequenced bacterial genomes range from 13% to 75%. Despite several decades of research aimed at understanding this wide variation, the forces controlling GC content are not well understood. Recent work has suggested that a universal adenine + thymine (AT) mutational bias exists in all bacteria and that the elevated GC contents found in some bacterial genomes is due to genome-wide selection for increased GC content. These results are generally consistent with the low GC contents observed in most strict endosymbiotic bacterial genomes, where the loss of DNA repair mechanisms combined with the population genetic effects of small effective population sizes and decreased recombination should lower the efficacy of selection and shift the equilibrium GC content in the mutationally favored AT direction. Surprisingly, the two smallest bacterial genomes, Candidatus Hodgkinia cicadicola (144 kb) and Candidatus Tremblaya princeps (139 kb), have the unusual combination of highly reduced genomes and elevated GC contents, raising the possibility that these bacteria may be exceptions to the otherwise apparent universal bacterial AT mutational bias. Here, using population genomic data generated from the Hodgkinia genome project, we show that Hodgkinia has a clear AT mutational bias. These results provide further evidence that an AT mutational bias is universal in bacteria, even in strict endosymbionts with elevated genomic GC contents.
Keywords: GC content, endosymbiont, mutational bias
The Smallest Bacterial Genomes Tend to Be Strongly AT Biased, with the Exception of Hodgkinia and Tremblaya
Genome reduction in bacteria is usually associated with a genome-wide shift toward increased AT content (Moran 2002; Bentley and Parkhill 2004; McCutcheon et al. 2009). This pattern is especially pronounced in bacteria that live exclusively in the cytoplasm of host cells; for example, the two most extremely AT-biased bacterial genomes yet reported are from the insect nutritional endosymbionts Candidatus Zinderia insecticola (13.5% GC) (McCutcheon and Moran 2010) and Candidatus Carsonella ruddii (16.5% GC) (Nakabachi et al. 2006). Two mechanisms are thought to explain the reduced GC content of endosymbiont genomes. First, endosymbionts tend to lose genes involved in DNA repair and recombination during genome reduction (Dale et al. 2003; Moran et al. 2008), which increases the load of unrepaired DNA damage. Second, endosymbionts have small effective population sizes and decreased rates of recombination, which reduces the efficacy of selection and allows more slightly deleterious mutations to be fixed by random genetic drift (Moran 1996; Woolfit and Bromham 2003). Combined with what seems to be an AT mutational bias in bacteria lacking DNA repair enzymes (Lind and Andersson 2008), these forces are thought to shift the GC–AT equilibrium toward AT in endosymbiont genomes. Until recently, empirical data from complete bacterial genomes universally supported this hypothesis. Remarkably, the only known exceptions to this trend are from the two bacteria with the smallest reported genomes: Candidatus Hodgkinia cicadicola (hereby referred to as Hodgkinia for simplicity, 144 kb, 58.4% GC; McCutcheon et al. 2009) and Candidatus Tremblaya princeps (Tremblaya, 138 kb, 58.8% GC; McCutcheon and von Dohlen 2011; López-Madrigal et al. 2011). Hodgkinia is a member of the Alphaproteobacteria, a group in which most free-living members have GC-rich genomes and most obligate intracellular members have reduced genomes that show the expected decrease in GC content (McCutcheon et al. 2009). These observations led to the hypothesis that the high GC content of Hodgkinia resulted from the retention of a GC mutational bias that was present in its free-living alphaproteobacterial ancestor (McCutcheon et al. 2009). That the GC content at third positions of 4-fold degenerate codons (GC4) in Hodgkinia is higher than the overall GC content in the genome (62.5% vs. 58.4%) seemed to support this hypothesis, as these positions are expected to be under little or no selection for protein-coding sequence and were therefore thought to more clearly reflect the mutational biases inherent in Hodgkinia's replication machinery (McCutcheon et al. 2009).
Recent Work Suggests that all Bacteria Have an Inherent AT Mutational Bias
Two recent reports provide evidence that an AT mutational bias exists in all bacteria (Hershberg and Petrov 2010; Hildebrand et al. 2010). The authors of both papers conclude that selection for increased GC content, or a selection-like process such as biased gene conversion, is the most likely explanation for the diverging patterns of AT-biased mutation and GC-biased substitution observed in most bacterial genomes (Hershberg and Petrov 2010; Hildebrand et al. 2010). Both papers also single out Hodgkinia as an outlier and a possible exception to this rule (Hershberg and Petrov 2010; Hildebrand et al. 2010). To help clarify the roles of mutational biases and selection on the GC content of the Hodgkinia genome, we sought to determine the direction of Hodgkinia's mutational bias (if any) from existing population data generated during genome sequencing.
Single Nucleotide Polymorphisms in the Hodgkinia Genome Reveal an AT Mutational Bias
The published Hodgkinia genome was generated by combining samples from 10 wild-caught individuals of the cicada Diceroprocta semicincta (McCutcheon et al. 2009). We reasoned that it might be possible to calculate mutational patterns from these population genomic data. We first reconfirmed that the pooled sample was from a single species of cicada by verifying a low level of sequence polymorphism in its mitochondrial cytochrome c oxidase I (COI) gene (about 0.6% of 815 sites were polymorphic, well within the 1–2% divergence levels typically seen in conspecific pairs of animal COI sequences; Hebert et al. 2003). We then calculated the number of single nucleotide polymorphisms (SNPs) in the Hodgkinia genome falling into all possible nucleotide change categories and found that the majority of the mutations (115 of 179 or 64%) were in the GC to AT direction. (The Tremblaya genome was generated from only three lab-reared insects, and no high-quality SNPs were observed in these data.)
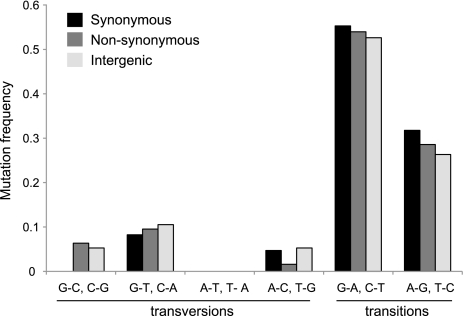
To unambiguously assign a mutational direction to these SNPs, we used a draft Hodgkinia genome assembly from a closely related but undescribed cicada species (referred to here as the cryptic species) as an outgroup to verify the ancestral state of each position where a SNP was identified (for a complete description of the methods, see Supplementary Materials online). The pairwise nucleotide divergence between partial mitochondrial COI sequences from Diceroprocta semicincta and the cryptic species was 3.5%. Of the 179 SNPs initially identified, 12 were not covered by contigs from the cryptic species. These 12 were removed from the data set, resulting in 167 SNPs in which the direction of mutation could be confidently determined (fig. 1 and table 1; table S1, Supplementary Materials online). The expected equilibrium GC content (GCeq) given the mutational patterns observed in the polarized data is 42%, significantly lower than the observed genomic value of 58% (table 1).
FIG. 1.—
The majority of SNPs in the Hodgkinia genome are G to A or C to T transitions and collectively show a pronounced AT mutational bias. SNPs are shown as a percentage of the total number in each category (synonymous, nonsynonymous, and intergenic sites).
Table 1.
Raw SNP counts, mutation rates, and expected GC equilibrium values for synonymous (S), nonsynonymous (NS), and intergenic (IG) sites
| Counts |
Rates |
Current GC | GCeq | |||
| Number of GC → AT | Number of AT → GC | rGC→AT | rAT→GC | |||
| S | 54 (43–67) | 31 (22–40) | 1.9 × 10−3 | 1.9 × 10−3 | 63 | 50 (40–58) |
| NS | 40 (29–51) | 19 (12–26) | 8.0 × 10−4 | 4.9 × 10−4 | 56 | 38 (27–50) |
| IG | 12 (7–19) | 6 (2–10) | 2.6 × 10−3 | 1.7 × 10−3 | 56 | 39 (17–56) |
| All | 106 (89–122) | 56 (44–68) | 1.3 × 10−3 | 9.5 × 10−4 | 58 | 42 (36–49) |
Note.—The numbers in parentheses are 95% confidence intervals; significant values are bolded. The values do not sum to 167 because five SNPs did not alter the GC content (i.e., a G to C mutation).
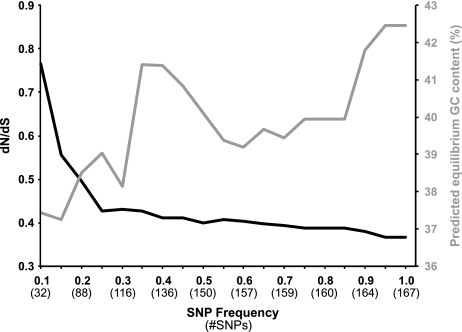
To estimate the strength of selection acting on these SNPs, we calculated the ratio of nonsynonymous and synonymous polymorphisms per nonsynonymous and synonymous site (dN/dS) and found evidence for weak purifying selection (dN/dS = 0.37). This value is slightly lower but consistent with values reported previously for populations of clonal bacterial pathogens, which range from 0.45 to 0.64 (Hershberg and Petrov 2010). Differences in the magnitude of dN/dS need to be interpreted with caution in this situation, as this measure assumes that sequence polymorphisms are fixed substitutions between species and not intraspecific mutations segregating in a population (Kryazhimskiy and Plotkin 2008). Some SNPs in the pooled data set include those at high frequencies, and we assume that these SNPs have been segregating in the population for some time and may have been exposed to significant levels of purifying selection. To assess whether we could measure differences in 1) the levels of purifying selection and 2) the magnitude of the AT mutational bias for SNPs partitioned into different frequency bins, we calculated dN/dS and GCeq values for SNPs binned at 0.1 frequency intervals (fig. 2). As 10 individuals were pooled for sequencing, an ideal experiment would reveal SNPs clustering at frequencies of 0.1, 0.2, 0.3, and so on up to 0.9. We did not observe an increased number of SNPs near these expected frequencies, and attribute this nonideal behavior to numerous potential experimental and computational artifacts (for a full discussion, see the Supplementary Materials online). Nevertheless, these results confirm that the SNPs present in the population at lower frequencies have been exposed to less purifying selection (indicated by a higher dN/dS value) and are more strongly AT biased than SNPs present at higher frequencies (fig. 2). For example, the GCeq content of the Hodgkinia genome is calculated to be 37% using only SNPs called at a frequency of 0.1 or less, lower than the 42% calculated when all SNPs are included. The true Hodgkinia GCeq is therefore probably closer to 37%, or perhaps even lower. From these data, we conclude that Hodgkinia has an AT mutational bias.
FIG. 2.—
Plotting GCeq (gray line) and dN/dS (black) at different SNP frequency cutoffs shows that SNPs present at lower frequencies (which are likely more recent mutations) have been subjected to less selection and are more AT biased.
Why Does Hodgkinia Have an Elevated Genomic GC Content?
Although our data clearly show an AT mutational bias in Hodgkinia, they do not directly implicate the force(s) responsible for the disparity between the observed patterns of mutation and substitution. Hershberg and Petrov (2010) and Hildebrand et al. (2010) suggest selection, or a selection-like process such as biased gene conversion, as the force driving the difference in bacteria. In bacteria, biased gene conversion involves horizontal gene transfer, recombination, and DNA repair-based mechanisms (Rocha and Feil 2010). As Hodgkinia encodes no gene homologs capable of these processes (McCutcheon et al. 2009), biased gene conversion seems unlikely to be responsible for Hodgkinia's elevated GC content. Therefore, it appears that an unidentified selective force (or forces) is the most likely explanation for the GC compositional bias in the Hodgkinia genome, although other explanations cannot be ruled out given the present data. For example, it is possible that the GC content in Hodgkinia is mostly driven by mutational patterns, and that it recently underwent a shift from a GC to an AT mutational bias. Were this true, we would have had to have measured the mutational pattern soon after the change from a GC to an AT bias, but before this shift would have had the chance to alter the genome-wide nucleotide composition. This seems unlikely based simply on parsimony. Rather, given the results of Hershberg, Hildebrand, and coworkers, we favor the explanation that Hodgkinia has, and has always had, an inherent AT mutational bias (Hershberg and Petrov 2010; Hildebrand et al. 2010).
Our results seem to present a paradox in the way that the population genetics of endosymbionts are normally considered. The prevailing view that endosymbionts have less efficacious selection, resulting from reduced effective population sizes (Moran 1996; Andersson and Kurland 1998; Woolfit and Bromham 2003; Fares et al. 2004), fits well with some features of Hodgkinia genome, in particular with its tiny size and overall rapid rate of sequence evolution. The disparity between the Hodgkinia's AT-biased mutational pattern and GC-biased genome does not fit easily into this framework because these results seem to require either an atypically large effective population size for Hodgkinia or an unusually large selection coefficient for each individual AT–GC polymorphism in the population, or some combination of the two. It is possible that the population size of the host cicada is large and thus inflates the effective population size of Hodgkinia; theoretical work has shown that host population size can have large effects on mutation accumulation in Buchnera aphidicola in the context of its symbiosis with aphids (Rispe and Moran 2000). Why G or C nucleotides would be globally favored over A or T nucleotides is unclear and is an interesting area of future study.
Hodgkinia is found as a symbiont throughout the cicada lineage (data not shown), and it will be of interest to examine the GC contents and mutational biases of Hodgkinia across the diversity of cicadas. If GC-poor lineages of Hodgkinia are found, then it may be possible to narrow the list of possible selective forces responsible for the elevated GC levels in Hodgkinia from D. semicincta, by considering factors such as the environmental conditions and population structures of the insect hosts. The mutational results reported here would predict that a lineage of Hodgkinia in which the selective restraints on elevated GC content were severely reduced or eliminated would have a genomic GC content as low as, or possibly lower than, 37%.
Materials and Methods
SNPs were identified using both Roche 454 GSmapper and SWAP454 (Brockman et al. 2008) software. Genome-wide dN/dS and GCeq calculations were performed as described (Hershberg and Petrov 2010). The confidence intervals in table 1 were estimated using the rpois and quantile functions of R (2.13.0) by generating 1,000 value Poisson distributions with means equal to the mutation counts; rate, equilibrium, and 95% confidence interval calculations were performed on these distributions. Full Materials and Methods, including a detailed description of how the SNPs were polarized, are included in the Supplementary Materials online.
Supplementary Materials
Supplementary Materials, figure S1, and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The Hodgkinia genome sequencing project was supported by the National Science Foundation Microbial Genome Sequencing award 0626716 to Nancy A. Moran. The reanalysis of these data presented in this paper was supported by the National Science Foundation Montana EPSCoR grant EPS-0701906.
References
- Andersson SGE, Kurland CG. Reductive evolution of resident genomes. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- Bentley SD, Parkhill J. Comparative genomic structure of prokaryotes. Annu Rev Genet. 2004;38:771–791. doi: 10.1146/annurev.genet.38.072902.094318. [DOI] [PubMed] [Google Scholar]
- Brockman W, et al. Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res. 2008;18:763–770. doi: 10.1101/gr.070227.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale C, Wang B, Moran N, Ochman H. Loss of DNA recombinational repair enzymes in the initial stages of genome degeneration. Mol Biol Evol. 2003;20:1188–1194. doi: 10.1093/molbev/msg138. [DOI] [PubMed] [Google Scholar]
- Fares MA, Moya A, Barrio E. GroEL and the maintenance of bacterial endosymbiosis. Trends Genet. 2004;20:413–416. doi: 10.1016/j.tig.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Hebert PDN, Ratnasingham S, de Waard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond B Biol Sci. 2003;270:S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 2010;6:e1001115. doi: 10.1371/journal.pgen.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand F, Meyer A, Eyre-Walker A. Evidence of selection upon genomic GC-content in bacteria. PLoS Genet. 2010;6:e1001107. doi: 10.1371/journal.pgen.1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S, Plotkin JB. The population genetics of dN/dS. PLoS Genet. 2008;4:e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Andersson DI. Whole-genome mutational biases in bacteria. Proc Natl Acad Sci U S A. 2008;105:17878–17883. doi: 10.1073/pnas.0804445105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Madrigal S, Latorre A, Porcar M, Moya A, Gil R. Complete genome sequence of “Candidatus Tremblaya princeps” strain PCVAL, an intriguing translational machine below the living-cell status. J Bacteriol. 2011;193:5587–5588. doi: 10.1128/JB.05749-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, McDonald BR, Moran NA. Origin of an alternative genetic code in the extremely small and GC–rich genome of a bacterial symbiont. PLoS Genet. 2009;5:e1000565. doi: 10.1371/journal.pgen.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol. 2010;2:708–718. doi: 10.1093/gbe/evq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, von Dohlen CD. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol. 2011;21:1366–1372. doi: 10.1016/j.cub.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci U S A. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. Microbial minimalism: genome reduction in bacterial pathogens. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, et al. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- Rispe C, Moran NA. Accumulation of deleterious mutations in endosymbionts: Muller's ratchet with two levels of selection. Am Nat. 2000;156:425–441. doi: 10.1086/303396. [DOI] [PubMed] [Google Scholar]
- Rocha EPC, Feil E. Mutational patterns cannot explain genome composition: are there any neutral sites in the genomes of bacteria? PLoS Genet. 2010;6:e1001104. doi: 10.1371/journal.pgen.1001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfit M, Bromham L. Increased rates of sequence evolution in endosymbiotic bacteria and fungi with small effective population sizes. Mol Biol Evol. 2003;20:1545–1555. doi: 10.1093/molbev/msg167. [DOI] [PubMed] [Google Scholar]