Abstract
One of the key goals of structural DNA nanotechnology is to build highly ordered structures self-assembled from individual DNA motifs in 1D, 2D, and finally 3D. All three of these goals have been achieved with a variety of motifs. Here, we report the design and characterization of 1D nanotubes and 2D arrays assembled from three novel DNA motifs, the 6-helix bundle (6HB), the 6-helix bundle flanked by two helices in the same plane (6HB+2), and the 6-helix bundle flanked by three helices in a trigonal arrangement (6HB+3). Long DNA nanotubes have been assembled from all three motifs. Such nanotubes are likely to have applications in structural DNA nanotechnology, so it is important to characterize their physical properties. Prominent among these are their rigidities, described by their persistence lengths, which we report here. We find large persistence lengths in all species, around 1-5 μm. The magnitudes of the persistence lengths are clearly related to the designs of the linkages between the unit motifs. Both the 6HB+2 and the 6HB+3 motifs have been successfully used to produce well-ordered 2D periodic arrays via sticky-ended cohesion.
Keywords: DNA, Nanotechnology, Self-Assembly, Multi-Helix Bundle, Nanotubes, Arrays, Persistence Length
INTRODUCTION
The unique interaction between the purines and pyrimidines of DNA (G pairing with C, and A pairing with T) is central to the ability of DNA to serve as material for genetic information storage. In a related fashion, its simplicity, predictability of structure, specificity of molecular recognition, and programmability also make DNA an outstanding candidate for structural and mechanical elements in nanotechnology. Structural DNA nanotechnology entails the design, construction, and characterization of objects, devices and arrays from branched DNA molecules on the nanometer scale.1a It has attracted an increasing amount of attention from scientists and engineers in the past decade.1b
One of the most successful aspects of DNA nanotechnology is the construction of 1D nanotubes and 2D periodic/aperiodic arrays (both finite and infinite).2-5 The chemical and structural properties of DNA nanotubes make them an interesting material, with potential applications in biology and engineering. For instance, DNA nanotubes can be functionalized by attaching other biomacromolecules (proteins) and nanoparticles to them.6 In addition, stiffer nanotubes have the potential to be used as replacements for actin filaments and other cytoskeletal components, both for biological and technological purposes. To implement those applications, it is important to characterize their physical properties. Rigidity is a key property, described by the persistence length. To date, various DNA nanotubes7-17 have been constructed, either by folding 2D arrays into cyclic species or by assembling designed closed species of DNA helix bundles. Some of these nanotubes have been metalized to fabricate nanowires or used as templates for the 3D geometrical arrangement of nanoparticles.18 Persistence lengths of nanotubes have been measured and reported in only a few cases8-9. Moreover, tube-opening has been observed for most of the cases in which DNA nanotubes result from the cyclization of 2D arrays. For some nanotube designs, enzymatic ligation can be used to prevent tube-opening.9 Alternatively, it is possible to produce intrinsically cyclic species, most prominently the six-helix bundle (6HB) motif. 1D tubes of 6HB have been reported elsewhere12 but the persistence length of the 6HB motif has not been characterized previously.
Here we report the design, construction and characterization of three DNA nanotubes consisting of three DNA motifs: 6HB, 6HB+2, and 6HB+3. Our cyclic DNA six-helix bundle (6HB) design consists of six DNA duplexes, in which every two adjacent duplexes are connected to each other at two crossover points; our 6HB+2 and 6HB+3 motifs contain, respectively, two and three extra double helices fused to the surface of the 6HB motif.
These new nanotubes are of novel and significant interest. First, they extend what is fundamentally a 1D motif into two and three dimensions. Second, they prototype the reinforcement of DNA motifs by the addition of extra domains. Third, they prototype the addition of other species of DNA domains (e,g., ref. 19a-c) to a motif that we show here is structurally robust, similar to the motivation for ref. 19. Fourth, they are clearly shown to be extendable to two and three dimensions.
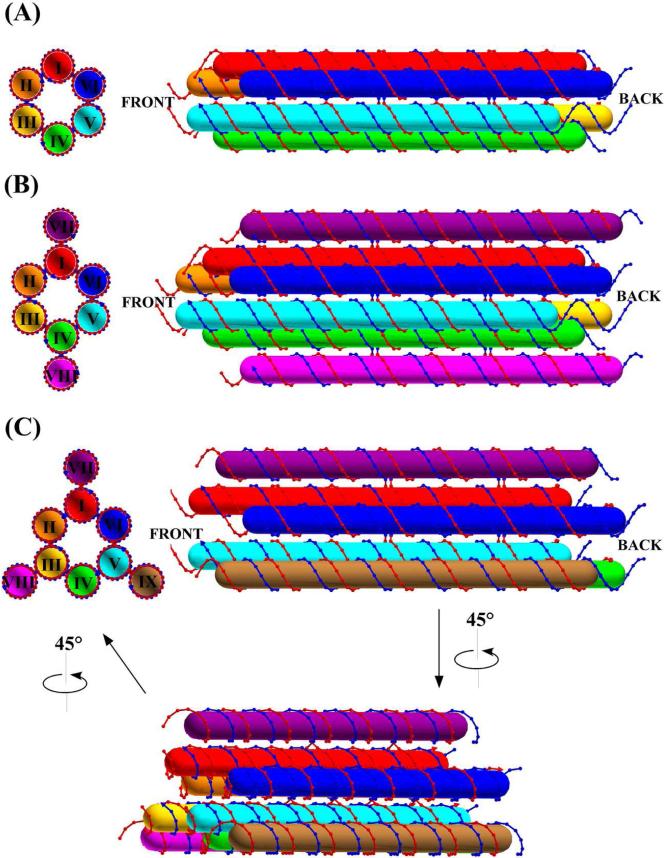
6HB is a designed cyclic arrangement of six DNA duplexes (Figure 1A). Two adjacent duplexes are held together through two crossovers to form a planar DAE double crossover20 molecule with 21 nucleotide pairs between crossovers.12 Thus, there are six DAE components in each 6HB. The dihedral angle of two adjacent DAE planes is 120°, which is achieved by designing 7 base pairs (2/3 of a 10.5-fold double helical turn), between the crossovers of two adjacent DAE components. Ideally, six duplexes form a hexagonal arrangement with a hollow channel (diameter ~2 nm) in the middle of the 6HB motif. In addition to the direct formation of the 6HB molecule from a group of DNA single strands, we recently reported the formation of 6HB molecules from the lateral cohesion of a pair of bent three-helix molecules (BTX), thus potentially facilitating the capture of a nanorod.20a,20b Adding two and three extra DNA double helices, respectively, to the 6HB motif results in motifs that we term 6HB+2 (Figure 1B) and 6HB+3 (Figure 1C). Two extra duplexes are attached to opposite duplexes in the 6HB motif to form the 6HB+2; three extra helices attached in a trigonal arrangement to the 6HB motif to form the 6HB+3 motif (Figure 1). The extra duplexes in the latter two motifs are meant to add rigidity to the 6HB motif. Ideally the four helices in the vertical plane of the 6HB+2 motif (Figure 1B) are coplanar, and the three extra helices in the 6HB+3 motif (Figure 1C) are organized with 3-fold symmetry about the hollow channel. Thus, in principle, the individual 6HB, 6HB+2 and 6HB+3 motifs have 6-fold, 2-fold and 3-fold symmetry axes, respectively, if the sequences and crossover positions are not taken into account (Figure 1).
Figure 1. Schematic Drawings of the Helix Bundle Motifs.
Cross-sectional views of DNA motifs, in which the duplexes are indicated by Roman numerals, are shown on the left in each panel, and side views are shown on the right in each panel. Cylinders in different colors represent different DNA duplex domains in each motif. The ‘Back’ and ‘Front’ are indicated as the ends to be used for sticky-ended cohesion. (A) 6HB. (B) 6HB+2. (C) 6HB+3. 6HB+3 rotates 45° from the side view as indicated to generate the view shown in the bottom of (C); the relationship of this rotated view to the cross-sectional image is also indicated. Duplexes overlap in side views, for example, duplex II is further from the viewer than duplex VI. Thus, duplex II is invisible to viewer in the side view of (C).
It is easy to assemble long nanotubes from bundles with a specific number of components, by adding complementary sticky ends to every helix: those at the front end are designed to be complementary to those at the back end, as shown in structural schematics in Figure 2A-C and sequence schematics in Figure S1A-C. The most straightforward design is to make the two sticky ends on the same duplex complementary to each other, without rotating them through a 'phase angle'. Thus, helix bundles repeat along helical axes by translating the length of one motif (73 nucleotide pairs or ~ 24 nm) in both directions without rotation (Figure 2A-C). In all three cases, sticky ends are 5’ overhangs consisting of 6 nucleotides. The specific sequences of the three motifs used in 1D nanotubes are illustrated in Figure S1A-C. In principle, other designs than simple translation of 1D nanotube repeats are possible, leading to two-fold (21) screw axes (6HB or 6HB+2), three-fold (31 or 32) screw axes (6HB or 6HB+3)) or six-fold (61 or 65) screw axes (6HB), but these have not been built. None of the 1D nanotubes treated here result from cyclic closure of 2D arrays. Nevertheless, all three motifs can interact with themselves to form 2D periodic arrays by programming the sticky ends on both ends of each helix. A large number of DNA 2D arrays have been assembled from branched DNA motifs to date2-4, 12, 21, 22 and many of them contain DNA motifs in which all DNA duplexes are coplanar, 2-4 that is, the directions of propagation associated with sticky-ended cohesion share the same plane. All of the motifs reported here are inherently 3D motifs because they can be linked to copies of themselves via three non-coplanar helices. It is only a little more complicated to design 2D arrays than 1D nanotubes. 2D arrays of 6HB have been published elsewhere.12, 23 By assigning proper sticky ends to four duplex domains I, IV, VII and VIII (Figures 1B and S2A), the 6HB+2 motif can self-assemble into a 2D array through double cohesion.22-23 Ideally those four duplex domains are coplanar. In the design, sticky ends are four nucleotides in length and their specificities are as follows: sticky ends at the front ends of domains I and VII are designed to complement those on the back ends of domains VIII and IV respectively; similarly sticky ends at the front ends of domains VIII and IV are designed to complement those on the back ends of I and VII respectively. We also designed a 2D array of 6HB+2 through single cohesion. In this case, only two duplex domains contain sticky ends: sticky ends on the front ends of domains VII and VIII complement those on the back ends of domains VIII and VII respectively (Figures 1B and S2B). Duplex domains that are not involved in the self-assembly have blunt ends on both their front and back ends. The presence of stacked blunt ends has been shown to stabilize cohesion.24 The specific sequences of 6HB+2 used in 2D arrays are illustrated in Figure S2. Sticky end A on helix VII is complementary to A’ on the other end of helix VII, B and B’ are related in an analogous fashion, and so on. The repeat of 6HB+2 in either of the two dimensions is a combination of two simple translations: one is along the helix axis of the duplex; the other is along the direction normal to the helical axis in the plane formed by the four coplanar helices (Figure 1B)
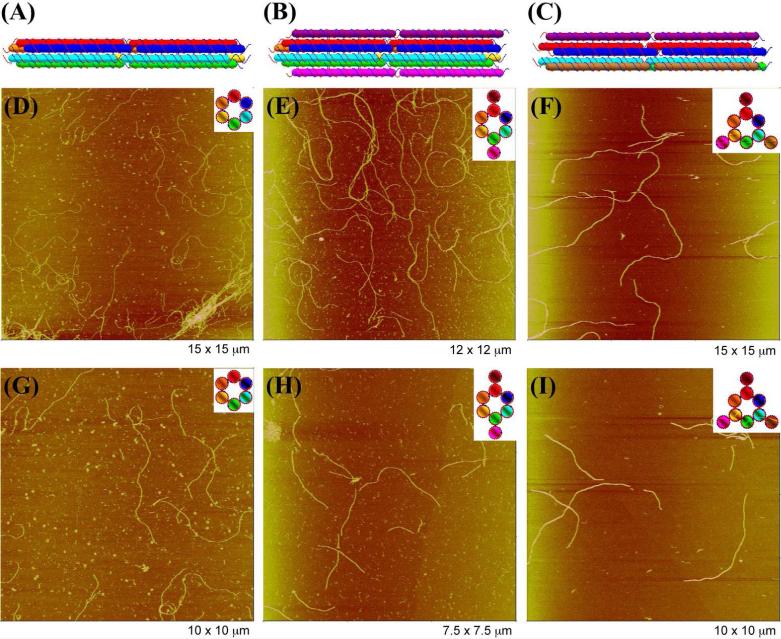
Figure 2. Design and AFM images of 1D Linear Nanotubes.
Two copies of each DNA motif are shown in panels (A)-(C). Six nucleotide-pair sticky ends on the front of each duplex are complementary to those on the back of the same duplex. Through sticky-ended cohesion, individual DNA motifs interact with each other to form linear DNA nanotubes in a head-to-tail manner. (A) 6HB. (B) 6HB+2 (C) 6HB+3. AFM images of 1D linear nanotubes are shown in (D)-(I). Schematic drawings of cross-sectional views of each DNA motif are inserted in the upper corner of each panel. (D) 6HB. (E) 6HB+2. (F) 6HB+3. (G) Zoomed image of 6HB nanotubes. (H) Zoomed image of 6HB+2 nanotubes. (F) Zoomed image of 6HB+3 nanotubes. The sizes of each image are shown at the bottom right corner of each panel.
Similar to 6HB+2, 6HB+3 is derived from 6HB by attaching three DNA double-helices (VII, VIII and IX) to 6HB (Figure 1C). Ideally the three planes defined by helices I and VII, by III and VIII, and by V and IX, form dihedral angles of 120° with each other. Each plane contains one of the three independent vectors (directions of propagation). In the first plane, the sticky end at the front end of duplex I pairs with that at the back end of duplex VII, while the sticky end at the front end of duplex domain VII pairs with the back end of duplex I (Figure 1C and S3). The same arrangement applies to the other two planes. Thus, there is a 2-fold screw axis (21), which is absent in the designs of 2D arrays of 6HB and 6HB+2, in each of the three planes mentioned above (Figure 1C). In principle, a 3D crystalline array would result if sticky ends in all three planes interact with each other in the way described above. If the sticky ends in one of the three planes are made to be blunt, then a 2D array will result. Three different such planes can be organized. The specific sequences of 6HB+3 are illustrated in Figure S3. Sticky end A interacts with A’, B interacts with B’, and so on. The repeat of 6HB+3 in each of the three directions is achieved by a combination of translating along the helix axis of the duplex and a 180° rotation around the 2-fold screw axis (21), as shown in Figure 1C.
Here, we show that the stiffness of nanotubes constructed from these motifs depends critically on the relative positions of sticky ends. When the sticky ends are staggered, the duplexes external to the basic 6HB motif enhance the stiffness as expected. In addition to 1D arrangements, we show that the new motifs are capable of forming 2D arrays.
MATERIALS AND METHODS
Design, Synthesis, and Purification
DNA strand sequences were designed using the program SEQUIN.25 The DNA strands with Fluorescein (FAM) were synthesized on an Applied Biosystems 394, removed from the support, and deprotected using routine phosphoramidite procedures. Other strands were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). All strands have been purified by denaturing gel electrophoresis (PAGE); bands were cut out of 15–20% denaturing gels and eluted in a solution containing 500 mM ammonium acetate, 11 mM magnesium acetate, and 1 mM EDTA. All strands and buffers were finally filtered by 0.22 μm Ultrafree®-MC centrifugal filter units (Millipore Corporation).
Formation of Hydrogen-Bonded Complexes and Helix Bundle Arrays
1D Nanotubes
A single tile was used in each 6-helix bundle system. The strands of each tile were mixed stoichiometrically as estimated by OD260 and dissolved to 0.05–0.1 μM in either 1X TAE/Mg2+ buffer (40 mM Tris-HCl, 20 mM Acetic Acid, 2 mM EDTA, 12.5 mM magnesium acetate, pH 8.0) or in 1X HEPES-Na buffer (10 mM HEPES - Na, 11 mM magnesium chloride, 1 mM EDTA, pH 7.8). The solutions were annealed from 90 °C to room temperature (RT) over 48 hours in a 2-liter water bath insulated in a Styrofoam box.
2D Arrays
Helix bundles were formed by mixing a stoichiometric quantity of each strand as estimated by OD260. The final DNA concentration for the 6HB+2 motif in the double cohesion system was 0.5 μM in 10 mM HEPES - Na, 26 mM magnesium chloride, 1 mM EDTA, pH 7.8. The final DNA concentration for the 6HB+2 motif in the single cohesion system was 0.5 μM in 10 mM HEPES - Na, 11 mM magnesium chloride, 1 mM EDTA, pH 7.8. The final DNA concentration for the 6HB+3 motif was 0.5 μM in 10 mM HEPES - Na, 11 mM magnesium chloride, 1 mM EDTA, pH 7.8. Mixtures were annealed from 90°C to RT during 48 hours in a 2-litre water bath insulated in a Styrofoam box. They were then put in a 1-liter water bath in a stainless steel thermos cup and slowly cooled down from 45°C to RT over 4 days.
Preparation of PVP-Coated Glass8,26
Microscope slides (3 × 1 in) and cover slips (18 mm sq) were immersed in 1L of 1M NaOH solution at room temperature for 1 hour, washed thoroughly with de-ionized (DI) H2O and immersed in 1L of 1 % v/v acetic acid solution at room temperature for 2 hours. These treated slides and cover slips were rinsed again with DI H2O, then, silanized at room temperature for at least 36 hours in 1L of a solution containing 1% v/v 3-(trimethoxysilyl)propylmethacrylate (Aldrich) in 1% v/v acetic acid; the cover slips were then rinsed with DI H2O. Finally, slides and coverslips were incubated in 1L of polymer solution [4% w/v polyvinylpyrrolidone (PVP) (MW=360,000) (USB Corp.) in water (the solution was heated to encourage dissolution of PVP) with 2.5 mL of 10 % w/v ammonium persulfate (APS) (Fisher Scientific) and 250 μl of N,N,N’,N’,-tetramethylethylenediamine (TEMED) (Fisher Scientific)] at 80 ± 1°C for 18 hours. PVP-coated slides and cover slips were rinsed with DI H2O to remove the uncrosslinked portion of the polymer and stored in DI H2O. PVP Coating was stable for at least two weeks.
Preparation of Oxygen Scavenging System27
For live fluorescence microscopy it is essential to scavenge oxygen so as to limit photodamage. The most convenient method for doing this is using a glucose oxidase/glucose/catalase mixture (OS mix). The component enzymes are stored as 100X stocks at - 80 °C and used for ~ 2 hours after mixing. It is important to keep the OS mix in a sealed tube on ice. The principle by which the mix scavenges oxygen is as follows:
A 10X OS mix (0.35 mg/ml catalase, 2 mg/ml glucose oxidase, 45 mg/ml glucose, 5% β-mercaptoethanol) is prepared on ice and stored in a sealed tube on ice. For optimal results, a fresh 10X stock is prepared after ~ 2 hours.
Preparation of Samples for Fluorescence Microscopy
Annealed DNA samples were stored at room temperature. Before microscope imaging, fresh OS mix was added to the DNA sample. 3 μL of the DNA sample in 1X OS mix (0.035 mg/ml catalase, 0.2 mg/ml glucose oxidase, 4.5 mg/ml glucose, 0.5% β-mercaptoethanol) was then deposited onto a slide, covered with a cover slip and sealed with epoxy or paraffin. The distance between the surfaces of the slide and the cover slip was ~ 10 μm. When nanotube length (L > 3 m) and stiffness (p ≥ 1 m) allowed, PVP-coated slides and coverslips were used to constrain nanotubes to diffuse in 2D. The thickness of the sample solution betweenthe PVP coatingswas 2-3 μm.
Fluorescence Microscopy
Samples were imaged on an inverted microscope (IX 70, Olympus) with a 100X/1.40 NA oil immersion objective. Blue light was filtered from a mercury arc lamp through an interference filter (475nm) for excitation. The emitted fluorescence passed through a dichroic mirror (505 DRLP) and a green interference filter (525nm). Raw images were captured and stored to a Mac mini via a cooled RETIGA EXi fast 1394 CCD camera (QImaging Corp.) using the corresponding software Qcapture (QImaging Corp.). Images were processed in ImageJ (NIH Image in Java: http://rsb.info.nih.gov/ij/).
Persistence Length Measurement
Persistence length, p, was measured in two different ways. In most cases it was possible to confine the nanotubes to 2D (see Preparation of Samples, above). Then, the persistence length was determined by extracting the end-to-end distance, R, and contour length, L, from fluorescence images of several different nanotubes (N>4) in many distinct conformations (<n>=23) and fitting the data to the 2D Kratky-Porod Model:9, 28
Each point on the <R2> versus L plot was derived from a set of n conformations observed for a single DNA nanotube. A complete data set for a given tube type consisted of N such points.
Uncertainty in the persistence length thus determined was estimated using a bootstrap method: From every set of n conformations of a given nanotube, k conformations were selected at random with replacement (thus a particular conformation might be picked twice or not at all). The average end-to-end distance of the selected conformations, <R2>k, was computed. This value typically differed only slightly from the end-to-end distance, <R2>n, of the “original” set. A least-squares fit of the Kratky-Porod model to a plot of <R2>k versus L was performed and, accordingly, resulted in a slightly different estimate of the persistence length. For each type of nanotube, this process was repeated 1000 times using randomly selected sets of the same size as the original (that is, k = n). The resulting persistence length estimates were normally distributed about the one derived from the original data set and their standard deviation was taken as the uncertainty (67% confidence interval) in the measurement.
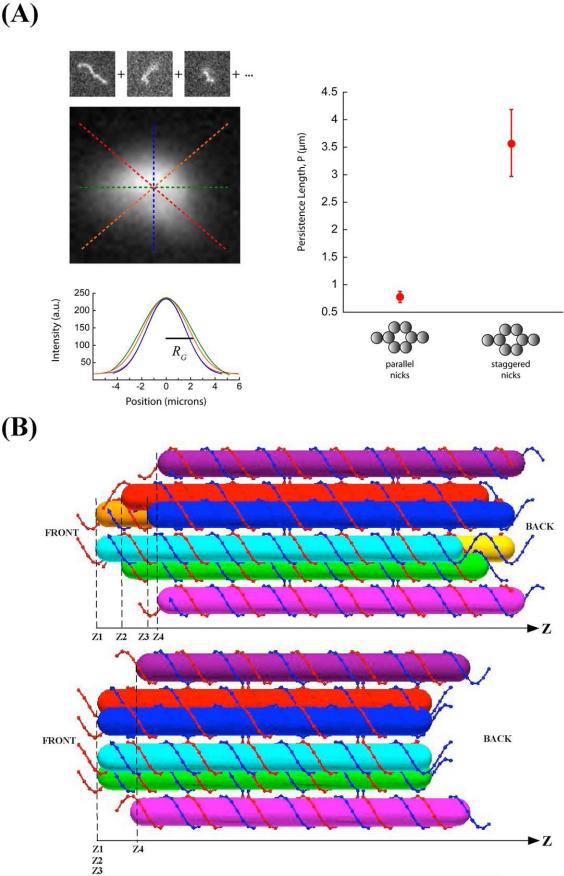
It was not possible to confine the 6HB+2 (parallel) nanotube (in which six out of eight sticky ends were at the same Z position along the helical axis) to 2D, so a different approach to estimating persistence lengths was taken. Movies containing several hundreds of frames were taken as a freely diffusing nanotube was manually tracked in the focal plane of a microscope. All independent images of a given nanotube were aligned according to their centers of mass and added together to generate a single composite image of the nanotube. The intensity profile along a straight line through the center of mass of the composite image fit well to a normalized Gaussian distribution, the standard deviation, σ, of which was taken as a measure of the nanotube's radius of gyration (RG). The contour length (L) was measured from the most extended conformation observed. These parameters were related to the persistence length (p) of the nanotube using the following two equations: RG=nvp/(6)1/2 and L=np, where v=0.6 for a 3D self-avoiding random walk. The average persistence length of the 6HB+2 nanotube design estimated in this manner was 0.78 ± 0.1 um, based on analysis of two different nanotubes using 400 independent images of each. These data are discussed below, where they are displayed in Figure 6.
Figure 6. Nanotubes made of molecules with parallel and staggered sticky-ends are significantly less stiff than those made of molecules with staggered sticky-ends.
(A) The right panel shows the persistence lengths of the two versions of 6HB+2 (parallel and staggered). The left panel illustrates the measurement of 6HB+2 (parallel) nanotubes, which could not be confined to 2D. Images of independent conformations of such a nanotube diffusing in 3D were aligned about their center of fluorescence and superimposed. The composite image was used to estimate the radius of gyration by averaging the width of gaussian fits to line profiles through the center. See Materials and Methods for details. (B) Positions of Sticky Ends in the Design of Nanotubes. Cylinders in different colors represent different DNA duplex domains in each motif. Top panel: The “front” end of 6HB+2 contains eight sticky ends that are at four different positions along the Z axis (helix axis), indicated by Z1, Z2, Z3, and Z4. This is 6HB+2 (staggered). Bottom panel: The “front” end of 6HB+2 contains eight sticky ends which are at two different positions along the Z axis (helix axis), in this design, Z1 = Z2 = Z3. This is 6HB+2 (parallel).
AFM Imaging
Helix bundles arrays were imaged in tapping-mode AFM in buffer. 5-7 μL of the annealed sample was spotted on freshly cleaved mica (Ted Pella, Inc.). An additional 25 μL of fresh buffer was added to both the mica and to the liquid cell. All AFM imaging was performed on a NanoScope IV MultiMode SPM (Digital Instruments), using commercial cantilevers with Silicon Nitride (Si3N4) tips (sharpened).
RESULTS
1D Nanotubes
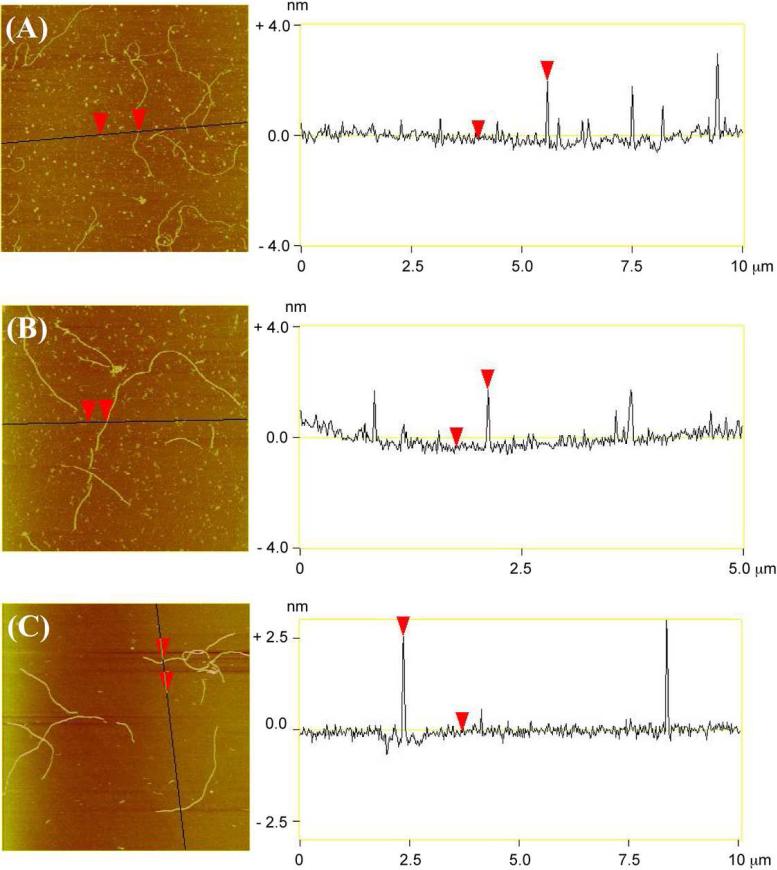
Figure 2 shows AFM images of 1D nanotubes self-assembled from individual 6HB motifs and from its two variants. It is evident that DNA helix bundles themselves self-assemble readily into 1D nanotubes. Figures 2D-F show evidence that all three motifs form linear 1D nanotubes. Typical nanotubes are shown in the zoomed pictures (Figure 2G-I). Those pictures show clearly that all three motifs have been assembled to form unbranched linear nanotubes. The average contour length of the 6HB, 6HB+2 and 6HB+3 DNA nanotubes are 2.8, 2.5 and 4.5 μm respectively (Figure S4). The measured heights of the 6HB, 6HB+2 and 6HB+3 DNA nanotubes are 2.1 ± 0.2, 2.1 ± 0.2, 2.6 ± 0.3 nm respectively. The height analysis of the 1D nanotubes is shown in Figure 3. The flattening of nanotubes when deposited on the mica surface and the force applied to nanotubes by AFM tips during imaging lead to the measured heights of nanotubes being smaller than the calculated ones. However, according to our design, the heights of nanotubes assembled from 6HB and 6HB+2 motifs are in principle the same and smaller than that of nanotube assembled from 6HB+3 when lying on the mica surface (Figure 1). Our observations are consistent with this design.
Figure 3. Cross-Sectional Analysis of 1D arrays.
The vertical distance between the two red arrows is the measured height of nanotubes. (A) 6HB. The average height of 6HB 1D arrays is 2.1 ± 0.2 nm. (B) 6HB+2. The average height of 6HB+2 1D arrays is 2.1 ± 0.2 nm. (C) 6HB+3. The average height of 6HB+3 1D arrays is 2.6 ± 0.3 nm. The averaged heights are based on five measurements of each nanotube.
Figure 4 shows fluorescence images of all three types of nanotubes. Several DNA single strands in three motifs are labeled with fluorescein (FAM). Nucleotides that are modified with fluorescein are shown in magenta and in larger font (Figure S1A-C). Snapshots and movies were taken as the nanotubes diffused freely, while confined to the focal plane of a microscope by two polymer-coated pieces of glass. Fluorescence images further confirm that multi-helix bundles self-assemble to yield unbranched linear DNA nanotubes.
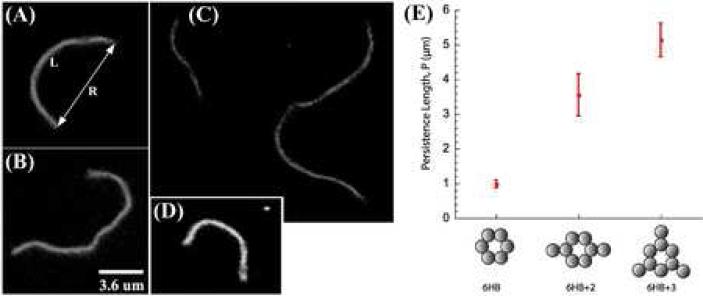
Figure 4. Fluorescence Micrographs of Nanotubes.
(A) and (B) 6HB+2. (C) 6HB+3. (D) 6HB. (E) The measured persistence lengths and associated uncertainties for the three different types of nanotubes are shown. Persistence length was measured by extracting the end-to-end distance, R, and contour length, L, from fluorescence images of several different nanotubes (N>4) in many distinct conformations (<n>=23), and fitting the data to the 2D Kratky-Porod Model.
Nanotube stiffness was measured by extracting the end-to-end distance, R, and contour length, L, from fluorescence images of several different nanotubes (N>4) in many distinct conformations (<n> = 23) and fitting the data to the 2D Kratky-Porod Model:9, 28
The only free parameter in this fit, p, is the persistence length – the length scale over which the orientation of the nanotube is randomized due to thermal fluctuations – a standard measure of polymer stiffness. We measured persistence lengths of 1.0 ± 0.1 μm, 3.6 ± 0.6 μm and 5.0 ± 0.5 μm for 6HB, 6HB+2 and 6HB+3, respectively (Figure 4E), where the uncertainty represents one standard deviation in the parameter values derived from a bootstrap analysis (see Materials and Methods). Extracting R and L of dynamic nanotubes from fluorescence micrographs (rather than AFM data) ensured that our measurement was not affected by adhesion events and allowed us to exclude data from nanotubes with folding errors, which are visible as permanent kinks or hyper-flexible hinges.
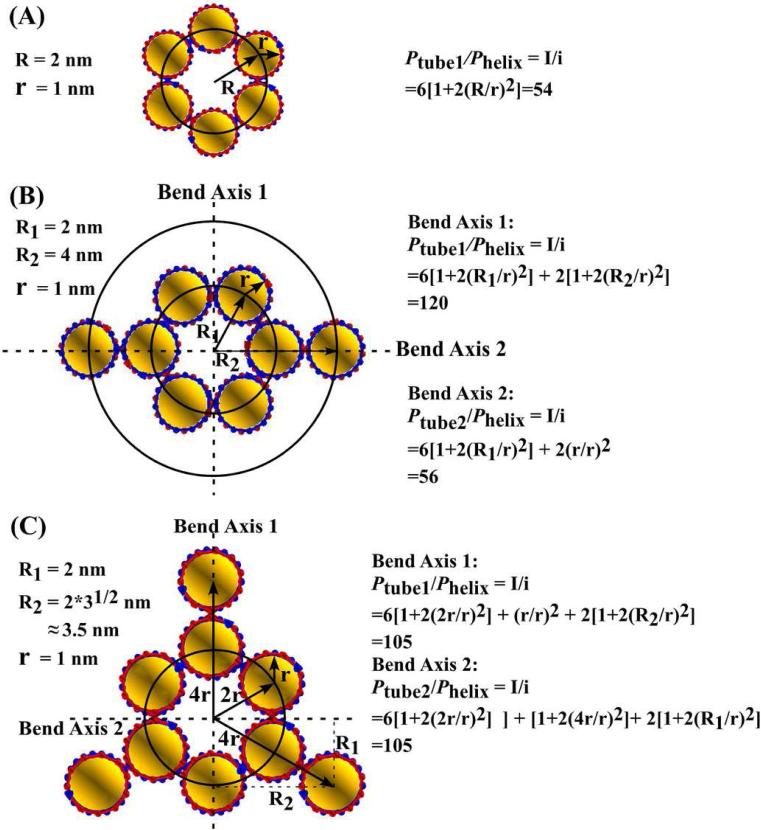
We compare these persistence lengths to the estimated values of all three types of nanotubes using a naïve model8, 9 in which (i) the 6HB nanotube is represented by a ring of rigidly linked rods (DNA helices), (ii) there are no gaps between the rods, and (iii) the Young's modulus of the rods is the same as that of duplex DNA (phelix = 50 nm). In this model, the ratio of the tube persistence length to the helix persistence length is the same as the ratio of their moments of inertia (I):8, 9
where r is the radius of DNA duplex, R is the radius of the nanotube measured from the center of the nanotube to the center of the DNA duplex and N is the number of helices in the nanotube circumference. The estimated persistence lengths for 6HB, 6HB+2, and 6HB+3 are 2.7, 4.4, 5.25 μm respectively (Figure 5).
Figure 5. Estimation of the Persistence Lengths of Nanotubes.
(A) 6HB. (B) 6HB+2. (C) 6HB+3. In the cases of 6HB+2 and 6HB+3, assuming two perpendicular bend axes, as indicated by the dotted lines, any bending of nanotubes can be treated as a combination of bends about those two axes. Thus, the overall persistence length was estimated to be the average of the persistence lengths calculated for each of the bend axes. For 6HB+2, estimates were calculated using r = 1 nm, R1 = 2 nm and R2 = 4 nm. For 6HB+3, estimates were calculated using r = 1 nm, R1 = 2 nm and R2 ≈ 3.5 nm.
As expected, the motifs with extra duplexes (6HB+2, 6HB+3) are more rigid than the 6HB motif. Their calculated and measured persistence lengths agree to within the experimental error. The measured persistence length of the 6HB nanotube, however, is significantly smaller than expected. This low stiffness might be due, in part, to the phasing of nicks in the DNA backbone. The sticky-ended cohesion that mediates the self-assembly of our nanotube motifs necessitates a pair of nicks in the phosphodiester backbones of each of the DNA duplexes (one on each strand). These nicks may be weak points, where thermal forces can bend the DNA duplex most easily.
Consistent with this idea, an earlier design of 6HB+2 (Figure S1D) where the sticky ends are parallel yielded nanotubes with a measured persistence smaller than the 6HB+2 nanotubes with staggered stick ends reported above (Figure 6A). Figure 6B (top) shows the positions of the middles of the sticky ends (Z1, Z2, Z3, and Z4) along the Z axis of the DNA duplex in the ultimate 6HB+2 motifs. In the earlier design (Figure 6B bottom) Z1 = Z2 = Z3. Staggering the sticky ends such that Z1 = Z2 - 5 = Z3 - 10 increased markedly the rigidity of the nanotube structure..
2D Arrays
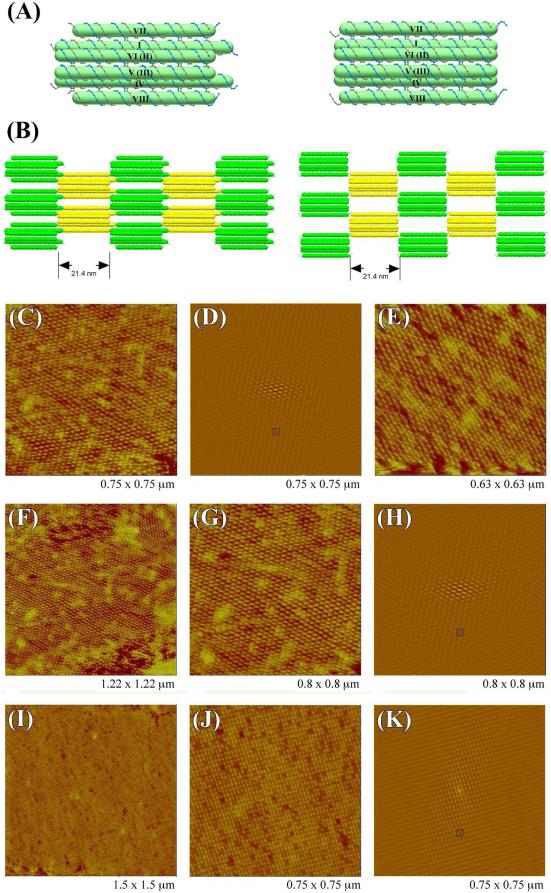
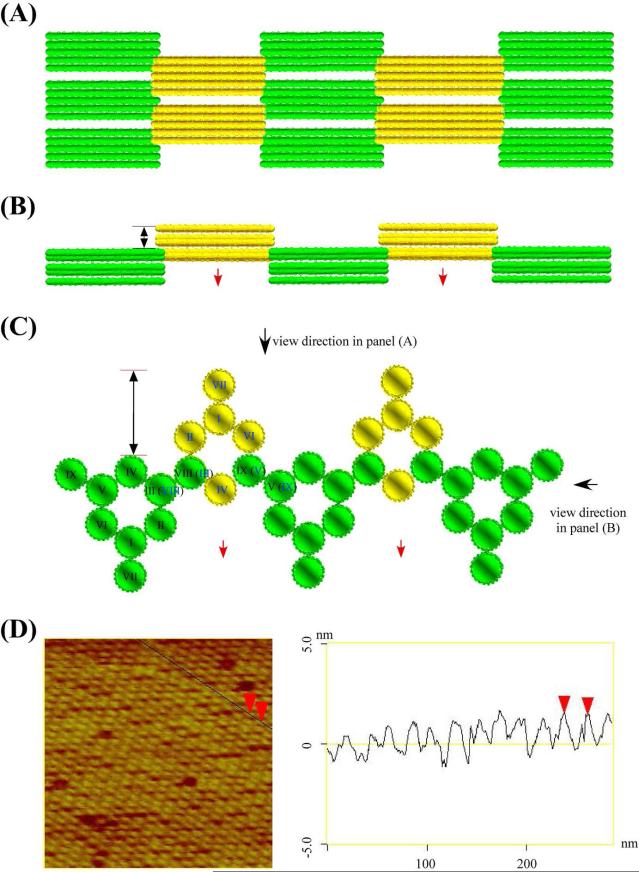
The diagrams of the 6HB+2 2D system tiles are shown in Figure 7A, both for double cohesion (left) and single cohesion (right). The designed lattices are shown in Figure 7B. Figures 7C-K show highly-ordered and flat 2D periodic arrays self-assembled from 6HB+2 through both double (Figures 7C and 7G) and single cohesion (Figure 7J); two directions of ordering are evident in its autocorrelation pattern, which is shown in Figures 7D, 7H & 7K respectively. Figures 7C & 7G show the alternating arrangement of individual 6HB+2 units via double cohesion, which is consistent with the design (Figure 7B). In the double cohesion system, the calculated length of the tile along helical axis is 21.4 nm, which is similar to the AFM measurement (~ 22 nm). However, the measured distance between two adjacent tiles in the direction perpendicular to the helical axis is ~ 17.5 nm, which is larger than the estimated distance of 12 nm. Figure 7J shows that the arrangement of individual 6HB+2 motifs in a single cohesion system is apparently an orthogonal lattice, rather than the alternating array that it was designed to be. This discrepancy between the experiment and the design may result from the deposition of two distorted interdigitated layers, where the gaps visible in the right panel of Figure 7B are filled in by the other layer. The large number of light red units and the few intense red units suggest the presence of single and double defects, respectively. Such a possible structure may be stabilized by the blunt-end stacking of the 6HB components of the motif. Distortion perpendicular to the direction of the helix axis is well-known in single-cohesion systems.8 The measured distance normal to the helical axis, around 15 nm, is consistent with this suggestion; the measured distance along the helix axis is ~ 20 nm, consistent with the 21.4 nm design.
Figure 7. Designs and AFM Images of 2D Periodic Arrays of 6HB+2.
(A) Schematic drawings of 6HB+2 motifs. The left panel shows 6HB+2 used in double cohesion system, and the right panel shows 6HB+2 used in the single cohesion system. Duplexes are labeled by Roman numerals in the same way as in Figure 1B, some duplexes are labeled by two numerals because of overlap in this view: duplexes with numerals in parenthesis are further from viewer. (B) Schematic drawing of the 2D arrangement of 6HB+2 motifs. The left panel shows a 2D array in the double cohesion system. The right panel shows a 2D array in the single cohesion system. AFM images of 2D arrays with double cohesion are shown in (C)-(H). A zoom is shown in panel (C), its autocorrelation function is shown in (D), and another two views are shown in (E) and (F). A zoom of (F) is shown in (G) with its autocorrelation function in (H). Three zooms (C), (E) and (G) show how the individual 6HB+2 motif forms an alternating arrangement. AFM images of 2D arrays with single cohesion are shown in (I)-(K). A zoom of (I) is shown in (J); its autocorrelation function is shown in (K). The zoom in (J) shows that individual 6HB+2 motif forms an orthogonal array.
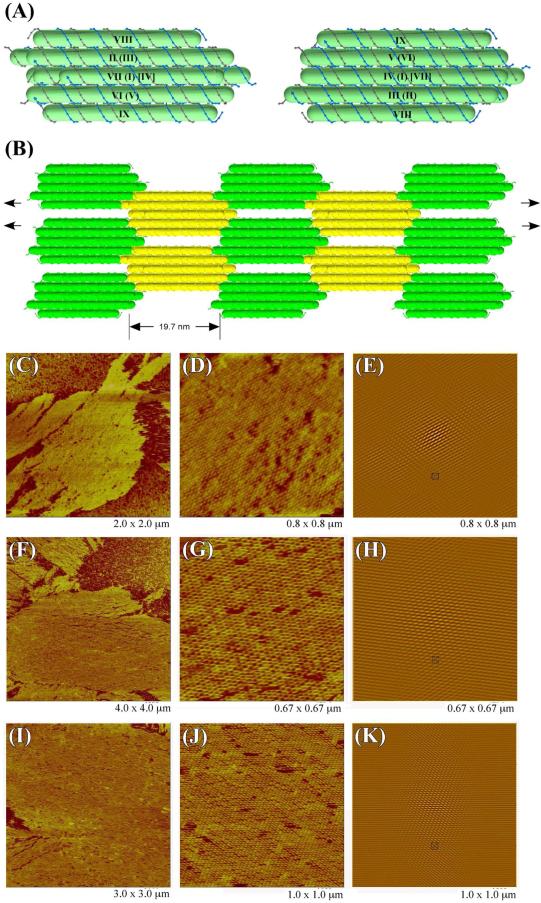
Figure 8A shows the tiles that form the 6HB+3 2D lattice. Two copies of one 6HB+3 molecule used in a 2D array are shown in the drawing. The motif on the right, which has the same orientation as the green copies in panel (B), is rotated 180° around a horizontal axis from the one on the left, which has the same orientation as the yellow copies in panel (B). Figures 8C-K show that all three different 2D sections of 6HB+3 doubly cohesive arrays form large, flat, and well-ordered arrays under AFM imaging. Potentially sticky ends can occur on helices I, III, V, VII, VIII and IX, while helices II, IV and VI are always blunt. Panels C, D, and E show the array structure, the zoom and the autocorrelation function of the array formed without sticky ends C and A on helices I and VII (Figure S3). Panels F, G and H show the analogous results when helices III and VIII lack sticky ends D and B (Figure S3); likewise, panels I, J and K show the results when sticky ends E and F on helices V and IX are made blunt (Figure S3). The zoomed images show clearly that all three rotational isomers of the 6HB+3 motif form an alternating arrangement, consistent with the design in Figure 8B. The autocorrelation patterns show similar degrees of ordering. The calculated length of the tile along the helical axis is 19.7 nm, which is similar to the AFM measurements of the three 2D lattices (21 ± 1 nm, 20.7 ± 0.7 nm, 20.2 ± 0.5 nm). However the measured distances (14 ± 0.8 nm, 14.2 ± 0.5 nm, 14.5 ± 0.4 nm) between two adjacent tiles in the direction perpendicular to the helical axis are larger than estimated distance of 10.4 nm.
Figure 8. Designs and AFM Images of 2D Periodic Arrays of the 6HB+3 Motif.
(A) Schematic drawings of 6HB+3 motifs. Two copies of one 6HB+3 molecule used in a 2D array are shown here. The motif on the right, which has the same orientation as the green copies in panel (B), rotates 180° around a horizontal axis from the one on the left, which has the same orientation as the yellow copies in panel (B). Duplexes are labeled by Roman numerals in the same way as in Figure 1C; some duplexes are labeled by two or three numerals because of overlapping in this view: The duplexes with numerals in parentheses are further from viewer; those with numerals in brackets are furthest from viewer. (B) Schematic drawing of a top view that is perpendicular to the 2D array plane. Pairs of black arrows represent the 2-fold screw axes in the design of the 2D array. AFM images of three sets of 2D arrays are shown in panel (C), (F), (I) respectively. Zoomed views of (C), (F), (I) are shown in (D), (G), (J) respectively with their autocorrelation functions are shown in (E), (H), (K). Panels (C), (F), (J) show that the individual 6HB+3 motif forms an alternating arrangement in all three sets of 2D arrays.
DISCUSSION
We have demonstrated that 6HB and its two variants are able to form unbranched linear nanotubes, and that the external helices enhance the stiffness of the 6HB structure in the order 6HB+3 (5.0 ± 0.5 m) > 6HB+2 (3.6 ± 0.6 μm) > 6HB (1.0 ± 0.1 μm). The stiffening of the 6HB motif by the addition of external helices suggests that it might be possible to strengthen it further by further reinforcement. The measured and calculated persistence lengths are in good agreement for 6HB+2 and 6HB+3. In addition, we found that the relative placement of sticky ends affects the rigidity of nanotubes self-assembled from cyclic DNA motifs: Sticky ends at various positions along the Z axis of the DNA duplex produce more rigid nanotubes than those at the same Z position (Figure 6).
We also demonstrate that 2D periodic arrays can be built from 6HB+2 and 6HB+3 motifs. In both cases the arrangements of individual motifs in 2D arrays through double cohesion are consistent with the designs, which further demonstrates that double cohesion is a powerful tool for the assembly of 2D arrays.22, 23 In the cases of double cohesion described (6HB+2 and 6HB+3), the measured distance between two adjacent tiles in the direction normal to the DNA helical axis is always seen to be larger than expected. Similar phenomena have been observed in other studies where antiparallel duplexes are often seen to be separated in this direction by much more than the estimated distance when observed by tapping mode AFM on mica, when the sample is in buffer.8, 12 Another important factor leading to this discrepancy may be the flattening of bundles when in contact with the mica surface.12
Ideally in a 2D array, two adjacent 6HB+3 motifs along the helix axis have height differences (Figure 9); black double-headed arrows in Figures 9B and 9C indicate the difference. Our AFM images show sagging of the tiles drawn in yellow (red arrows in Figures 9B and 9C) and do not show obvious height differences (Figure 9D). The cause of this discrepancy is unlikely to be the motif itself, since all three 2D sections form large well-ordered arrays in much the same way. This uniformity indicates three-fold symmetry in the individual motif if one ignores the differences of sequences and crossover locations. The flattening of the bundles on the mica surface, mentioned above, is likely the major reason for the height difference elimination. Another contribution may be from the electrostatic attraction between the backbones of DNA and mica, to which the tiles drawn in yellow seem to adhere when examined by AFM (Figures 9A-9C).
Figure 9. 6HB+3 2D Arrays in Different Views.
The 6HB+3 motif is shown in two different colors (green and yellow), representing two different orientations of 6HB+3 in a 2D array. (A) Top View. 6HB+3 is in an alternating arrangement along the helical axis in this view. (B) Side View. In principle, two adjacent 6HB+3 motifs (green and yellow) along the helical axis have different heights in the 2D array, as indicated by double black arrows. (C) Cross-Sectional View. Motifs in green and yellow are at different levels along helical axis: Motifs in green are closer to viewer than those in yellow. Because of the overlapping of duplexes in the cross-sectional view some duplexes in yellow are invisible to viewer. Duplexes in green are labeled by Roman numerals in black; duplexes in yellow are labeled by Roman numerals in blue. Duplexes with blue numerals in parenthesis are further from viewer. It is also clear in this view that two 6HB+3 motifs have different heights. The scales of the drawings are different in the three views. (D) Cross-Sectional Analysis of 6HB+3 2D Array. No obvious height differences are observed through AFM imaging.
We expect the three designed motifs can be put to a number of applications in DNA nanotechnology, especially the 6HB+3 motif, which prototypes a surface with a designed curvature. In its current form, it appears to have the potential to act as a nanomechanical strut, because it appears to be relatively stiff.
The PX-JX2 sequence-dependent device is based on the PX molecule, which has a pseudo-two-fold symmetry axis.29 The 6HB motif has a pseudo-six-fold symmetry element, while the 6HB+3 has a pseudo-three-fold axis, which enables these motifs to be potential candidates for nanomechanical devices capable of assuming multiple superimposable states. In addition, the extra double helices in the 6HB+2 and 6HB+3 motifs provide a group of duplexes that could be used to mount and orient other motifs, further nanomechanical devices or even modified nucleotides.30
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Ruojie Sha for assistance with DNA synthesis. This research has been supported by NSF grants CCF-0622257 and CHE-0848375 to DKF and by the following grants to NCS: GM-29554 from the National Institute of General Medical Sciences, CTS-0608889 and CCF-0726378 from the National Science Foundation, 48681-EL and W911NF-07-1-0439 from the Army Research Office, N000140910181 and N000140911118 from the Office of Naval Research and a grant from the W.M. Keck Foundation.
REFERENCES
- 1.a Seeman NC. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]; b Seeman NC. Ann. Rev. Biochem. 2010;79:65–87. doi: 10.1146/annurev-biochem-060308-102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winfree E, Liu F, Wenzler LA, Seeman NC. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 3.LaBean T, Yan H, Kopatsch J, Liu F, Winfree E, Reif JH, Seeman NC. J. Am. Chem. Soc. 2000;122:1848–1860. [Google Scholar]
- 4.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 5.Rothemund PWK. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 6.Li H, Park SH, Reif JH, LaBean TH, Yan H. J. Am. Chem. Soc. 2004;126:418–419. doi: 10.1021/ja0383367. [DOI] [PubMed] [Google Scholar]
- 7.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. Science. 2006;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 8.Rothemund PWK, Ekani-Nkodo A, Papadakis N, Kumar A, Fygenson DK, Winfree E. J. Am. Chem. Soc. 2004;126:16344–16352. doi: 10.1021/ja044319l. [DOI] [PubMed] [Google Scholar]
- 9.O'Neill P, Rothemund PWK, Kumar A, Fygenson DK. Nanolett. 2006;6:1379–1383. doi: 10.1021/nl0603505. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell JC, Harris JR, Malo J, Bath J, Turberfield AJ. J. Am. Chem. Soc. 2004;126:16342–16343. doi: 10.1021/ja043890h. [DOI] [PubMed] [Google Scholar]
- 11.Liu D, Park SH, Reif JH, LaBean TH. Proc. Natl. Acad. Sci. U.S.A. 2004;101:717–722. doi: 10.1073/pnas.0305860101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathieu F, Liao S, Kopatsch J, Wang T, Mao C, Seeman NC. NanoLett. 2005;5:661–665. doi: 10.1021/nl050084f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park SH, Barish R, Li H, Reif JH, Finkelstein G, Yan H, LaBean TH. NanoLett. 2005;5:693–696. doi: 10.1021/nl050108i. [DOI] [PubMed] [Google Scholar]
- 14.Liu H, Chen Y, He Y, Ribbe AE, Mao C. Angew. Chem. Int. Ed. 2006;45:1942–1945. doi: 10.1002/anie.200504022. [DOI] [PubMed] [Google Scholar]
- 15.Ke Y, Liu Y, Zhang J, Yan H. J. Am. Chem. Soc. 2006;128:4414–4421. doi: 10.1021/ja058145z. [DOI] [PubMed] [Google Scholar]
- 16.Douglas SM, Chou JJ, Shih WM. Proc. Natl. Acad. Sci. U.S.A. 2007;104:6644–6648. doi: 10.1073/pnas.0700930104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin P, Hariadi RF, Sahu S, Choi HMT, Park SH, LaBean TH, Reif JH. Science. 2008;321:824–826. doi: 10.1126/science.1157312. [DOI] [PubMed] [Google Scholar]
- 18.Sharma J, Chhabra R, Cheng A, Brownell J, Liu Y, Yan H. Science. 2009;323:112–116. doi: 10.1126/science.1165831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a Kuzuya A, Wang R, Sha R, Seeman NC. NanoLett. 2007;7:1757–1763. doi: 10.1021/nl070828k. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Wang R, Liu W, Seeman NC. Chem. & Biol. 2009;16:862–867. doi: 10.1016/j.chembiol.2009.07.008. [DOI] [PubMed] [Google Scholar]; c Clever GH, Shionoya M. Coord. Chem. Revs. 2010;254:2391–2402. [Google Scholar]
- 20.Fu T-J, Seeman NC. Biochem. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 21.Liu D, Wang M, Deng Z, Walulu R, Mao C. J. Am. Chem. Soc. 2004;126(8):2324–2325. doi: 10.1021/ja031754r. [DOI] [PubMed] [Google Scholar]
- 22.Ding B, Sha R, Seeman NC. J. Am. Chem. Soc. 2004;126:10230–10231. doi: 10.1021/ja047486u. [DOI] [PubMed] [Google Scholar]
- 23.Constantinou P, Wang T, Kopatsch J, Israel LB, Mao C, Ding B, Sha R, Zhang X, Yang X, Seeman NC. Org. Biomol. Chem. 2006;4:3414–3419. doi: 10.1039/b605212f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang R, Kuzuya A, Liu W, Seeman NC. Chem. Commun. 2010;46:4905–4907. doi: 10.1039/c0cc01167c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seeman NC. J. Biomol. Str. & Dyns. 1990;8:573–581. doi: 10.1080/07391102.1990.10507829. [DOI] [PubMed] [Google Scholar]
- 26.Srinivasan K, Pohl G, Avdalovic N. Analytical Chemistry. 1997;69:2798–2805. [Google Scholar]
- 27. http://mitchison.med.harvard.edu/protocols/flowcell.html.
- 28.Doi M, Edwards SF. The Theory of Polymer Dynamics. Clarendon Press; Oxford: 1986. [Google Scholar]
- 29.Yan H, Zhang X, Shen Z, Seeman NC. Nature. 2002;415:62–65. doi: 10.1038/415062a. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka K, Shionoya M. J. Org. Chem. 1999;64:5002–5003. doi: 10.1021/jo990326u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.