Abstract
In recent years, sugars with a unique chemical handle have been used to detect and elucidate the function of glycoconjugates. Such chemical handles have generally been part of an N-acetyl moiety of a sugar. We have previously developed several applications using the single mutant Y289L-β1,4-galactosyltransferase I (Y289L-β4Gal-T1) and the wild-type polypeptide-α-GalNAc-T enzymes with UDP-C2-keto-Gal. Here, we describe for the first time that the GlcNAc-transferring enzymes—R228K-Y289L-β4Gal-T1 mutant enzyme, the wild-type human β1,3-N-acetylglucosaminyltransferase-2 and human Maniac Fringe—can also transfer the GlcNAc analog C2-keto-Glc molecule from UDP-C2-keto-Glc to their respective acceptor substrates. Although the R228K-Y289L-β4Gal-T1 mutant enzyme transfers the donor sugar substrate GlcNAc or its analog C2-keto-Glc only to its natural acceptor substrate, GlcNAc, it does not transfer to its analog C2-keto-Glc. Thus, these observations suggest that the GlcNAc-transferring glycosyltransferases can generally accommodate a chemical handle in the N-acetyl-binding cavity of the donor sugar substrate, but not in the N-acetyl-binding cavity of the acceptor sugar.
Keywords: N-acetylglucosaminyltransferase, UDP-C2-keto-Gal, UDP-C2-keto-Glc, UDP-GlcNAc
Introduction
Sugars with a chemical handle, such as the azido, alkene, sulfo or keto groups attached to the N-acetyl moiety, have been increasingly used to study the function of glycoconjugates (Prescher and Bertozzi 2006; Laughlin and Bertozzi 2009; Wang et al. 2009). These studies exploit the ability of the glycosyltransferase to accommodate sugars with such chemical handles in their N-acetyl group binding pocket. Structural studies on the N-acetyl-hexosamine sugar-binding proteins, such as glycosyltransferases and lectins, have shown that a hydrophobic cavity is found in the sugar-binding site that facilitates the binding of the N-acetyl rest (Unligil et al. 2000; Qasba and Ramakrishnan 2007; Schwefel et al. 2010). Creation of such a hydrophobic cavity in β1,4-galactosyltransferase I (β4Gal-T1) and in α1,3-galactosyltransferase by mutating residue(s) in the vicinity of the O2 hydroxyl group of the donor sugar Gal enables these enzymes to transfer GalNAc from UDP-GalNAc (Ramakrishnan and Qasba 2002; Tumbale et al. 2008; Pasek et al. 2009). Studies have shown that these mutant enzymes, and the wild-type α-polypeptidyl-N-acetylgalactosaminyltransferase (αppGalNAc-T), which transfers GalNAc from UDP-GalNAc to a polypeptide sequence in a protein, can all transfer Gal analogs with a chemical handle at the C2 position, such as C2-keto-Gal or GalNAz [Gal-C2-NH-C(O)-CH2-N3], to their respective acceptor substrates (Khidekel et al. 2003; Boeggeman et al. 2007, 2009; Ramakrishnan et al. 2007; Pasek et al. 2009).
In the metabolic labeling of cell surface glycans using modified sugars, it has been observed that only galactose with a chemical handle such as C2-keto-Gal gets incorporated into the surface glycans, whereas no measurable quantity of C2-keto-Glc was observed on the cell surface (Hang and Bertozzi 2001). It was reasoned that the lack of incorporation of 2-keto-Glc may be due to the competition with the endogenous GlcNAc molecule (Hang and Bertozzi 2001). This observation also raises the possibility that the glycosyltransferases that utilize GlcNAc as either donor or acceptor sugar substrates could not use glucose with chemical handles that are isosteres of the GlcNAc molecule as their substrate molecule. To elucidate this possibility, in the present study, we have tested the mutant and wild-type glycosyltransferase enzymes that use UDP-GlcNAc as a donor substrate with UDP-C2-keto-Glc.
The β4Gal-T1 enzyme transfers Gal from UDP-Gal to GlcNAc present at the non-reducing end of an acceptor substrate, and its double mutant R228K-Y289L-β4Gal-T1 exhibits better GlcNAc-transferase activity where it transfers GlcNAc from UDP-GlcNAc to the same acceptor substrate (Ramakrishnan et al. 2005). In the present study, we find that this double-mutant enzyme can transfer C2-keto-Glc from UDP-C2-keto-Glc, however, only to GlcNAc, not to its analog, C2-keto-Glc. Furthermore, we also show that the two wild-type N-acetylglucosaminyltransferases, human β1,3-N-acetylglucosaminyltransferase-2 (β3GN-T2; Togayachi et al. 2006; Seko and Yamashita 2008; Togayachi et al. 2010) and human Maniac Fringe (hum-MFng; Haltiwanger and Stanley 2002; Rampall et al. 2005; Stanley and Okajima 2010), which accommodate the N-acetyl group of the donor sugar GlcNAc, can also transfer the C2-modified Glc, C2-keto-Glc, to their corresponding acceptors, LacNAc on the N-glycans of Asialofetuin (ASF) and O-fucosylated epidermal growth factor (EGF) repeat from Factor VII, respectively.
Results
Double mutation in the sugar-donor-binding site of β4Gal-T1, Arg288 to Lys288 and Tyr289 to Leu289, resulted in N-acetylglucosaminyltransferase and N-acetylgalactosaminyltransferase activities
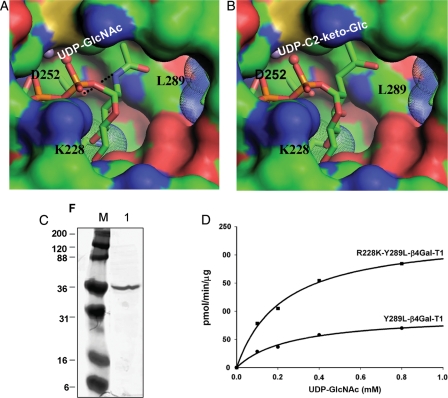
It was previously observed that the mutation of Tyr289 to Leu289 in the wild-type β4Gal-T1 creates a cavity around the binding site of the C2 position of the donor sugar Gal (Ramakrishnan et al. 2002). This cavity accommodates the N-acetyl group of the donor sugar GalNAc (Table I). This single-mutant enzyme is also shown to transfer GlcNAc from the donor substrate UDP-GlcNAc (Table I). The lesser GlcNAc transferase activity reported here (65 pmol/min/µg), compared with the one previously reported value (8.2 pmol/min/ng; Table 2 in Ramakrishnan et al. 2002), could have been due to an error in the previous protein estimation (Table I). Also, it has been shown that mutation of Arg228 to Lys228 creates a cavity at the C4 position of the donor sugar Gal, which enhances glucosyltransferase activity in the mutant enzyme R228K-β4Gal-T1 with no GlcNAc transferase activity (Table I). When these two single mutations are combined, resulting in a double mutant, R228K-Y289L-β4Gal-T1, the UDP-GlcNAc and UDP-C2-keto-Glc molecules are expected to bind better to the double-mutant enzyme as donor sugars (Figure 1A and B).
Table I.
Specific activity of the wild-type and mutant enzymes of β4Gal-T1
| Enzyme | UDP-GalNAc → GlcNAc (pmol/min/ng)# | UDP-GlcNAc → GlcNAc (pmol/min/μg)* |
|---|---|---|
| WT-β4Gal-T1 | 0.02 | ND |
| Y289L-β4Gal-T1 | 22 ± 5 | 65 ± 1 |
| R228K-β4Gal-T1 | 0.67 ± 0.04 | ND |
| R228K-Y289L–β4Gal-T1 | 29 ± 6 | 207 ± 8 |
ND, none detected. The GalNAc-T activity is expressed as per nanograms of protein (#) compared with GlcNAc activity (*), which is per micrograms of protein.
Fig. 1.
Modeling of UDP-GlcNAc (A) and UDP-C2-keto-Glc (B) binding to mutant R228K-Y289L- β4Gal-T1 molecule. The modeling was based on the UDP-GalNAc and UDP-Glc binding to the wild-type β4Gal-T1 molecule (PDB 1OQM and 1O23, respectively) and the structure of the R228K-β4Gal-T1 molecule (PDB 1YRO). The hydrogen bond between the amino group of the donor GlcNAc molecule and the side-chain carboxylate oxygen atom of the Asp252 residue is shown in a black dotted line in (A), and this interaction is absent in (B). The modeling was performed without any energy minimization and the C2-keto-Glc geometry was generated by ChemSketch program. (C) PAGE showing the purified R228K-Y289L-β4Gal-T1 protein after affinity column purification. (D) The catalytic activity of Y289L-β4Gal-T1 and the R228K-Y289L-β4Gal-T1 enzymes measured at 37°C for 10 min, using UDP-GlcNAc as the donor substrate with 25 mM β-benzyl-GlcNAc as the acceptor substrate.
The double-mutant enzyme Y289L-R228K-β4Gal-T1 was expressed in Escherichia coli, in vitro refolded the protein from inclusion bodies and purified using a UDP-agarose column as described (Ramakrishnan and Qasba 2002), yielding 30 mg of folded protein per 1 L of bacterial culture. The purified double-mutant enzyme in the radioactivity assay showed a 3-fold better GlcNAc-transferase activity, compared with the single-mutant enzyme Y289L-β4Gal-T1 (Table I, Figure 1C). The apparent Km value for UDP-GlcNAc binding to the Y289L-β4Gal-T1 and the R288K-Y289L-β4Gal-T1 mutant enzymes is 0.26 ± 0.05 and 0.22 ± 0.03 mM, respectively, and these values are comparable with the true Km value for the UDP-GalNAc binding to the Y289L-β4Gal-T1 enzyme (Ramakrishnan et al. 2002).
Transfer of GlcNAc and C2-keto-Glc residue on dichitotriose and on N-glycans of Ovalbumin
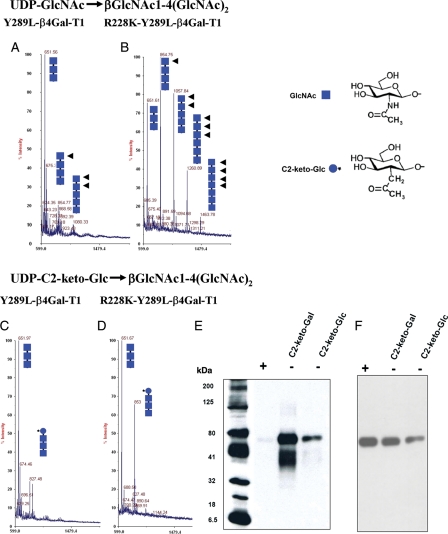
The transfer of GlcNAc and 2-keto-Glc from their UDP derivatives to the acceptor dichitotriose [GlcNAcβ1-4-(GlcNAcβ1-4-)2] by the Y289L-β4Gal-T1 and the R228K-Y289L-β4Gal-T1 mutant enzymes was followed by matrix-assisted laser desorption/ionization (MALDI) mass profiling (Figure 2). The mass spectrometry analysis shows that under identical conditions, the single mutant Y289L-β4Gal-T1 extends with less efficiency the chitotriose acceptor to a chitotetrose and chitopentose (Figure 2A), compared with the double-mutant enzyme R228K-Y289L-β4Gal-T1, which extends the acceptor chitotriose to chitotetrose, chitopentose, chitohexose and chitoheptose (Figure 2B). Furthermore, the double mutant transfers the C2-keto-Glc much better (Figure 2D), compared with the Y289L-β4Gal-T1 enzyme (Figure 2C). However, these enzymes do not elongate the acceptor substrate beyond the addition of a single C2-keto-Glc residue. The double mutant R228K-Y289L-β4Gal-T1 transferred two modified sugars, C2-keto-Glc and C2-keto-Gal, to the terminal GlcNAc residue on the glycan moiety of the acceptor Ovalbumin (Ova), as detected by western blotting after conjugation with amino-oxy-biotin (Figure 2E). Chemiluminescence was not detected when Ova was treated with PNGase F before western blotting (Figure 2E, lane + sample).
Fig. 2.
MALDI mass spectra of GlcNAc-T catalytic reaction using the acceptor chitotriose [(GlcNAcβ1,4-(GlcNAcβ1,4)2], by the enzymes Y289L-β4Gal-T1 (A) and R228K-Y289L-β4Gal-T1 (B). The chitotriose in both the reactions is elongated by the enzymes, because in the GlcNAc-T reaction, the product with GlcNAc at the non-reducing end (squares with arrows) is an acceptor substrate for the enzyme; thus, the initial acceptor substrate is elongated by these enzymes. Under similar reaction conditions and enzyme concentrations, the double mutant is better than the single-mutant enzyme in the GlcNAc-T catalytic reaction. The MALDI mass spectra of the catalytic reaction, using the same acceptor substrate chitotriose with UDP-C2-keto-Glc as the donor substrate, by the mutant enzymes Y289L-β4Gal-T1and R228K-Y289L-β4Gal-T1 are shown in (C) and (D), respectively. The double mutant shows a major peak at m/z 853, which corresponds to the product after the transfer of the C2-keto-Glc (blue circle with star) to the acceptor sugar chitotriose, which is absent in the transfer with the single mutant. However, no elongation of the acceptor substrate is seen with C2-keto-Glc, indicating that C2-keto-Glc, though it is an isostere of GlcNAc, is not an acceptor sugar for these enzymes. (E) The chemoenzymatic detection of C2-keto-Glc or C2-keto-Gal transferred to Ova. The transfer of C2-keto-Glc or C2-keto-Gal to GlcNAc residues on the N-glycans chains of Ova by the R228K-Y289L-β4Gal-T1 was monitored by linking the product with AOB, followed by western blotting and chemiluminescence detection. Chemiluminescence was detected only in the samples that contained C2-keto-Glc or C2-keto-Gal enzyme and 25 ng of Ova. After the transfer of C2-keto-Glc or C2-keto-Gal, a portion of Ova samples was treated with PNGase F, which removes the N-glycan chains from the protein (+). In contrast to the PNGase F-treated samples (+), the untreated samples (−) exhibited chemiluminescence, indicating that C2-keto-Glc or C2-keto-Gal is selectively transferred to the glycan portion of the Ova molecule. (F) The amount of Ova samples loaded in each lane was monitored by treating the membranes with the monoclonal antibody against Ova after western blot analysis.
Expression, purification and activity of galectin-1-hum-β3GN-T2 and galectin-1-hum-MFng
The hum-β3GN-T2 and hum-MFng proteins, when expressed in E. coli, produced inclusion bodies that could not be folded in vitro, in contrast to the inclusion bodies of the single mutants (Y289L-β4Gal-T1 or R228K-β4Gal-T1) or the double mutant (R228K-Y289L-β4Gal-T1) of β4Gal-T1, which can be folded in vitro. We have previously shown that the human β4Gal-T7 inclusion bodies also could not be folded in vitro, whereas, when fused to galectin-1, the fusion protein galectin-hum-β4Gal-T7 was expressed as soluble folded protein in E. coli. Thus, we attempted here to express hum-β3GN-T2 and hum-MFng proteins as the galectin-1 fusion protein. We show here that these proteins are also expressed as soluble folded proteins.
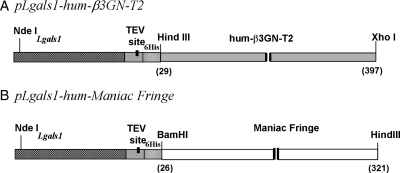
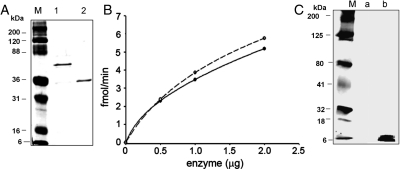
The DNA sequences of β3GN-T2 (corresponding to residues 29–397 amino acids) and hum-MFng (corresponding to residues 26–321 amino acids) were inserted into the pLgals1-Tev (tobacco etch virus) vector, resulting in the expression vector pLgals1-hum-β3GN-T2 and pLgals1-hum-MFng (Figure 3A and B). These constructs expressed in E. coli, Rosetta (DE3) pLysS cells, the soluble fusion proteins of galectin-1-hum-β3GN-T2 and galectin-1-hum-MFng. After sonication of the cells, the supernatant, which contained soluble proteins, was loaded onto an α-lactose column. After purification on the lactose column, the fusion protein galectin-1-hum-β3GN-T2 yielded ∼1.5 mg of protein from a liter of bacterial culture. It was purified on the UDP-agarose column as an active soluble fusion protein with a yield of ∼1 mg, which showed a single band of ∼60 kDa on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE; Figure 4). The fusion protein galectin-1-hum-β3GN-T2 could not be cleaved with the Tev protease. However, the fusion protein showed a specific activity of 0.3 pmol/min/mg of protein toward the LacNAc moiety of N-glycans on the acceptor ASF (Figure 4B, insert).
Fig. 3.
Schematic representation of the expression constructs (A) pLgals1-hum-β3GN-T2 and (B) pLgals1-hum-MFng. The PCR-amplified DNA sequences were inserted in the pLgals1-Tev vector construct at the multicloning site, which is preceded by the 6HisTag and Tev cleavage site.
Fig. 4.
(A) SDS–PAGE of the fusion protein galectin-1-hum-β3GN-T2, purified on a UDP-agarose column (1) and marker (M). (B) The catalytic activity of the galectin-1-hum-β3GN-T2 (0.5–2 µg of enzyme) using 50 µM UDP-GlcNAc as the sugar donor substrate. The insert shows the catalytic activity of galectin-hum-β3GN-T2 with UDP-GlcNAc as the donor substrate at different concentrations (10–40 µg), using ASF as an acceptor substrate. Each assay was carried out for 1 h with 2 µg of enzyme. (C) The chemoenzymatic detection of the transferred C2-keto-Glc on ASF after linking the product with AOB, followed by western blotting and chemiluminescence detection. Marker (M) and lanes (+) and (−) represent PNGase F-treated and -untreated samples, respectively. (D) SDS–PAGE analysis of the loading samples of ASF, with and without PNGase F treatment. Arrow points to the PNGase F protein band.
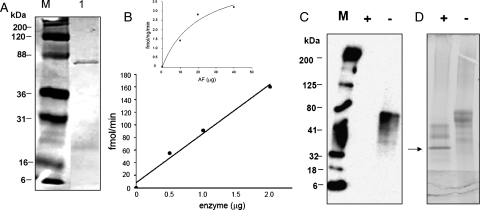
Following purification on the lactose column, the fusion protein galectin-1-hum-MFng yielded ∼1 mg from 1 L of bacterial culture and was detected as a 53 kDa band (Figure 5A, lane 1). This fusion protein was cleaved with Tev protease to release the MFng protein. This protein was purified on a UDP-agarose column, yielding ∼500 µg of purified MFng protein that showed a single band of ∼33 kDa on SDS–PAGE (Figure 5A, lane 2). Free MFng has shown a specific activity of 0.5 pmol/min/mg of protein with 100 mM fucose as an acceptor and 0.6 pmol/min/mg of protein with 4 mM p-nitrophenyl-α-fucose (pNP-α-Fuc) as an acceptor (Figure 5B).
Fig. 5.
(A) SDS–PAGE of (1) galectin-1-hum-MFng and (2) hum-MFng after Tev protease cleavage of the fusion protein and purification on a UDP-agarose column. (B) The catalytic activity of the hum-MFng with 500 µM UDP-GlcNAc as the sugar donor substrate and fucose (continuous line) or pNP-fucose (dashed line) as acceptor substrates. Each assay was carried out for 1 h at 37°C. (C) The chemoenzymatic detection of the transferred C2-keto-Glc on O-fucosylated EGF repeat from Factor VII (a kind gift from Dr. R. Haltiwanger). Reaction samples without enzyme (a) and with enzyme (b).
Transfer of C2-keto-Glc to LacNAc residues on N-glycans of ASF by hum-β3GN-T2
The transfer of C2-keto-Glc to the LacNAc moiety of N-glycans on the ASF by the enzyme hum-β3GN-T2 was followed by the conjugation with N-aminooxymethylcarbonylhydrazino-d-biotin (AOB) and western blot analysis of the protein (Figure 4C). Chemiluminescence of the protein band was detected only when samples were not treated with PNGase F, which removes N-glycans from glycoproteins, prior to western blot analysis (Figure 4C, lane −, sample). The sample treated with PNGase F before western blot analysis showed no chemiluminescence protein bands (Figure 4C, lane +).
Transfer of C2-keto-Glc to fucose residues on EGF repeat from factor VII by the hum-MFng
Chemiluminescence was used to detect the transferred C2-keto-Glc to the fucose moiety present on the EGF repeat from factor VII (mass of >6 kDa) by the wild-type MFng (Figure 5C, lane b). After C2-keto-Glc was transferred to fucose residues and the keto moiety at the C2 position of Glc was coupled with AOB, the biotinylated product was analyzed on western blot and detected by chemiluminescence with streptavidin conjugated to horseradish peroxidase (HRP). As a negative control, the O-fucosylated EGF repeat from factor VII was treated with C2-keto-Glc without enzyme (Figure 5C, lane a).
Discussion
The double-mutant enzyme R228K-Y289L-β4Gal-T1 transfers not only GalNAc but also GlcNAc from their respective UDP derivatives to the acceptor substrate that has GlcNAc at the non-reducing end. Interestingly, in the GlcNAc-T catalytic reaction, the product itself is an acceptor substrate for the enzyme, resulting in the elongation of the acceptor substrate by GlcNAc residues (Figure 2B). The N-acetyl-binding cavity in the donor sugar-binding site of the double mutant can also facilitate the binding of a chemical handle like a ketone or an azide functional group attached at the C2 position of either Gal of UDP-Gal or Glc of UDP-Glc. The double mutant transferred the C2-keto-Gal or the C2-keto-Glc to the glycan chains of Ova, as detected by conjugation of an aminooxy-biotin that carries an orthogonal reactive group (Figure 2). In the donor substrate binding site of β4Gal-T1, when a UDP-GalNAc or a UDP-Gal molecule is bound, the N2 or O2 atom of the donor sugar is hydrogen-bonded to the side-chain carboxylate oxygen atom of the Asp252 residue (Figure 1A; Ramakrishnan et al. 2002). This hydrogen bond would be absent when a UDP-C2-keto-Glc molecule is bound to the R228K-Y289L-β4Gal-T1 enzyme (Figure 1B).
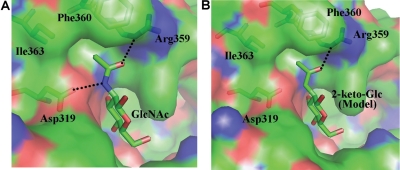
Although the double-mutant enzyme R228K-Y289L-β4Gal-T1 transfers C2-keto-Glc, which is analogous to the acceptor substrate GlcNAc, no elongation of the acceptor substrate is observed. This suggests that the initial product with the terminal C2-keto-Glc is not an acceptor substrate for the enzyme to further transfer C2-keto-Glc to it. Since the binding of the acceptor sugar GlcNAc to the β4Gal-T1 enzyme (Ramakrishnan and Qasba 2001) is well known at the molecular level, a structure-based explanation for C2-keto-Glc not being an acceptor substrate can be obtained. When the acceptor GlcNAc is bound to the β4Gal-T1 enzyme, the amino group of the N-acetyl group forms a hydrogen bond with the side-chain carboxylate oxygen atom of the Asp319 residue, and this is similar to the donor sugar binding, where the same amino group forms a hydrogen bond with the Asp252 residue (Figures 1A and 6A). In addition, the N-acetyl group of the acceptor GlcNAc molecule binds to a hydrophobic cavity created by the residues comprising Arg359 to Ile363, where the side-chain Nδ atom of the Arg359 forms a hydrogen bond with the carbonyl-oxygen atom of the acceptor GlcNAc molecule (Figure 6A). When a C2-keto-Glc molecule is modeled in the acceptor binding site similar to GlcNAc binding, due to the absence of the amino group in the C2-keto-Glc molecule, the hydrogen bond between the Asp319 residue and the acceptor substrate is absent (Figure 6B). Loss of this hydrogen bond is similar to C2-keto-Glc sugar binding in the donor sugar binding, where a hydrogen bond with the Asp252 residue is absent (Figure 1B). Thus, the loss of these hydrogen bond interactions of the C2-keto-Glc molecule with the β4Gal-T1 molecule, either as a donor or as an acceptor sugar, seems to be similar; yet, C2-keto-Glc is utilized only as a donor sugar substrate by the enzyme. This difference may lay in the way the donor and the acceptor sugars are presented to the enzyme, where the donor sugar is always presented to the enzyme as a UDP-sugar as the donor substrate, while the acceptor substrate is often an extended oligosaccharide with the acceptor sugar at the non-reducing end (Ramasamy et al. 2005). However, such an oligosaccharide acceptor substrate that is without the acceptor sugar moiety does not bind to the enzyme, while the UDP moiety alone binds to the enzyme in the presence of Mn2+, although with reduced affinity, compared with the UDP-sugar. This indicates that the binding of the acceptor sugar residue is essential for the binding of the oligosaccharide acceptor substrate to the enzyme, while the donor sugar only enhances the donor substrate binding. Therefore, any loss of interactions between the acceptor sugar and the protein molecule may have greater consequences than the donor sugar losing similar interactions with the protein molecule. For example, although a Glc molecule could still bind in the acceptor binding site of the β4Gal-T1 enzyme retaining the hydrogen bond with the Asp319 residue through its O2 atom, the lack of the N-acetyl moiety and its interaction with the hydrophobic pocket of the protein results in loss of its binding to the enzyme. Thus, its Km value is 1000-fold higher than that of the GlcNAc molecule (Ramakrishnan and Qasba 2001). Also, it has been shown that the 4-deoxy-GlcNAc molecule does not bind to the β4Gal-T1 molecule as an acceptor sugar substrate (Hindsgaul et al. 1991).
Fig. 6.
(A) The binding of GlcNAc molecule to β4Gal-T1 enzyme. The β4Gal-T1 enzyme is shown in a molecular surface diagram; the GlcNAc molecule and the protein residues that are interacting with its N-acetyl moiety are shown in a stick diagram (PDB 1NQI). The hydrogen bonds are shown in dotted black lines. (B) Based on the GlcNAc binding to β4Gal-T1 interactions, the 2-keto-Glc binding has been modeled without any energy minimization. Due to the absence of the amino group in the C2-keto-Glc molecule, the hydrogen bond between the amino group and the side-chain carboxylate oxygen atom of Asp319 residue is absent here.
In this study, we have also investigated whether two wild-type N-acetylglucosaminyltransferases, human β3GN-T2 and hum-MFng, which transfer GlcNAc from UDP-GlcNAc to their respective acceptors, can also accommodate and transfer a Glc analog with a chemical handle at the C2 position that replaces the N-acetyl group of GlcNAc, such as C2-keto-Glc. The human β3GN-T2 belongs to one of the eight members of the β3GN-T family that, in association with β4Gal-T1, has been shown to synthesize the linear poly-N-acetyllactosamine glycan chain, which has repeating N-acetyllactosamine units (Galβ1-4GlcNAcβ1-3)n (Togayachi et al. 2006; Seko and Yamashita 2008). These structures occur in glycosphingolipids and N-linked/O-linked glycan chains of specific glycoproteins, which, in some cases, are modified at the 2-OH and/or 6-OH of Gal and GlcNAc residues with sialic acid, fucose and/or sulfate residues and are identified as carcinoembryonic antigens or ligands for various cell–cell recognition antigens (Togayachi et al. 2007; Varki 2009). The MFng transfers GlcNAc in β1-3-linkage to O-fucose residues present on the EGF repeats of the extracellular domains of the Notch receptor (Haltiwanger and Stanley 2002; Rampall et al. 2005; Stanley and Okajima 2010; Togayachi et al. 2010). Notch receptors are activated by the ligands that are cell surface proteins on adjacent cells. Fringe has been shown to be a regulator of the Notch signaling pathway, which plays a crucial role in cell proliferation and differentiation events of metazoan development. The structure of the catalytic domain of mouse MFng shows a cavity in the sugar-donor-binding site, where UDP-GlcNAc binding has been modeled (Jinek et al. 2006).
Since both β3GN-T2 and MFng, when expressed in E. coli, produced inclusion bodies that could not be folded by the in vitro folding method used for folding the wild-type and mutant β4Gal-T1, we used the galectin-1 as a fusion partner to produce soluble folded protein, as described recently from our laboratory (Pasek et al. 2010). Both galectin-1 proteins were purified on lactose and UDP-agarose affinity columns. The galectin-1-hum-MFng was cleaved with Tev protease to release MFng, which could be purified on a UDP-agarose affinity column. In contrast to galectin-1-hum-MFng, the galectin-1-hum-β3GN-T2 fusion protein could not be cleaved with Tev protease to release β3GN-T2, suggesting that the linker sequence in this construct is not accessible to the protease enzyme. However, galectin-1-hum-β3GN-T2 fusion protein purified on a UDP-agarose column (Figure 5) was an active enzyme that transferred GlcNAc to the acceptor substrate. In contrast to the galectin-1-hum-β3GN-T2 fusion protein, the galectin-1-MFng fusion protein was cleaved with Tev protease. The released MFng was purified on an UDP-agarose column (Figure 2). Furthermore, both fusion protein galectin-1-β3GN-T2 and MFng transferred C2-keto-Glc from its UDP derivative to their respective acceptors; ASF and O-fucosylated EGF repeat from factor VII, respectively (Figures 5 and 6).
We have previously shown that the mutant enzymes, β4Gal-T1-Y289L (Khidekel et al. 2003; Boeggeman et al. 2007, 2009) and α3Gal-T-280S/AGG282 (Pasek et al. 2009), and the wild-type αppGalNAc-T2, which transfer GalNAc (Ramakrishnan et al. 2007) to their respective acceptors, accommodate a chemical handle in the N-acetyl-binding cavity. The chemical handle, similar to a ketone or an azide functional group, is attached at the C2 position of Gal of UDP-Gal and is transferred by the enzymes to their respective acceptors. To determine whether in the GlcNAc-transferring enzyme the N-acetyl-binding site of the donor sugar GlcNAc can also accommodate a chemical handle such as the C2-keto or the C2-azido group of Glc, in the present study, we have tested the double-mutant enzyme R228K-Y289L-β4Gal-T1 and other wild-type N-acetylglucosaminyltransferases with C2-keto-Glc as the donor substrate. In the past, metabolic labeling of cells showed that the surface glycans incorporate C2-keto-Gal or GalNAz but not C2-keto-Glc or GlcNAz (Hang and Bertozzi 2001). Authors reasoned that this may be due to the competition with the endogenous GlcNAc molecule. This observation also raises the possibility that the glycosyltransferases that utilize GlcNAc as either a donor or an acceptor sugar substrate could not use glucose with chemical handles that are isosteres of the GlcNAc molecule as their substrate molecule. Our present study indicates that the N-acetylglucosaminyltransferases could transfer C2-keto-Glc as a donor sugar substrate. However, the same modified sugar analog fails to function as an acceptor substrate for the double-mutant enzyme. A study of the metabolic labeling of the O-GlcNAc modification of cytosolic proteins using GlcNAz has clearly shown that the mammalian cells could not biosynthesize UDP-GlcNAz and this was why researchers were unable to detect GlcNAz on the cell surface (Boyce et al. 2011). Although failure to detect the C2-keto-Glc on the cell surface through metabolic labeling could be due to a similar cause, not all the glycosyltransferases may accept these modified sugars as acceptor substrates, as we have seen in the present study. Therefore, it is possible that in the metabolic labeling of cell surface glycans with the modified sugar, at least some of the glycans with the modified sugar may be present without the extended sugar residues.
In conclusion, we have shown that the N-acetyl groups of the donor sugars GlcNAc and GalNAc of the N-acetylglucosaminyltransferase and N-acetylgalactosaminyltransferase are generally embedded in a cavity or a hydrophobic pocket which can also accommodate a ketone group in the N-acetyl-binding pocket, making it possible to attach to the chemical handle affinity probes for detection, isolation and characterization of the product.
Materials and methods
Materials
The DNA clones were obtained from Open Biosystems, Huntsville, Alabama and contained the coding sequence for hum-β3GN-T2 (accession number BC030579) and hum-MFng (accession number BC094814); the pLgals1 vector was constructed at our laboratory (25); Rosetta (DE3)pLysS cells were obtained from Novagen; XL2 blue ultra-competent cells from Stratagene; Taq-DNA polymerase, polymerase chain reaction (PCR) nucleotide mix and rapid DNA ligation kit from Roche Pharmaceuticals; DNA miniprep spin columns, PCR purification and low-melting agarose extraction kits from Qiagen; restriction enzymes from New England Biolabs, Inc.; Ampicillin, UDP-GlcNAc, UDP-GalNAc, free GlcNAc and pNP-α-Fuc from Sigma-Aldrich; UDP-[6-3H]-GalNAc and UDP-[6-3H]-GlcNAc from American Radiolabeled Chemicals; AG 1-X8 chloride resin 200–400 mesh from Bio-Rad; UDP-agarose gel from Calbiochem. DNA primers were synthesized by Integrated Technologies, Inc. Tev protease was obtained from Invitrogen. UDP-C2-keto-Gal and UDP-C2-keto-Glc were synthesized at the Chemical Biology Laboratory, Imaging Probe Development Center, National Institutes of Health, as described (Dulcey et al. 2011). The AOB, an aldehyde-reacting probe, was purchased from Dojindo Laboratories. Bovine ASF was purchased from Sigma-Aldrich. O-Fucosylated EGF repeat from factor VII was a gift from Dr. Robert S Haltiwanger, Department of Biochemistry and Cell Biology, State University of New York at Stony Brook, NY.
Cloning, expression and purification of mutant β4GalT1-Y289L-R228K
Site-directed mutagenesis was performed using the PCR method. Construction of the mutant was done using plasmid with the Y289L-β4Gal-T1 mutant, described previously (Ramakrishnan and Qasba 2002), to insert the second mutation R228K. Since the intended mutation site was present between the BamHI site and the unique restriction site, StuI, these sites were utilized for the construction of the mutants. The mutation primer corresponding to the lower DNA strand is as follows: R228K 5′-AAGGCCTCTTTAAAGCCAACATTGAGGAGCTTTGCTTTGTTGAACATGGA-3′. The StuI restriction site is shown in italics and the mutation codon for K228 in bold letters. The 5′-terminal fragment of Y289L-β4Gal-T1 mutant was PCR-amplified using the 5′-terminal cloning primer containing the BamHI site and the mutagenesis primer R228K. Then the PCR fragment carrying the mutation was digested with BamHI and StuI and ligated to the vector fragment obtained from the plasmid with Y289L-β4GalT1 mutant DNA cut with the same enzymes. The sequence of mutant R228K-Y289L-β4GalT1 DNA was confirmed by sequence analysis.
For protein expression, Rosetta (DE3) pLysS-competent cells were transformed with the identified cDNA clones according to the manufacturer's protocols. The transformed cells were grown in Luria–Bertani (LB) containing 100 µg/mL ampicillin to OD600nm = 0.7 and were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cultures were harvested after 3–4 h by centrifugation at 3000 × g for 10 min. Inclusion bodies were purified from the bacterial pellet, and the in vitro folding and purification of the enzymes were carried out as described previously (Ramakrishnan and Qasba 2002).
Enzyme assay of R228K-Y289L-β4Gal-T1 mutant
The activities were measured using UDP-GlcNAc or UDP-GalNAc as sugar nucleotide donors and GlcNAc as the acceptor sugar. For the specific activity measurements, a 100-µL incubation mixture containing 25 mM β-benzyl-GlcNAc as an acceptor, 10 mM MnCl2, 10 mM Tris–HCl (pH 8.0), 500 µM UDP-GalNAc or 1 mM UDP-GlcNAc and 0.5 µCi of UDP-[6-3H]-GlcNAc or UDP-[6-3H]-GalNAc was used for each GlcNAc-T or GalNAc-T reaction, respectively. A 100 ng and 2 μg of either the single- or the double-mutant protein were used in the above GalNAc-T and the GlcNAc-T reactions, respectively. The incubation was carried out at 37°C for 10 min. The reaction was terminated by adding 200 µL of cold water and then analyzed as described earlier (Ramakrishnan and Qasba 2002). The enzyme activity curve is fitted with SIGMA plot using an equation defining ligand binding to one site saturation without inhibition.
MALDI time-of-flight analyses of chitotriose derivatives
An Applied Biosystems Voyager-DE Pro time-of-flight mass spectrometer was utilized for analyses. The accelerating voltage was 20 kV, guide wire 0.05% and grid voltage 94%. The instrument was operated in linear mode under positive ion conditions. A nitrogen laser was used at 337 nm, with 150 laser shots averaged per spectrum. 2,5-Dihydroxybenzoic acid (Aldrich Chemicals) was used as the matrix for all experiments. The matrix concentration was 20 mg/mL in water. Samples were prepared using a modified “dried droplet” procedure, in which 0.3 µL of sample was spotted onto the MALDI target, followed by 0.3 µL of matrix. The mixture was then allowed to air-dry prior to analysis. Calibration was performed using instrument default settings, and data analysis was carried out using the Data Explorer software residing on the Voyager mass spectrometer.
Cloning of hum-MFng and human β3GN-T2 with human galectin-1 as the fusion partner
For the construction of plasmid containing hum-β3GN-T2 DNA or hum-MFng sequence fused with galectin-1 in the pLgals1-Tev vector (Pasek et al. 2010), the following DNA fragments were synthesized.
For the PCR amplification of the DNA sequence corresponding to the coding residues 29–397 of the hum-β3GN-T2 DNA, the following primers were used:
Upper primer: 5′-CCCAAGCTTAAAAGCAGTAGCCAAGAAAAAAATGGAAAA-3′
Lower primer: 5′-CCGCTCGAGTTAGCATTTTAAATGAGCACTCTGCAACTG-3′
For the PCR amplification of the DNA sequence corresponding to the coding residues 26–321 of the hum-MFng DNA, the following primers were used:
Upper primer: 5′-CGGGGATCCTACCACTTGAACCTGTCCCCGCAGCGGGTA-3′
Lower primer: 5′-GGCCGCAAGCTTTCATCGGGCACCCAGCTGGGGACACCA-3′
The restriction sites of enzymes are shown in italics. The PCR fragment of MFng was digested with BamHI and HindIII and that of β3GNT2 was digested with HindIII and XhoI enzymes, and the fragments were subcloned into the pLgals1-Tev vector, respectively. The DNA sequences of galectin-1-hum-β3GN-T2 and galectin-1-hum-MFng were confirmed by sequence analysis.
Expression and purification of galectin-1-hum-β3GN-T2 and galectin-1-hum-MFng fusion proteins
To express the soluble proteins, Rosetta (DE3)pLysS cells were transformed with the plasmid containing galectin-1-hum-β3GN-T2 or galectin-1-hum-MFng sequence and grown at 37°C in LB supplemented with ampicillin (100 μg/mL) to OD600 nm = 0.6–0.8. IPTG was then added to a final concentration of 1 mM, and the cells were further incubated overnight at 23°C. Cells were harvested by centrifugation at 3000 × g for 10 min and resuspended in 100 mM phosphate-buffered saline (PBS) buffer (pH 7.4), containing 0.5 M NaCl; cells were then disrupted by sonication (6 × 30 s) in ice. The solution was centrifuged at 15,000 × g for 20 min at 4°C. The resulting supernatant was loaded onto a 2 mL bed volume of the α-lactose column, which was first equilibrated with 100 mM PBS buffer (pH 7.4), containing 0.5 M NaCl (equilibrate/wash buffer). Next, the column was washed with equilibrate/wash buffer five times the bed volume of the column, and the bound protein was eluted with 100 mM lactose. The eluted fractions were analyzed by SDS–PAGE.
Purification of hum-β3GN-T2
The fusion protein galectin-1-hum-β3GN-T2 was purified using UDP-agarose columns (Figure 5A). This protein could not be cleaved with Tev protease, suggesting that the linker sequence containing the Tev cleavage sequence is not accessible to Tev protease. However, the fusion protein was enzymatically active.
Purification of free hum-MFng
About 1 mg of galectin-1-hum-MFng fusion protein was cleaved with 20 units of Tev protease in 1 mL of 50 mM Tris–HCl (pH 8.0), 0.5 mM ethylenediaminetetraacetic acid and 1 mM dithiothreitol at room temperature for 16 h. After adjusting the MnCl2 concentration to 25 mM, the protein was loaded on a UDP-agarose column, and free hum-MFng was purified and activity was measured.
Enzyme activity assay of fusion protein galectin-1-hum-β3GN-T2
For specific activity measurements, reactions were carried out at 37°C for 1 h in 100 μL of final volume containing 50 mM HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]-NaOH (pH 7.2), 10 mM MnCl2, 50 µM UDP-GlcNAc and 50 µM UDP-[3H]GlcNAc, 0.05 μCi of 3H-labeled sugar nucleotide, 2 µg of galectin-hum-β3GN-T2 and 10–40 µg of ASF as an acceptor. The reactions were terminated by adding 200 µL of ice-cold water to the incubation mixture, and they were analyzed as described above. The enzyme activity curve is fitted with the SIGMA plot using an equation defining ligand binding to one site saturation without inhibition.
Enzyme activity assay of hum-MFng
The assays were performed for 1 h at 37°C in 50 mM HEPES (pH 6.8), 10 mM MnCl2 and 500 μM UDP-GlcNAc in 100 μL of final volume. For acceptors, 4 mM pNP-α-Fuc or 100 mM fucose were used at concentrations of the MFng enzyme ranging from 0.5 to 2 µg per assay. The reactions were terminated by the addition of 200 µL ice-cold water to the incubation mixture, which was then passed through 0.5 mL of AG 1-X8 anion exchange resin in a scintillation vial, as described previously (Ramakrishnan and Qasba 2002).
Transfer of modified sugars from their UDP derivatives on a glycoprotein, ASF, Ova and O-fucosylated EGF repeat from factor VII and oligosaccharide chitotetrose
Reactions were carried out at 37°C overnight in a 25 µL volume for each enzyme in the following conditions.
Bovine Y289L-R228K-β4Gal-T1 mutant: 10 mM MnCl2, 10 mM Tris–HCl (pH 8.0), 1 mM UDP-Glc-2-keto or UDP-Gal-2-keto, 200 µg of Ova or 1 mM chitotriose (GlcNAc(β1-4GlcNAc)2) and 4 µg of enzyme. The chitotetrose samples were analyzed by MALDI-time of flight as described above and Ova samples by western blot as described below.
Hum-β3GN-T2 and hum-MFng: 50 mM HEPES-NaOH (pH 7.2), 10 mM MnCl2, 1 mM UDP-C2-keto-Glc, 2 µg of the enzyme and either 200 µg of ASF or 2 µg of O-fucosylated-EGF repeat from factor VII. The C2-keto-Glc-labeled proteins were biotinylated for western blot analysis, subsequently diluted to 30 µL in a mixture containing 50 mM NaOAc (pH 3.9) and 3 mM AOB. The biotinylation reactions were incubated with gentle shaking for 12–16 h at 25°C.
Western blotting of Ova, ASF and O-fucosylated EGF repeat from factor VII
The SDS–PAGE analyses of biotinylated Ova and ASF, before and after PNGase F treatment and O-fucosylated EGF from factor VII, were performed in 14% Tris–glycine gels. For western blot analysis, the proteins from the gels were then transferred by electrophoresis to nitrocellulose paper (0.45 μm pore size) for 2 h at 25 V, and biotinylated protein bands were identified as described previously (Boeggeman et al. 2009; Pasek et al. 2009). For loading control of Ova, after western blot analysis, membrane was washed with TBS-T (Tris-buffered saline with Tween 20) and incubated 2 h at room temperature with 1:500 monoclonal anti-Ova antibodies (Santa Cruz Biotechnology, sc-65984) in TBS-T with 5% non-fat dry milk. After three washes with TBS-T buffer, the membrane was incubated for 1 h at room temperature with rabbit anti-mouse-HRP (Santa Cruz Biotechnology, sc-358914) with a dilution of 1:2000 in TBS-T buffer with 5% non-fat dry milk. Following 3× washings with TBS-T, membrane was incubated for 2 min with the HRP substrate and exposed to film for 30 s.
Funding
This project has been funded with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. government. This research was supported in part by the Intramural Research Program of NIH, Center for Cancer Research, National Cancer Institute.
Conflict of interest
None declared.
Abbreviations
AOB, N-aminooxymethylcarbonylhydrazino-d-biotin; αppGalNAc-T, α-polypeptidyl-N-acetylgalactosaminyltransferase; ASF, Asialofetuin; β4Gal-T1, β1,4-galactosyltransferase I; β3GN-T2, β1,3-N-acetylglucosaminyltransferase-2; EGF, epidermal growth factor; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; HRP, horseradish peroxidase; IPTG, isopropyl-β-d-thiogalactopyranoside; LB, Luria–Bertani; MALDI, matrix-assisted laser desorption/ionization; MFng, Maniac Fringe; Ova, Ovalbumin; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; pNP-α-Fuc, p-nitrophenyl-α-fucose; SDS–PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; TBS-T, Tris-buffered saline with Tween 20; Tev, Tobacco etch virus.
Acknowledgements
We thank Dr. Robert Haltiwanger for providing O-fucosylated EGF repeat from Factor VII and Drs. Jack Simpson and Simona Colantonio for MS analysis.
References
- Boeggeman E, Ramakrishnan B, Kilgore C, Khidekel N, Hsieh-Wilson LC, Simpson JT, Qasba PK. Direct identification of nonreducing GlcNAc residues on N-glycans of glycoproteins using a novel chemoenzymatic method. Bioconjug Chem. 2007;18:806–814. doi: 10.1021/bc060341n. doi:10.1021/bc060341n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeggeman E, Ramakrishnan B, Pasek M, Manzoni M, Puri A, Loomis KH, Waybright TJ, Qasba PK. Site-specific conjugation of fluoroprobes to the remodeled Fc N-glycans of monoclonal antibodies using mutant glycosyltransferases: Application for cell surface antigen detection. Bioconjug Chem. 2009;20:1228–1236. doi: 10.1021/bc900103p. doi:10.1021/bc900103p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce M, Carrico IS, Ganguli AS, Yu S-H, Hangauer MJ, Hubbard SC, Kohler JJ, Bertozzi CR. Metabolic cross-talk allows labeling of O-linked β-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc Nat Acad Sci USA. 2011;108:3141–3146. doi: 10.1073/pnas.1010045108. doi:10.1073/pnas.1010045108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulcey AE, Qasba PK, Lamb J, Griffiths GL. Improved synthesis of UDP-2-(2-ketopropyl) galactose and a first synthesis of UDP-2-(2-ketopropyl) glucose for the site-specific linking of biomolecules via modified glycan residues using glycosyltransferases. Tetrahedron. 2011;67:2013–2017. doi: 10.1016/j.tet.2011.01.081. doi:10.1016/j.tet.2011.01.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltiwanger RS, Stanley P. Modulation of receptor signaling by glycosylation: Fringe is an O-fucose- β1,3-N-acetylglucosaminyltransferase. Biochem Biophys Acta. 2002;1573:328–335. doi: 10.1016/s0304-4165(02)00400-2. [DOI] [PubMed] [Google Scholar]
- Hang HC, Bertozzi CR. Ketone isosteres of 2-N-acetamidosugars as substrates for metabolic cell surface engineering. J Am Chem Soc. 2001;123:1242–1243. doi: 10.1021/ja002962b. doi:10.1021/ja002962b. [DOI] [PubMed] [Google Scholar]
- Hindsgaul O, Kaur KJ, Srivastava G, Blaszczyk-Thurin M, Crawley SC, Heerze LD, Palcic MM. Evaluation of deoxygenated oligosaccharide acceptor analogs as specific inhibitors of glycosyltransferases. J Biol Chem. 1991;266:17858–17862. [PubMed] [Google Scholar]
- Jinek M, Chen YW, Clausen H, Cohen SM, Conti E. Structural insights into the Notch-modifying glycosyltransferase Fringe. Nat Struct Mol Biol. 2006;13:945–946. doi: 10.1038/nsmb1144. doi:10.1038/nsmb1144. [DOI] [PubMed] [Google Scholar]
- Khidekel N, Arndt S, Lamarre-Vincent N, Lippert A, Poulin-Kerstien KG, Ramakrishnan B, Qasba PK, Hsieh-Wilson LC. A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications. J Am Chem Soc. 2003;125:16162–16163. doi: 10.1021/ja038545r. doi:10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- Laughlin ST, Bertozzi CR. Imaging the glycome. Proc Natl Acad Sci USA. 2009;106:12–17. doi: 10.1073/pnas.0811481106. doi:10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasek M, Boeggeman E, Ramakrishnan B, Qasba PK. Galectin-1 as a fusion partner for the production of soluble and folded human β-1,4-galactosyltransferase-T7 in E. coli. Biochem Biophys Res Commun. 2010;394:679–684. doi: 10.1016/j.bbrc.2010.03.051. doi:10.1016/j.bbrc.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasek M, Ramakrishnan B, Boeggeman E, Manzoni M, Waybright TJ, Qasba PK. Bioconjugation and detection of lactosamine moiety using α1,3-galactosyltransferase mutants that transfer C2-modified galactose with a chemical handle. Bioconjug Chem. 2009;20:608–618. doi: 10.1021/bc800534r. doi:10.1021/bc800534r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescher JA, Bertozzi CR. Chemical technologies for probing glycans. Cell. 2006;126:851–854. doi: 10.1016/j.cell.2006.08.017. doi:10.1016/j.cell.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Qasba PK, Ramakrishnan B. X-ray crystal structures of glycosyltransferas. In: Kamerling JP, Boons GJ, Lee YC, Suzuki A, Taniguchi N, Voragen AGJ, editors. Comprehensive Glycosciences: From Chemistry to Systems Biology. Burlington, MA: Elsevier Ltd; 2007. pp. 251–281. [Google Scholar]
- Ramakrishnan B, Balaji PV, Qasba PK. Crystal structure of β1,4-galactosyltransferase complex with UDP-Gal reveals an oligosaccharide acceptor binding site. J Mol Biol. 2002;318:491–502. doi: 10.1016/S0022-2836(02)00020-7. doi:10.1016/S0022-2836(02)00020-7. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan B, Boeggeman E, Qasba PK. Mutation of arginine 228 to lysine enhances the glucosyltransferase activity of bovine β-1,4-galactosyltransferase I. Biochemistry. 2005;44:3202–3210. doi: 10.1021/bi0479454. doi:10.1021/bi0479454. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan B, Boeggeman E, Qasba PK. Novel method for in vitro O-glycosylation of proteins: Application for bioconjugation. Bioconjug Chem. 2007;18:1912–1918. doi: 10.1021/bc7002346. doi:10.1021/bc7002346. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan B, Qasba PK. Crystal structure of lactose synthase reveals a large conformational change in its catalytic component, the β1,4-galactosyltransferase-I. J Mol Biol. 2001;310:205–218. doi: 10.1006/jmbi.2001.4757. doi:10.1006/jmbi.2001.4757. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan B, Qasba PK. Structure-based design of β1,4-galactosyltransferase I (β4Gal-T1) with equally efficient N-acetylgalactosaminyltransferase activity: Point mutation broadens β4Gal-T1 donor specificity. J Biol Chem. 2002;277:20833–20839. doi: 10.1074/jbc.M111183200. doi:10.1074/jbc.M111183200. [DOI] [PubMed] [Google Scholar]
- Ramasamy V, Ramakrishnan B, Boeggeman E, Ratner DM, Seeberger PH, Qasba PK. Oligosaccharide preferences of β1,4-galactosyltransferase-I: Crystal structures of Met340His mutant of human β1,4-galactosyltransferase-I with a pentasaccharide and trisaccharides of the N-glycan moiety. J Mol Biol. 2005;353:53–67. doi: 10.1016/j.jmb.2005.07.050. doi:10.1016/j.jmb.2005.07.050. [DOI] [PubMed] [Google Scholar]
- Rampall R, Li ASY, Moloney DJ, Georgiou SA, Luther KB, Nita-Lazar A, Haltiwanger RS. Lunatic fringe, manic fringe, and radical fringe recognize similar specificity determinants in O-fucosylated epidermal growth factor-like repeats. J Biol Chem. 2005;280:42454–42463. doi: 10.1074/jbc.M509552200. doi:10.1074/jbc.M509552200. [DOI] [PubMed] [Google Scholar]
- Schwefel D, Maierhofer C, Beck JG, Seeberge S, Diederichs K, Möller HM, Welte W, Wittmann V. Structural basis of multivalent binding to wheat germ agglutinin. J Am Chem Soc. 2010;132:8704–8719. doi: 10.1021/ja101646k. doi:10.1021/ja101646k. [DOI] [PubMed] [Google Scholar]
- Seko A, Yamashita K. Activation of β1,3-N-acetylglucosaminyltransferase-2 (β3Gn-T2) by β3Gn-T8. Possible involvement of β3Gn-T8 in increasing poly-N-acetyllactosamine chains in differentiated HL-60 cells. J Biol Chem. 2008;283:33094–33100. doi: 10.1074/jbc.M806933200. doi:10.1074/jbc.M806933200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P, Okajima T. Roles of glycosylation in Notch signaling. Curr Top Dev Biol. 2010;92:131–164. doi: 10.1016/S0070-2153(10)92004-8. doi:10.1016/S0070-2153(10)92004-8. [DOI] [PubMed] [Google Scholar]
- Togayachi A, Kozono Y, Ishida H, Abe S, Suzuki N, Tsunoda Y, Hagiwara K, Kuno A, Ohkura T, Sato N, et al. Polylactosamine on glycoproteins influences basal levels of lymphocyte and macrophage activation. Proc Nat Acad Sci USA. 2007;104:15829–15834. doi: 10.1073/pnas.0707426104. doi:10.1073/pnas.0707426104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togayachi A, Kozono Y, Kuno K, Ohkura T, Sato T, Hirabayashi J, Ikehara Y, Narimatsu H. β3GnT2 (B3GNT2), a major polylactosamine synthase: Analysis of B3GNT2-deficient mice. Meth Enzymol. 2010;479:185–204. doi: 10.1016/S0076-6879(10)79011-X. doi:10.1016/S0076-6879(10)79011-X. [DOI] [PubMed] [Google Scholar]
- Togayachi A, Sato T, Narimatsu H. Comprehensive enzymatic characterization of glycosyltransferases with a β3GT or β4GT motif. Meth Enzymol. 2006;416:91–102. doi: 10.1016/S0076-6879(06)16006-1. doi:10.1016/S0076-6879(06)16006-1. [DOI] [PubMed] [Google Scholar]
- Tumbale P, Jamaluddin H, Thiyagarajan N, Acharya KR, Brew K. Screening a limited structure-based library identifies UDP-GalNAc-specific mutants of α-1,3-galactosyltransferase. Glycobiology. 2008;18:1036–1043. doi: 10.1093/glycob/cwn083. doi:10.1093/glycob/cwn083. [DOI] [PubMed] [Google Scholar]
- Unligil UM, Zhou S, Yuwaraj S, Sarkar M, Schachter H, Rini JM. X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: Catalytic mechanism and a new protein superfamily. EMBO J. 2000;19:5269–5280. doi: 10.1093/emboj/19.20.5269. doi:10.1093/emboj/19.20.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Kannagi R, Toole BP. Glycosylation Changes in Cancer. In: Varki A, Cummings R, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2009. 2nd Ed. Chapter 44. [PubMed] [Google Scholar]
- Wang Z, Du J, Che PL, Meledeo MA, Yarema KJ. Hexosamine analogs: From metabolic glycoengineering to drug discovery. Curr Opin Chem Biol. 2009;13:565–572. doi: 10.1016/j.cbpa.2009.08.001. doi:10.1016/j.cbpa.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]