Abstract
Phage display technology is an emerging drug discovery tool. Using that approach, short peptides that mimic part of a carbohydrate's conformation are selected by screening a peptide-displaying phage library with anti-carbohydrate antibodies. Chemically synthesized peptides with an identified sequence have been used as an alternative ligand to carbohydrate-binding proteins. These peptides represent research tools useful to assay the activities of glycosyltransferases and/or sulfotransferases or to inhibit the carbohydrate-dependent binding of proteins in vitro and in vivo. Peptides can also serve as immunogens to raise anti-carbohydrate antibodies in vivo in animals. Phage display has also been used in single-chain antibody technology by inserting an immunoglobulin's variable region sequence into the phage. A single-chain antibody library can then be screened with a carbohydrate antigen as the target, resulting in a recombinant anti-carbohydrate antibody with high affinity to the antigen. This review provides examples of successful applications of peptide-displaying phage technology to glycobiology. Such an approach should benefit translational research by supplying carbohydrate-mimetic peptides and carbohydrate-binding polypeptides.
Keywords: bacterial capsule saccharides, cancer, chondroitin sulfate, heparan sulfate, selectin
Introduction
A significant advantage of gene engineering technology is that DNA and by extension recombinant proteins can be amplified, an approach opposite to that of old-fashioned biochemistry, in which experimenters generally began with large amounts of material and ended up with a very small amount of purified sample. Although we can now produce recombinant, and thus amplifiable, glycosyltransferases by gene engineering techniques, we cannot produce the carbohydrate itself. In this regard, peptide-displaying phage technology provides carbohydrate-mimetic peptides, which are equivalent to amplifiable carbohydrates (Oldenburg et al. 1992; Scott et al. 1992; Taki et al. 1997, 2008; Fukuda 2006). Carbohydrate-mimetic peptides can be used as immunogens to raise anti-carbohydrate antibodies for cancer therapy (Kieber-Emmons et al. 1999; Kozbor 2009; Monzavi-Karbassi et al. 2001) and infectious diseases (Pincus et al. 1998; Hou and Gu 2003; Gnanasekar et al. 2004; Bua et al. 2009; Menendez et al. 2010; Wu et al. 2010). In addition, phage display technology provides peptide sequences that bind to carbohydrates, enabling us to produce recombinant humanized monoclonal anti-carbohydrate IgG antibodies against cancer cells (Kubota et al. 2010) and infectious agents (Berdichevsky et al. 1999; Haidaris et al. 2001).
Phage display: how it works
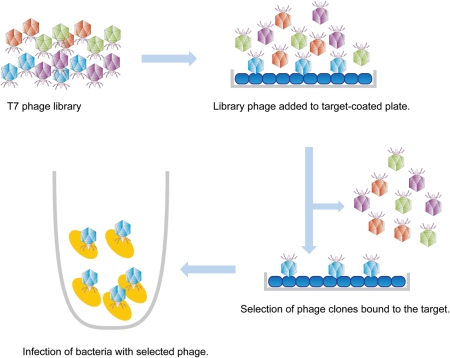
Phages or bacteriophages are viruses that infect bacteria. Currently, two types of phages are available for phage display. One is an M13-based filamentous phage. M13 infects bacteria by the interaction of the phage coat protein 3 (pIII) with the F pili of host bacteria (Smith 1985; Smith and Scott 1993). Following the insertion of a foreign gene encoding a short peptide at the 5′ end of pIII gene, each phage displays 4–5 copies of the short peptide as C-terminal fusion. A second type of phage is the T7-based phage with a round head plus tail fiber (Rosenberg et al. 1996; Figure 1). T7 can display 200–400 copies of peptides as N-terminal fusions on the head (Rosenberg et al. 1996). Inserted genes are made from oligonucleotides encoding 4–15 peptides that can be chemically synthesized in a completely random manner. A circular peptide library can also be produced by adding cysteine residues at both N- and C-termini (Pasqualini et al. 1995). Phage libraries constructed in this manner can display more than 109 peptides in an aqueous volume of <1 mL.
Fig. 1.
Biopanning of T7-based peptide-displaying phage library. A phage library consisting of as many as 109–10 clones is added to a plastic well coated with a target, such as a monoclonal anti-carbohydrate antibody. Phage clones that do not bind the target will be washed away from the plate. Competent host bacteria are then added to the well, so that phages remaining in the well will infect bacteria. Following the growth of infected bacteria, the phages are amplified. By repeating this procedure 3–4 rounds, phage clones displaying peptides with high target-binding affinity are isolated. Biopanning of the filamentous M13-based phage can be done in a similar manner.
Following the infection of the host bacterium, the phage proliferates in host cells. In the M13-based filamentous fd phage, infected bacteria become resistant to tetracycline as phages carry a tetracycline-resistance gene and form colonies on tetracycline-containing agar plates. In the host cell, phage DNA replicates produce coat proteins and assemble into new particles, which leak from the host bacterium and can be collected from a liquid culture of infected bacteria. In T7-based phages, the phage amplified in bacteria burst from host bacterial cells, creating a plaque on soft agar plates. Importantly, each colony or plaque appearing on a plate is a genetically identical clone.
The most commonly used method for library screening is biopanning (Figure 1), in which phage clones displaying a peptide sequence are selected using a target molecule, such as an antibody (Hoess et al. 1993; Taki et al. 1997; Fukuda et al. 2000; Fukuda 2006; Popa et al. 2006). Antibodies are coated on a plastic plate. Media containing phage particles is applied to the plate, and phages that do not bind to the antibody-coated plate are washed away. The inserts of the antibody-binding phage are sequenced to reveal epitopes recognized by the antibody. In this way, the peptide-displaying phage has been used for epitope mapping of antibodies (Scott and Smith 1990; Smith and Scott 1993; Cortese et al. 1995; Irving et al. 2001). However, the peptide displayed on the phage may not necessarily represent a “peptide”, as shown by experiments employing streptavidin as a target: streptavidin-binding peptides were isolated and their binding to streptavidin was inhibited by biotin (Devlin et al. 1990). As biotin is not a peptide, this demonstrated that it is possible to identify a molecular mimetic for non-peptide compounds.
Use of phage libraries to identify carbohydrate mimetics
In 1992, Goldstein's group screened peptide-displaying phage libraries using the lectin concanavalin A (Con A) as a target and identified Con A ligand-mimetic peptides (Oldenburg et al. 1992; Scott et al. 1992). Identified peptides contained the consensus sequence YPY and bound Con A with high affinity comparable with that of methyl α-d-mannopyranoside, a known carbohydrate ligand. This pioneering work led to the development of carbohydrate-mimetic peptides in glycobiology.
In subsequent studies, monoclonal anti-carbohydrate antibodies served as targets for phage display library screening to identify carbohydrate-mimetic peptides. Pastan's group screened a peptide-displaying phage library by a monoclonal anti-Lewis Y antibody (clone B3) and found that phage clones selected by this antibody encode the sequence APWLYGPA (Hoess et al. 1993). Alanine-scanning mutagenesis of this sequence indicated that PWLY is critical for B3 binding. The authors also showed that PWLY peptide can induce an immune response to the Lewis Y antigen in rabbits and mice, demonstrating that carbohydrate-mimetic peptides provide a novel strategy to elicit immune responses for carbohydrate antigens (Lou and Pastan 1999).
Peptides identified by anti-carbohydrate antibodies and lectins are generally assumed to be carbohydrate mimetics, or moieties behaving like carbohydrates. However, it is unclear whether peptides mimic a carbohydrate structure or merely provide an epitope that fits the antigen-binding site of the antibody. Harris et al. (1997) addressed this question by identifying a series of peptides using closely related monoclonal antibodies directed against the cell wall polysaccharide of group A Streptococcus. All identified peptides bound at or near the carbohydrate-binding site. The group found that each monoclonal antibody binds each specific peptide sequence but could not identify a common sequence among peptides selected by multiple monoclonal antibodies. These findings indicate that peptides likely do not mimic the carbohydrate structure.
Taki et al. (1997) successfully identified 9-mer peptides apparently mimicking the non-reducing terminal galactose structure. The consensus sequence VPPXFXXXY was recognized by two monoclonal antibodies directed to lactotetraosylceramide and neolactotetraosylceramide. These peptides also bound to Ricinus lectin and inhibited Jack bean β-galactosidase activity toward neolactotetraosylceramide. The authors concluded that the 9-mers are sufficient to mimic the carbohydrate epitope structure. These studies suggest that peptides function in carbohydrate mimicry by interacting with carbohydrate-binding proteins at the epitope recognition site. However, the peptide identified by a monoclonal antibody may not necessarily mimic the carbohydrate structurally, particularly when an epitope is part of a complex structure (Dharmasena et al. 2007). A study of three anti-Lewis A monoclonal antibodies showed that each antibody recognizes a part of Lewis A oligosaccharide (Young et al. 1983). It is likely that each peptide selected by a monoclonal antibody represents a three-dimensional structure that uniquely fits the antigen-binding site of the antibody, but the peptide may not represent the entire carbohydrate structure.
Carbohydrate-mimetic peptides as immunogens
Since many anti-carbohydrate antibodies were raised by immunizing mice by cancer cells and tissues (Solter and Knowles 1978; Gooi et al. 1981; Anger et al. 1982; Shevinsky et al. 1982; Kannagi et al. 1983; Hakomori 1984, 1989; Kannagi 2000), complex carbohydrate structures presented on the cell surface often serve as potent immunogens. However, oligosaccharides or monomeric forms of carbohydrates are frequently poor immunogens, as they are T-cell-independent antigens (Milich and McLachlan 1986; Dullforce et al. 1998). Investigators have explored the molecular mimicry of tumor-associated carbohydrate antigens by peptides in order to raise antibodies against tumor-associated carbohydrate antigens, as efforts to create cancer vaccines (Kieber-Emmons et al. 1999; Lou and Pastan 1999). The rationale behind these studies is that carbohydrate-mimetic peptides can elicit antibody responses that cross-react with representative human cancer-associated carbohydrate antigens. In one such study, the vaccination of mice with a cancer-associated antigenic-mimetic peptide reduced tumor growth and prolonged host survival in both murine sarcoma and breast tumor models (Kieber-Emmons et al. 1999).
Microbial polysaccharides are often poor immunogens. Pincus et al. (1998) developed a protective monoclonal antibody against the type III capsular polysaccharide of group B streptococci (GBS), using peptide phage display to identify epitope analogs. Among sequences identified, FDTGAFDPDWPA peptide inhibited antibody binding to GBS. When the peptide was conjugated to three different carriers and used to immunize mice, all mice produced a significant antibody response to GBS and to the purified capsular polysaccharide following a single immunization.
Other studies (Pincus et al. 1998; Kieber-Emmons et al. 1999) indicate that carbohydrate-mimetic peptides are both antigenic and immunogenic. Peptide mimetopes may enhance vaccine efficacy by enhancing immune responses to poorly immunogenic bacterial carbohydrate epitopes.
Peptides that function as ligands for carbohydrate-binding protein
Several carbohydrate-mimetic peptides that bind to carbohydrate-binding proteins have been identified. We screened a peptide-displayed phage library using the anti-Lewis A antibody clone 7LE and identified a series of 7-mer peptides (Fukuda et al. 2000). Among them, the peptide designated I-peptide, or IELLQAR, bound to E-selectin, P-selectin and L-selectin in a calcium-dependent manner. Synthetic I-peptide inhibited binding of the selectin ligand carbohydrate, sialyl Lewis X, to E-selectin. When I-peptide was injected intravenously into mice prior to the injection of sialyl Lewis X-expressing melanoma cells, it inhibited melanoma colonization in mouse lung, suggesting the existence of an I-peptide receptor in lung endothelial cell surfaces. We had hypothesized that I-peptide interacts with E- or P-selectin in the lung vasculature. However, I-peptide receptor activity was found in E-/P-selectin double-deficient mutant mice, suggesting the existence of a novel I-peptide receptor (Zhang et al. 2002). A subsequent study identified the I-peptide receptor as a pre-mRNA splicing factor (Hatakeyama et al. 2009). This surprising finding is reminiscent of reports that some galectins are nuclear and exhibit mRNA splicing activity (Dagher et al. 1995; Vyakarnam et al. 1997). Thus, both pre-mRNA splicing factors and members of the galectin family exhibit both nuclear pre-mRNA splicing activity and cell surface carbohydrate-binding activity (Haudek et al. 2010).
Ishikawa et al. (1998) isolated phage clones that bound to a monoclonal antibody against ganglioside GD1α. One of the phages displaying the sequence WHWRHRIPLQLAAGR inhibited the adhesion of mouse lymphoma cells to hepatic endothelial cells. The group identified WHW as the minimal sequence required for this function. Intravenous injection of WHW peptide-attached liposomes efficiently inhibited the metastasis of lymphoma RAW117-H10 cells to the liver, lung and spleen (Takikawa et al. 2000). Their results strongly suggest that GD1α mediates the adhesion of RAW117-H10 cells to endothelial cells in these organs. This group has identified other short peptides using anti-ganglioside antibodies (Taki et al. 1997; Matsubara et al. 1999; Popa et al. 2006) with the aim of developing novel therapeutics against infectious disease and cancer (Taki et al. 2008).
Polysialic acid (PSA) attached to the neural cell adhesion molecule is a permissive determinant for numerous morphogenetic and neural plasticity processes (Yang et al. 1992; Suzuki et al. 2005). PSA-mimetic peptides were identified by phage display library screening using a monoclonal antibody specific for PSA as the target (Marino et al. 2009). In cultures of neural progenitors, micromolar concentrations of a peptide designated PR-21 promoted axon growth, defasciculation and cell migration. When injected into the developing chicken retina, the peptides altered the pathfinding of retinal ganglion cell axons. Moreover, PR-21 peptide enhanced the migration of neuroblasts grafted into the mouse brain. These effects were selective and required the presence of PSA on transplanted cells in mice and in cultured neurons (Torregrossa et al. 2004; Marino et al. 2009). In an independent study, Schnaar's group showed that the injection of sialidase enhances recovery from spinal cord injury in rats (Mountney et al. 2010). These studies suggest that both PSA-mimetic peptides and sialidase could be useful as therapeutics for spinal cord injury.
N-deacetylase–N-sulfotransferase 1 (Ndst1) catalyzes the reaction required for the biosynthesis of heparan sulfate and heparin by the removal of acetyl groups from the subsets of N-acetylglucosamine units and the subsequent sulfation of resulting free amino groups (Aikawa and Esko 1999). When a phage library displaying 10-mer cyclic peptides was screened against Ndst1, two peptides were selected: CRGWRGEKIGNC and CNMQALSMPVTC. The former binds to heparan sulfate and blocks binding of enzyme to substrate, whereas the latter inhibits mNdst1 activity by a direct interaction at the active site. This study showed that phage display is useful for developing inhibitors of enzymes functioning in glycan synthesis (Gesteira et al. 2010).
Single-chain variable region antibody for carbohydrate antigen
The studies described in the previous sections indicate that peptides can be displayed on the bacteriophage surface in a way that can be recognized by antibodies (Smith 1985; Parmley and Smith 1988; Smith and Scott 1993). It was then shown that the reverse situation is also possible: a fragment of an antibody or the variable region of IgG displayed on a filamentous phage can be specifically enriched by selection on an immobilized antigen (McCafferty et al. 1990; Barbas et al. 1991). Phage display technology has advantages over the direct expression of antibody genes in the viral genome. By combining high cloning efficiency of the lambda phage with filamentous phage display (Hogrefe et al. 1993), large libraries allow an enhanced diversity of the antibody repertoire, making it possible to detect a wide range of antigens (Persson et al. 1991; Little et al. 1999).
The immunoglobulin-encoding DNA sequences cloned for antibody engineering are the variable (V) domains of both heavy (H) and light (L) chains, which constitute the antigen-binding site. If the humanized antibody is desired, cDNAs are prepared by reverse transcription of RNA extracted from human B-lymphocytes, and the VL and VH immunoglobulin regions are amplified by polymerase chain reaction (PCR). PCR primer sets are designed on the basis of conserved flanking base sequences found at the beginning of VH and VL genes (corresponding to the FR1 in the V region), at the end of the JH or JL segments (FR4 in the rearranged V region) and at the proximal CL or CH1 constant regions (Gavilondo and Larrick 2000; Rojas et al. 2004). Alternatively, mice can be immunized by an antigen and cDNAs prepared from spleen RNA, which can then serve as PCR templates (De Jaeger et al. 1997).
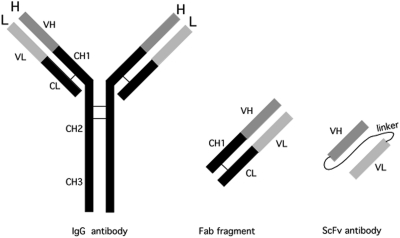
Phage clones displaying VH and VL genes with antigen-binding activities can be enriched by biopanning with an antigen. Identified peptide sequences are then incorporated into the bacterial expression vectors for single-chain variable fragments (scFv) to produce a single-chain antibody, which is composed of VH, VL and linker (Figure 2). Carbohydrate antigens, such as heparan sulfate and chondroitin sulfate, were coated on a polystyrene plastic surface and an scFv phage library was screened (van Kuppevelt et al. 1998; Smetsers et al. 2004; Smits et al. 2006). For the scFv anti-Tn antibody, Kubota et al. (2010) used avidin-conjugated beads with the biotinylated synthetic Tn antigen, GalNAcα1 → Ser/Thr, for positive selection and those with biotinylated bovine serum albumin conjugated with the blood group A trisaccharide, GalNAcα1 → 3(Fucα1 → 2)Galβ1→, for negative selection. Clones selected in the first round of biopanning were expanded in vitro by host bacterial cell infection and then subjected to three or four rounds of biopanning. Clones specifically binding to the antigen were progressively enriched (1000–10,000-fold) over each round. After three or four rounds, individual phage clones were then characterized in terms of V region sequence and antigen affinity. This method allows the isolation of human and mouse antibodies against any antigen and bypasses both hybridoma technology and in vivo immunization.
Fig. 2.
Schematic illustration of the IgG antibody, Fab fragment of IgG and single-chain (scFv) antibody.
Single-chain anti-carbohydrate antibodies were produced against bacterial cell wall (Deng et al. 1994, 1995; Hughes-Jones et al. 1994). Subsequently, van Kuppevelt et al. produced and characterized several anti-heparan sulfate scFv antibodies by screening a semi-synthetic scFv phage library by panning with heparan sulfate from the bovine kidney (van Kuppevelt et al. 1998; Jenniskens et al. 2000; Dennissen et al. 2002; van de Westerlo et al. 2002; ten Dam et al. 2004; Kurup et al. 2007; Uchimura et al. 2010). Each antibody, encoded by distinct VH and VL domain peptide sequences, reacted differently toward various heparan sulfate preparations and showed different staining patterns in tissue sections, suggesting that the antibodies recognize unique epitopes within the heparan sulfate chain. Since the scFv antibody designated RB4CD12 requires a fully sulfated epitope for binding, this antibody was used to measure the enzymatic activity of an endosulfatase, which removes sulfate residues from heparan sulfate (Uchimura et al. 2010).
Anti-chondroitin sulfate scFv antibodies were also produced (Smetsers et al. 2004). These antibodies show distinct immunostaining patterns in normal vs psoriasis skin, as well as in neural tissues and tumors, suggesting that each recognizes a unique structure within the chondroitin sulfate molecule (Smetsers et al. 2004). Those antibodies were used for determining functional microdomains within chondroitin sulfates and dermatan sulfates in metastatic cancer cells (Li et al. 2008): structural analysis of chondroitin/dermatan sulfates from mouse Lewis lung carcinoma-derived cell lines with different metastatic potentials suggested a potential role for chondroitin sulfate E-like structures, which was supported by immunocytochemistry with the chondroitin sulfate E epitope, which is specifically recognized by the phage display antibody GD3G7. Interestingly, both this antibody and a chondroitin sulfate E decasaccharide, the minimal structure recognized by GD3G7, antagonized metastasis likely by modifying proliferative and invasive behavior of metastatic tumor cells.
Phage display technology has also been used to produce antibodies against “difficult” antigens. For example, the Tn antigen or GalNAcα1 → Ser/Thr is highly expressed in carcinoma tumors in the gastric, colorectal, ovarian, breast and pancreatic tumors, whereas its expression is very limited in benign and normal tissues (Springer 1984). Tn antigen expression is reportedly an independent marker for poor prognosis in carcinomas (Iwasaki et al. 2000; Hiraoka et al. 2002; Rousseau et al. 2002; Engelsberg et al. 2003), suggesting that it could be an attractive candidate for cancer immunotherapy. However, due to the small size of the antigen, it has been difficult to generate an anti-Tn antibody using conventional hybridoma technology. To overcome this difficulty, Kubota et al. (2010) identified the anti-Tn scFv antibody by phage display technology. To do so, they immunized mice with Tn antigen-positive Jurkat cells, isolated mouse spleen, made cDNAs for PCR amplification of VH- and VL-encoding sequences and constructed their own scFv phage library. The library was screened by biopanning with the synthetic Tn antigen for positive selection and with the blood group A antigen for negative selection. Blood group A antigen contains a terminal GalNAcα1 → structure, which should be excluded from the Tn-binding phage pool. Tn antigen-specific scFv was then linked to the Fc constant region of human IgG to produce scFv-Fc fusion proteins. Since the same group previously found that ADCC (antigen-dependent cell-mediated cytotoxicity) activity is enhanced by IgG with fucose-free oligosaccharides in the Fc region (Shinkawa et al. 2003; Natsume et al. 2005), they produced anti-Tn scFv-Fc fusion in a Chinese hamster ovary cell line, in which FUT8 gene encoding α-1,6-fucosyltransferase was knocked out (Yamane-Ohnuki et al. 2004). Human scFv-Fc antibody produced in this manner exhibited high Tn-ADCC activity.
ScFv technology has been used to produce humanized antibodies against weakly immunogenic pathogens (Gerstenbruch et al. 2010; Hussack et al. 2011). Neisseria meningitidis is one of the most common causes of bacterial meningitis in infants and young adults (van Deuren et al. 2000; Thorburn et al. 2001; Munro 2002). Type B, which is responsible for up to 80% of the total cases of N. meningitidis in industrialized countries, is resistant to any attempt to develop a capsule-based vaccine due to poor immunogenicity of the capsule. Weak immunogenicity is probably due to the similarity between the type B capsule polysaccharide and PSA expressed on the neural cell adhesion molecule in mammalian tissues (Finne et al. 1983). Thus, type B capsular saccharide is a self-antigen that cannot stimulate an immune response but can potentially induce autoimmunity if used as a vaccine (Wildes and Tunkel 2002). Beninati et al. (2001, 2004) raised a mouse monoclonal antibody designated Seam 3 against a chemically modified form of the group B capsular polysaccharides. They then immunized mice with this antibody and produced an anti-idiotypic scFv phage library using spleen mRNA from immunized mice. The scFv specific to Seam 3 competed with group B capsular polysaccharides for binding to Seam 3. Moreover, when these scFv antibodies were injected to mice and rabbits, antibodies reacting with meningococci but not with human PSA were produced.
Additional examples of scFv antibodies against capsular lipopolysaccharides have been reported for Chlamydiaceae (Gerstenbruch et al. 2010), Clostridium thermocellum (Berdichevsky et al. 1999), Escherichia coli O157:H7 (Kanitpun et al. 2004) and E. coli endotoxin B (Chung et al. 2008), Burkholderia mallei and Burkholderia pseudomallei (Kim et al. 2010).
Fujita-Yamaguchi's group has produced scFv antibodies against N-glycans (Sakai et al. 2007; Zhang et al. 2007; Yuasa et al. 2010). Since the N-glycan core structure is shared by many glycoproteins, and N-glycans are expressed in eukaryotes, these carbohydrates are not immunogenic and an antibody for N-glycans is not available. Antibodies produced by scFv technology could provide useful reagents to study the localization and function of N-glycans in mammalian cells.
Future perspectives
Phage display is an emerging approach useful for combinatorial drug discovery (Ruoslahti 2000; Landon et al. 2004). Carbohydrate-mimetic peptides whose sequences have been identified by peptide-displaying phage library screening may have benefits over genuine carbohydrates because of their ease of synthesis. Peptides also have advantages over synthetic organic chemistry-based drugs, as peptides are degradable to amino acids in vivo, avoiding side effects in humans and environmental contamination. Carbohydrate-mimetic peptides and carbohydrate-binding single-chain recombinant antibodies should serve as valuable tools in both basic and translational glycobiology research.
Funding
The author has been supported by NIH grant CA71932.
Conflict of interest
None declared.
Abbreviations
A, Ala; ADCC, antigen-dependent cell-mediated cytotoxicity; C, Cys; Con A, concanavalin A; D, Asp; E, Glu; F, Phe; Fuc, fucose; G, Gly; Gal, galactose; GalNAc, N-acetylgalactosamine; GBS, group B streptococci; GlcNAc, N-acetylglucosamine; H, His; I, Ile; K, Lys; L, Leu; M, Met; Man, mannose; N, Asn; Ndst1, N-deacetylase–N-sulfotransferase 1; P, Pro; PCR, polymerase chain reaction; pIII, protein 3; PSA, polysialic acid; Q, Gln; R, Arg; S, Ser; scFv, single-chain variable fragments; T, Thr; V, Val; W, Trp; Y, Tyr.
Acknowledgements
The author greatly appreciates Dr. Elise Lamar for her excellent editing of the manuscript.
References
- Aikawa J, Esko JD. Molecular cloning and expression of a third member of the heparan sulfate/heparin GlcNAc N-deacetylase/N-sulfotransferase family. J Biol Chem. 1999;274:2690–2695. doi: 10.1074/jbc.274.5.2690. doi:10.1074/jbc.274.5.2690. [DOI] [PubMed] [Google Scholar]
- Anger BR, Lloyd KO, Oettgen HF, Old LJ. Mouse monoclonal IgM antibody against human lung cancer line SK-LC-3 with specificity for H(O) blood group antigen. Hybridoma. 1982;1:139–147. doi: 10.1089/hyb.1.1982.1.139. doi:10.1089/hyb.1.1982.1.139. [DOI] [PubMed] [Google Scholar]
- Barbas CF, 3rd, Kang AS, Lerner RA, Benkovic SJ. Assembly of combinatorial antibody libraries on phage surfaces: The gene III site. Proc Natl Acad Sci USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. doi:10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beninati C, Arseni S, Mancuso G, Magliani W, Conti S, Midiri A, Biondo C, Polonelli L, Teti G. Protective immunization against group B meningococci using anti-idiotypic mimics of the capsular polysaccharide. J Immunol. 2004;172:2461–2468. doi: 10.4049/jimmunol.172.4.2461. [DOI] [PubMed] [Google Scholar]
- Beninati C, Oggioni M, Mancuso G, Midiri A, Polonelli L, Pozzi G, Teti G. Anti-idiotypic vaccination against group B streptococci. Int Rev Immunol. 2001;20:263–273. doi: 10.3109/08830180109043038. doi:10.3109/08830180109043038. [DOI] [PubMed] [Google Scholar]
- Berdichevsky Y, Ben-Zeev E, Lamed R, Benhar I. Phage display of a cellulose binding domain from Clostridium thermocellum and its application as a tool for antibody engineering. J Immunol Methods. 1999;228:151–162. doi: 10.1016/s0022-1759(99)00096-4. doi:10.1016/S0022-1759(99)00096-4. [DOI] [PubMed] [Google Scholar]
- Bua A, Rosu V, Molicotti P, Das Gupta SK, Ahmed N, Zanetti S, Sechi LA. Phages specific for mycobacterial lipoarabinomannan help serodiagnosis of tuberculosis. New Microbiol. 2009;32:293–296. [PubMed] [Google Scholar]
- Chung WY, Sack M, Carter R, Spiegel H, Fischer R, Hirst TR, Williams NA, James RF. Phage-display derived single-chain fragment variable (scFv) antibodies recognizing conformational epitopes of Escherichia coli heat-labile enterotoxin B-subunit. J Immunol Methods. 2008;339:115–123. doi: 10.1016/j.jim.2008.08.005. doi:10.1016/j.jim.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Cortese R, Monaci P, Nicosia A, Luzzago A, Felici F, Galfre G, Pessi A, Tramontano A, Sollazzo M. Identification of biologically active peptides using random libraries displayed on phage. Curr Opin Biotechnol. 1995;6:73–80. doi: 10.1016/0958-1669(95)80012-3. doi:10.1016/0958-1669(95)80012-3. [DOI] [PubMed] [Google Scholar]
- Dagher SF, Wang JL, Patterson RJ. Identification of galectin-3 as a factor in pre-mRNA splicing. Proc Natl Acad Sci USA. 1995;92:1213–1217. doi: 10.1073/pnas.92.4.1213. doi:10.1073/pnas.92.4.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jaeger G, Buys E, Eeckhout D, Bruyns AM, De Neve M, De Wilde C, Gerats T, Van Montagu M, Fischer R, Depicker A. Use of phage display for isolation and characterization of single-chain variable fragments against dihydroflavonol 4-reductase from Petunia hybrida. FEBS Lett. 1997;403:116–122. doi: 10.1016/s0014-5793(97)00025-2. doi:10.1016/S0014-5793(97)00025-2. [DOI] [PubMed] [Google Scholar]
- Deng SJ, MacKenzie CR, Hirama T, Brousseau R, Lowary TL, Young NM, Bundle DR, Narang SA. Basis for selection of improved carbohydrate-binding single-chain antibodies from synthetic gene libraries. Proc Natl Acad Sci USA. 1995;92:4992–4996. doi: 10.1073/pnas.92.11.4992. doi:10.1073/pnas.92.11.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng SJ, MacKenzie CR, Sadowska J, Michniewicz J, Young NM, Bundle DR, Narang SA. Selection of antibody single-chain variable fragments with improved carbohydrate binding by phage display. J Biol Chem. 1994;269:9533–9538. [PubMed] [Google Scholar]
- Dennissen MA, Jenniskens GJ, Pieffers M, Versteeg EM, Petitou M, Veerkamp JH, van Kuppevelt TH. Large, tissue-regulated domain diversity of heparan sulfates demonstrated by phage display antibodies. J Biol Chem. 2002;277:10982–10986. doi: 10.1074/jbc.M104852200. doi:10.1074/jbc.M104852200. [DOI] [PubMed] [Google Scholar]
- Devlin JJ, Panganiban LC, Devlin PE. Random peptide libraries: A source of specific protein binding molecules. Science. 1990;249:404–406. doi: 10.1126/science.2143033. doi:10.1126/science.2143033. [DOI] [PubMed] [Google Scholar]
- Dharmasena MN, Jewell DA, Taylor RK. Development of peptide mimics of a protective epitope of Vibrio cholerae Ogawa O-antigen and investigation of the structural basis of peptide mimicry. J Biol Chem. 2007;282:33805–33816. doi: 10.1074/jbc.M707314200. doi:10.1074/jbc.M707314200. [DOI] [PubMed] [Google Scholar]
- Dullforce P, Sutton DC, Heath AW. Enhancement of T cell-independent immune responses in vivo by CD40 antibodies. Nat Med. 1998;4:88–91. doi: 10.1038/nm0198-088. doi:10.1038/nm0198-088. [DOI] [PubMed] [Google Scholar]
- Engelsberg A, Hermosilla R, Karsten U, Schulein R, Dorken B, Rehm A. The Golgi protein RCAS1 controls cell surface expression of tumor-associated O-linked glycan antigens. J Biol Chem. 2003;278:22998–23007. doi: 10.1074/jbc.M301361200. doi:10.1074/jbc.M301361200. [DOI] [PubMed] [Google Scholar]
- Finne J, Leinonen M, Makela PH. Antigenic similarities between brain components and bacteria causing meningitis. Implications for vaccine development and pathogenesis. Lancet. 1983;2:355–357. doi: 10.1016/s0140-6736(83)90340-9. doi:10.1016/S0140-6736(83)90340-9. [DOI] [PubMed] [Google Scholar]
- Fukuda MN. Screening of peptide-displaying phage libraries to identify short peptides mimicking carbohydrates. Methods Enzymol. 2006;416:51–60. doi: 10.1016/S0076-6879(06)16004-8. doi:10.1016/S0076-6879(06)16004-8. [DOI] [PubMed] [Google Scholar]
- Fukuda MN, Ohyama C, Lowitz K, Matsuo O, Pasqualini R, Ruoslahti E, Fukuda M. A peptide mimic of E-selectin ligand inhibits sialyl Lewis X-dependent lung colonization of tumor cells. Cancer Res. 2000;60:450–456. [PubMed] [Google Scholar]
- Gavilondo JV, Larrick JW. Antibody engineering at the millennium. Biotechniques. 2000;29:128–132. doi: 10.2144/00291ov01. 134–136, 138 passim. [DOI] [PubMed] [Google Scholar]
- Gerstenbruch S, Brooks CL, Kosma P, Brade L, Mackenzie CR, Evans SV, Brade H, Muller-Loennies S. Analysis of cross-reactive and specific anti-carbohydrate antibodies against lipopolysaccharide from Chlamydophila psittaci. Glycobiology. 2010;20:461–472. doi: 10.1093/glycob/cwp198. doi:10.1093/glycob/cwp198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteira TF, Coulson-Thomas VJ, Taunay-Rodrigues A, Oliveira V, Thacker BE, Juliano MA, Pasqualini R, Arap W, Tersariol IL, Nader HB, et al. Inhibitory peptides of the sulfotransferase domain of the heparan sulfate enzyme, N-deacetylase-N-sulfotransferase-1. J Biol Chem. 2010;286:5338–5346. doi: 10.1074/jbc.M110.100719. doi:10.1074/jbc.M110.100719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanasekar M, Rao KV, He YX, Mishra PK, Nutman TB, Kaliraj P, Ramaswamy K. Novel phage display-based subtractive screening to identify vaccine candidates of Brugia malayi. Infect Immun. 2004;72:4707–4715. doi: 10.1128/IAI.72.8.4707-4715.2004. doi:10.1128/IAI.72.8.4707-4715.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooi HC, Feizi T, Kapadia A, Knowles BB, Solter D, Evans MJ. Stage-specific embryonic antigen involves α1→3 fucosylated type2 blood group chains. Nature. 1981;292:156–158. doi: 10.1038/292156a0. doi:10.1038/292156a0. [DOI] [PubMed] [Google Scholar]
- Haidaris CG, Malone J, Sherrill LA, Bliss JM, Gaspari AA, Insel RA, Sullivan MA. Recombinant human antibody single chain variable fragments reactive with Candida albicans surface antigens. J Immunol Methods. 2001;257:185–202. doi: 10.1016/s0022-1759(01)00463-x. doi:10.1016/S0022-1759(01)00463-X. [DOI] [PubMed] [Google Scholar]
- Hakomori S. Tumor associated carbohydrate antigens. Ann Rev Immunol. 1984;2:103–126. doi: 10.1146/annurev.iy.02.040184.000535. doi:10.1146/annurev.iy.02.040184.000535. [DOI] [PubMed] [Google Scholar]
- Hakomori S. Aberrant glycosylation in tumors and tumor associated carbohydrate antigens. Adv Cancer Res. 1989;52:257–331. doi: 10.1016/s0065-230x(08)60215-8. doi:10.1016/S0065-230X(08)60215-8. [DOI] [PubMed] [Google Scholar]
- Harris SL, Craig L, Mehroke JS, Rashed M, Zwick MB, Kenar K, Toone EJ, Greenspan N, Auzanneau FI, Marino-Albernas JR, et al. Exploring the basis of peptide-carbohydrate crossreactivity: Evidence for discrimination by peptides between closely related anti-carbohydrate antibodies. Proc Natl Acad Sci USA. 1997;94:2454–2459. doi: 10.1073/pnas.94.6.2454. doi:10.1073/pnas.94.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S, Sugihara K, Nakayama J, Akama TO, Wong SM, Kawashima H, Zhang J, Smith DF, Ohyama C, Fukuda M, et al. Identification of mRNA splicing factors as the endothelial receptor for carbohydrate-dependent lung colonization of cancer cells. Proc Natl Acad Sci USA. 2009;106:3095–3100. doi: 10.1073/pnas.0810110106. doi:10.1073/pnas.0810110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudek KC, Patterson RJ, Wang JL. SR proteins and galectins: What's in a name? Glycobiology. 2010;20:1199–1207. doi: 10.1093/glycob/cwq097. doi:10.1093/glycob/cwq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka K, Hida Y, Miyamoto M, Oshikiri T, Suzuoki M, Nakakubo Y, Shinohara T, Itoh T, Shichinohe T, Kondo S, et al. High expression of tumor-associated antigen RCAS1 in pancreatic ductal adenocarcinoma is an unfavorable prognostic marker. Int J Cancer. 2002;99:418–423. doi: 10.1002/ijc.10381. doi:10.1002/ijc.10381. [DOI] [PubMed] [Google Scholar]
- Hoess R, Brinkmann U, Handel T, Pastan I. Identification of a peptide which binds to the carbohydrate-specific monoclonal antibody B3. Gene. 1993;128:43–49. doi: 10.1016/0378-1119(93)90151-r. doi:10.1016/0378-1119(93)90151-R. [DOI] [PubMed] [Google Scholar]
- Hogrefe HH, Mullinax RL, Lovejoy AE, Hay BN, Sorge JA. A bacteriophage lambda vector for the cloning and expression of immunoglobulin Fab fragments on the surface of filamentous phage. Gene. 1993;128:119–126. doi: 10.1016/0378-1119(93)90162-v. doi:10.1016/0378-1119(93)90162-V. [DOI] [PubMed] [Google Scholar]
- Hou Y, Gu XX. Development of peptide mimotopes of lipooligosaccharide from nontypeable Haemophilus influenzae as vaccine candidates. J Immunol. 2003;170:4373–4379. doi: 10.4049/jimmunol.170.8.4373. [DOI] [PubMed] [Google Scholar]
- Hughes-Jones NC, Gorick BD, Bye JM, Finnern R, Scott ML, Voak D, Marks JD, Ouwehand WH. Characterization of human blood group scFv antibodies derived from a V gene phage-display library. Br J Haematol. 1994;88:180–186. doi: 10.1111/j.1365-2141.1994.tb04994.x. doi:10.1111/j.1365-2141.1994.tb04994.x. [DOI] [PubMed] [Google Scholar]
- Hussack G, Arbabi-Ghahroudi M, van Faassen H, Songer JG, Ng KK, MacKenzie R, Tanha J. Neutralization of Clostridium difficile toxin A with single-domain antibodies targeting the cell receptor binding domain. J Biol Chem. 2011;286:8961–8976. doi: 10.1074/jbc.M110.198754. doi:10.1074/jbc.M110.198754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving MB, Pan O, Scott JK. Random-peptide libraries and antigen-fragment libraries for epitope mapping and the development of vaccines and diagnostics. Curr Opin Chem Biol. 2001;5:314–324. doi: 10.1016/S1367-5931(00)00208-8. doi:10.1016/S1367-5931(00)00208-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa D, Kikkawa H, Ogino K, Hirabayashi Y, Oku N, Taki T. GD1α-replica peptides functionally mimic GD1α, an adhesion molecule of metastatic tumor cells, and suppress the tumor metastasis. FEBS Lett. 1998;441:20–24. doi: 10.1016/s0014-5793(98)01511-7. doi:10.1016/S0014-5793(98)01511-7. [DOI] [PubMed] [Google Scholar]
- Iwasaki T, Nakashima M, Watanabe T, Yamamoto S, Inoue Y, Yamanaka H, Matsumura A, Iuchi K, Mori T, Okada M. Expression and prognostic significance in lung cancer of human tumor-associated antigen RCAS1. Int J Cancer. 2000;89:488–493. doi: 10.1002/1097-0215(20001120)89:6<488::aid-ijc4>3.0.co;2-d. doi:10.1002/1097-0215(20001120)89:6<488::AID-IJC4>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Jenniskens GJ, Oosterhof A, Brandwijk R, Veerkamp JH, van Kuppevelt TH. Heparan sulfate heterogeneity in skeletal muscle basal lamina: Demonstration by phage display-derived antibodies. J Neurosci. 2000;20:4099–4111. doi: 10.1523/JNEUROSCI.20-11-04099.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanitpun R, Wagner GG, Waghela SD. Characterization of recombinant antibodies developed for capturing enterohemorrhagic Escherichia coli O157:H7. Southeast Asian J Trop Med Public Health. 2004;35:902–912. [PubMed] [Google Scholar]
- Kannagi R. Monoclonal anti-glycosphingolipid antibodies. Methods Enzymol. 2000;312:160–179. doi: 10.1016/s0076-6879(00)12907-6. doi:10.1016/S0076-6879(00)12907-6. [DOI] [PubMed] [Google Scholar]
- Kannagi R, Cochran NA, Ishigami F, Hakomori S, Andrews PW, Knowles BB, Solter D. Stage-specific embryonic antigens (SSEA-3 and -4) are epitopes of a unique globo-series ganglioside isolated from human teratocarcinoma cells. EMBO J. 1983;2:2355–2361. doi: 10.1002/j.1460-2075.1983.tb01746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber-Emmons T, Luo P, Qiu J, Chang TY, O I, Blaszczyk-Thurin M, Steplewski Z. Vaccination with carbohydrate peptide mimotopes promotes anti-tumor responses. Nat Biotechnol. 1999;17:660–665. doi: 10.1038/10870. doi:10.1038/10870. [DOI] [PubMed] [Google Scholar]
- Kim HS, Tsai S, Zou N, Lo SC, Wear DJ, Izadjoo MJ. Construction and molecular characterization of mouse single-chain variable fragment antibodies against Burkholderia mallei and Burkholderia pseudomallei. J Immunol Methods. 2010;365:101–109. doi: 10.1016/j.jim.2010.12.004. doi:10.1016/j.jim.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Kozbor D. Cancer vaccine with mimotopes of tumor-associated carbohydrate antigens. Immunol Res. 2009;46:23–31. doi: 10.1007/s12026-009-8120-y. doi:10.1007/s12026-009-8120-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T, Matsushita T, Niwa R, Kumagai I, Nakamura K. Novel anti-Tn single-chain Fv-Fc fusion proteins derived from immunized phage library and antibody Fc domain. Anticancer Res. 2010;30:3397–3405. [PubMed] [Google Scholar]
- Kurup S, Wijnhoven TJ, Jenniskens GJ, Kimata K, Habuchi H, Li JP, Lindahl U, van Kuppevelt TH, Spillmann D. Characterization of anti-heparan sulfate phage display antibodies AO4B08 and HS4E4. J Biol Chem. 2007;282:21032–21042. doi: 10.1074/jbc.M702073200. doi:10.1074/jbc.M702073200. [DOI] [PubMed] [Google Scholar]
- Landon LA, Zou J, Deutscher SL. Is phage display technology on target for developing peptide-based cancer drugs? Curr Drug Discov Technol. 2004;1:113–132. doi: 10.2174/1570163043335108. doi:10.2174/1570163043335108. [DOI] [PubMed] [Google Scholar]
- Li F, Ten Dam GB, Murugan S, Yamada S, Hashiguchi T, Mizumoto S, Oguri K, Okayama M, van Kuppevelt TH, Sugahara K. Involvement of highly sulfated chondroitin sulfate in the metastasis of the Lewis lung carcinoma cells. J Biol Chem. 2008;283:34294–34304. doi: 10.1074/jbc.M806015200. doi:10.1074/jbc.M806015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little M, Welschof M, Braunagel M, Hermes I, Christ C, Keller A, Rohrbach P, Kurschner T, Schmidt S, Kleist C, et al. Generation of a large complex antibody library from multiple donors. J Immunol Methods. 1999;231:3–9. doi: 10.1016/s0022-1759(99)00164-7. doi:10.1016/S0022-1759(99)00164-7. [DOI] [PubMed] [Google Scholar]
- Lou Q, Pastan I. A Lewis(y) epitope mimicking peptide induces anti-Lewis(y) immune responses in rabbits and mice. J Pept Res. 1999;53:252–260. doi: 10.1034/j.1399-3011.1999.00025.x. doi:10.1034/j.1399-3011.1999.00025.x. [DOI] [PubMed] [Google Scholar]
- Marino P, Norreel JC, Schachner M, Rougon G, Amoureux MC. A polysialic acid mimetic peptide promotes functional recovery in a mouse model of spinal cord injury. Exp Neurol. 2009;219:163–174. doi: 10.1016/j.expneurol.2009.05.009. doi:10.1016/j.expneurol.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Matsubara T, Ishikawa D, Taki T, Okahata Y, Sato T. Selection of ganglioside GM1-binding peptides by using a phage library. FEBS Lett. 1999;456:253–256. doi: 10.1016/s0014-5793(99)00962-x. doi:10.1016/S0014-5793(99)00962-X. [DOI] [PubMed] [Google Scholar]
- McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies: Filamentous phage displaying antibody variable domains. Nature. 1990;348:552–554. doi: 10.1038/348552a0. doi:10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- Menendez T, Santiago-Vispo NF, Cruz-Leal Y, Coizeau E, Garay H, Reyes O, Batista Y, Cobas K, Carmenate T, Chinea G, et al. Identification and characterization of phage-displayed peptide mimetics of Neisseria meningitidis serogroup B capsular polysaccharide. Int J Med Microbiol. 2010;301:16–25. doi: 10.1016/j.ijmm.2010.04.020. doi:10.1016/j.ijmm.2010.04.020. [DOI] [PubMed] [Google Scholar]
- Milich DR, McLachlan A. The nucleocapsid of hepatitis B virus is both a T-cell-independent and a T-cell-dependent antigen. Science. 1986;234:1398–1401. doi: 10.1126/science.3491425. doi:10.1126/science.3491425. [DOI] [PubMed] [Google Scholar]
- Monzavi-Karbassi B, Cunto-Amesty G, Luo P, Shamloo S, Blaszcyk-Thurin M, Kieber-Emmons T. Immunization with a carbohydrate mimicking peptide augments tumor-specific cellular responses. Int Immunol. 2001;13:1361–1371. doi: 10.1093/intimm/13.11.1361. doi:10.1093/intimm/13.11.1361. [DOI] [PubMed] [Google Scholar]
- Mountney A, Zahner MR, Lorenzini I, Oudega M, Schramm LP, Schnaar RL. Sialidase enhances recovery from spinal cord contusion injury. Proc Natl Acad Sci USA. 2010;107:11561–11566. doi: 10.1073/pnas.1006683107. doi:10.1073/pnas.1006683107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro R. Meningococcal disease: Treatable but still terrifying. Intern Med J. 2002;32:165–169. doi: 10.1046/j.1444-0903.2001.00188.x. doi:10.1046/j.1444-0903.2001.00188.x. [DOI] [PubMed] [Google Scholar]
- Natsume A, Wakitani M, Yamane-Ohnuki N, Shoji-Hosaka E, Niwa R, Uchida K, Satoh M, Shitara K. Fucose removal from complex-type oligosaccharide enhances the antibody-dependent cellular cytotoxicity of single-gene-encoded antibody comprising a single-chain antibody linked the antibody constant region. J Immunol Methods. 2005;306:93–103. doi: 10.1016/j.jim.2005.07.025. doi:10.1016/j.jim.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Oldenburg KR, Loganathan D, Goldstein IJ, Schultz PG, Gallop MA. Peptide ligands for a sugar-binding protein isolated from a random peptide library. Proc Natl Acad Sci USA. 1992;89:5393–5397. doi: 10.1073/pnas.89.12.5393. doi:10.1073/pnas.89.12.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmley SF, Smith GP. Antibody-selectable filamentous fd phage vectors: Affinity purification of target genes. Gene. 1988;73:305–318. doi: 10.1016/0378-1119(88)90495-7. doi:10.1016/0378-1119(88)90495-7. [DOI] [PubMed] [Google Scholar]
- Pasqualini R, Koivunen E, Ruoslahti E. A peptide isolated from phage display libraries is a structural and functional mimic of an RGD-binding site on integrins. J Cell Biol. 1995;130:1189–1196. doi: 10.1083/jcb.130.5.1189. doi:10.1083/jcb.130.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson MA, Caothien RH, Burton DR. Generation of diverse high-affinity human monoclonal antibodies by repertoire cloning. Proc Natl Acad Sci USA. 1991;88:2432–2436. doi: 10.1073/pnas.88.6.2432. doi:10.1073/pnas.88.6.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus SH, Smith MJ, Jennings HJ, Burritt JB, Glee PM. Peptides that mimic the group B streptococcal type III capsular polysaccharide antigen. J Immunol. 1998;160:293–298. [PubMed] [Google Scholar]
- Popa I, Ishikawa D, Tanaka M, Ogino K, Portoukalian J, Taki T. GD3-replica peptides selected from a phage peptide library induce a GD3 ganglioside antibody response. FEBS Lett. 2006;580:1398–1404. doi: 10.1016/j.febslet.2006.01.063. doi:10.1016/j.febslet.2006.01.063. [DOI] [PubMed] [Google Scholar]
- Rojas G, Talavera A, Munoz Y, Rengifo E, Krengel U, Angstrom J, Gavilondo J, Moreno E. Light-chain shuffling results in successful phage display selection of functional prokaryotic-expressed antibody fragments to N-glycolyl GM3 ganglioside. J Immunol Methods. 2004;293:71–83. doi: 10.1016/j.jim.2004.07.002. doi:10.1016/j.jim.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Rosenberg A, Griffin K, Studier WF, McCormick M, James Berg J, Novy R, Robert Mierendorf R. T7Select phage display system: a powerful new protein display system based on bacteriophage T7. InNovation. 1996;6:1–6. [Google Scholar]
- Rousseau J, Tetu B, Caron D, Malenfant P, Cattaruzzi P, Audette M, Doillon C, Tremblay JP, Guerette B. RCAS1 is associated with ductal breast cancer progression. Biochem Biophys Res Commun. 2002;293:1544–1549. doi: 10.1016/S0006-291X(02)00401-1. doi:10.1016/S0006-291X(02)00401-1. [DOI] [PubMed] [Google Scholar]
- Ruoslahti E. Targeting tumor vasculature with homing peptides from phage display. Semin Cancer Biol. 2000;10:435–442. doi: 10.1006/scbi.2000.0334. doi:10.1006/scbi.2000.0334. [DOI] [PubMed] [Google Scholar]
- Sakai K, Shimizu Y, Chiba T, Matsumoto-Takasaki A, Kusada Y, Zhang W, Nakata M, Kojima N, Toma K, Takayanagi A, et al. Isolation and characterization of phage-displayed single chain antibodies recognizing nonreducing terminal mannose residues. 1. A new strategy for generation of anti-carbohydrate antibodies. Biochemistry. 2007;46:253–262. doi: 10.1021/bi061875e. doi:10.1021/bi061875e. [DOI] [PubMed] [Google Scholar]
- Scott JK, Loganathan D, Easley RB, Gong X, Goldstein IJ. A family of concanavalin A-binding peptides from a hexapeptide epitope library. Proc Natl Acad Sci USA. 1992;89:5398–5402. doi: 10.1073/pnas.89.12.5398. doi:10.1073/pnas.89.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science. 1990;249:386–390. doi: 10.1126/science.1696028. doi:10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- Shevinsky LH, Knowles BB, Damjanov I, Solter D. Monoclonal antibody to murine embryos defines a stage-specific embryonic antigen expressed on mouse embryos and human teratocarcinoma cells. Cell. 1982;30:697–705. doi: 10.1016/0092-8674(82)90274-4. doi:10.1016/0092-8674(82)90274-4. [DOI] [PubMed] [Google Scholar]
- Shinkawa T, Nakamura K, Yamane N, Shoji-Hosaka E, Kanda Y, Sakurada M, Uchida K, Anazawa H, Satoh M, Yamasaki M, et al. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J Biol Chem. 2003;278:3466–3473. doi: 10.1074/jbc.M210665200. doi:10.1074/jbc.M210665200. [DOI] [PubMed] [Google Scholar]
- Smetsers TF, van de Westerlo EM, ten Dam GB, Overes IM, Schalkwijk J, van Muijen GN, van Kuppevelt TH. Human single-chain antibodies reactive with native chondroitin sulfate detect chondroitin sulfate alterations in melanoma and psoriasis. J Invest Dermatol. 2004;122:707–716. doi: 10.1111/j.0022-202X.2004.22316.x. doi:10.1111/j.0022-202X.2004.22316.x. [DOI] [PubMed] [Google Scholar]
- Smith GP. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. doi:10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- Smith GP, Scott JK. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 1993;217:228–257. doi: 10.1016/0076-6879(93)17065-d. doi:10.1016/0076-6879(93)17065-D. [DOI] [PubMed] [Google Scholar]
- Smits NC, Lensen JF, Wijnhoven TJ, Ten Dam GB, Jenniskens GJ, van Kuppevelt TH. Phage display-derived human antibodies against specific glycosaminoglycan epitopes. Methods Enzymol. 2006;416:61–87. doi: 10.1016/S0076-6879(06)16005-X. doi:10.1016/S0076-6879(06)16005-X. [DOI] [PubMed] [Google Scholar]
- Solter D, Knowles BB. Monoclonal antibody defining a stage-specific mouse embryonic antigen (SSEA-1) Proc Natl Acad Sci USA. 1978;75:5565–5569. doi: 10.1073/pnas.75.11.5565. doi:10.1073/pnas.75.11.5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer GF. T and Tn, general carcinoma autoantigens. Science. 1984;224:1198–1206. doi: 10.1126/science.6729450. doi:10.1126/science.6729450. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Nakayama J, Suzuki A, Angata K, Chen S, Sakai K, Hagihara K, Yamaguchi Y, Fukuda M. Polysialic acid facilitates tumor invasion by glioma cells. Glycobiology. 2005;15:887–894. doi: 10.1093/glycob/cwi071. doi:10.1093/glycob/cwi071. [DOI] [PubMed] [Google Scholar]
- Taki T, Ishikawa D, Hamasaki H, Handa S. Preparation of peptides which mimic glycosphingolipids by using phage peptide library and their modulation on β-galactosidase activity. FEBS Lett. 1997;418:219–223. doi: 10.1016/s0014-5793(97)01386-0. doi:10.1016/S0014-5793(97)01386-0. [DOI] [PubMed] [Google Scholar]
- Taki T, Ishikawa D, Ogino K, Tanaka M, Oku N, Asai T, Popa I, Portoukalian J. A new approach for drug discovery from glycobiology and phage-displayed peptide library technology. Biochim Biophys Acta. 2008;1780:497–503. doi: 10.1016/j.bbagen.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Takikawa M, Kikkawa H, Asai T, Yamaguchi N, Ishikawa D, Tanaka M, Ogino K, Taki T, Oku N. Suppression of GD1α ganglioside-mediated tumor metastasis by liposomalized WHW-peptide. FEBS Lett. 2000;466:381–384. doi: 10.1016/s0014-5793(00)01110-8. doi:10.1016/S0014-5793(00)01110-8. [DOI] [PubMed] [Google Scholar]
- ten Dam GB, van de Westerlo EM, Smetsers TF, Willemse M, van Muijen GN, Merry CL, Gallagher JT, Kim YS, van Kuppevelt TH. Detection of 2-O-sulfated iduronate and N-acetylglucosamine units in heparan sulfate by an antibody selected against acharan sulfate (IdoA2S-GlcNAc)n. J Biol Chem. 2004;279:38346–38352. doi: 10.1074/jbc.M404166200. doi:10.1074/jbc.M404166200. [DOI] [PubMed] [Google Scholar]
- Thorburn K, Baines P, Thomson A, Hart CA. Mortality in severe meningococcal disease. Arch Dis Child. 2001;85:382–385. doi: 10.1136/adc.85.5.382. doi:10.1136/adc.85.5.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torregrossa P, Buhl L, Bancila M, Durbec P, Schafer C, Schachner M, Rougon G. Selection of poly-α 2,8-sialic acid mimotopes from a random phage peptide library and analysis of their bioactivity. J Biol Chem. 2004;279:30707–30714. doi: 10.1074/jbc.M403935200. doi:10.1074/jbc.M403935200. [DOI] [PubMed] [Google Scholar]
- Uchimura K, Lemjabbar-Alaoui H, van Kuppevelt TH, Rosen SD. Use of a phage display antibody to measure the enzymatic activity of the Sulfs. Methods Enzymol. 2010;480:51–64. doi: 10.1016/S0076-6879(10)80003-5. doi:10.1016/S0076-6879(10)80003-5. [DOI] [PubMed] [Google Scholar]
- van de Westerlo EM, Smetsers TF, Dennissen MA, Linhardt RJ, Veerkamp JH, van Muijen GN, van Kuppevelt TH. Human single chain antibodies against heparin: Selection, characterization, and effect on coagulation. Blood. 2002;99:2427–2433. doi: 10.1182/blood.v99.7.2427. doi:10.1182/blood.V99.7.2427. [DOI] [PubMed] [Google Scholar]
- van Deuren M, Brandtzaeg P, van der Meer JW. Update on meningococcal disease with emphasis on pathogenesis and clinical management. Clin Microbiol Rev. 2000;13:144–166. doi: 10.1128/cmr.13.1.144-166.2000. table of contents doi:10.1128/CMR.13.1.144-166.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kuppevelt TH, Dennissen MA, van Venrooij WJ, Hoet RM, Veerkamp JH. Generation and application of type-specific anti-heparan sulfate antibodies using phage display technology. Further evidence for heparan sulfate heterogeneity in the kidney. J Biol Chem. 1998;273:12960–12966. doi: 10.1074/jbc.273.21.12960. doi:10.1074/jbc.273.21.12960. [DOI] [PubMed] [Google Scholar]
- Vyakarnam A, Dagher SF, Wang JL, Patterson RJ. Evidence for a role for galectin-1 in pre-mRNA splicing. Mol Cell Biol. 1997;17:4730–4737. doi: 10.1128/mcb.17.8.4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildes SS, Tunkel AR. Meningococcal vaccines: A progress report. BioDrugs. 2002;16:321–329. doi: 10.2165/00063030-200216050-00001. doi:10.2165/00063030-200216050-00001. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang Q, Sales D, Bianco AE, Craig A. Vaccination with peptide mimotopes produces antibodies recognizing bacterial capsular polysaccharides. Vaccine. 2010;28:6425–6435. doi: 10.1016/j.vaccine.2010.07.049. doi:10.1016/j.vaccine.2010.07.049. [DOI] [PubMed] [Google Scholar]
- Yamane-Ohnuki N, Kinoshita S, Inoue-Urakubo M, Kusunoki M, Iida S, Nakano R, Wakitani M, Niwa R, Sakurada M, Uchida K, et al. Establishment of FUT8 knockout Chinese hamster ovary cells: An ideal host cell line for producing completely defucosylated antibodies with enhanced antibody-dependent cellular cytotoxicity. Biotechnol Bioeng. 2004;87:614–622. doi: 10.1002/bit.20151. doi:10.1002/bit.20151. [DOI] [PubMed] [Google Scholar]
- Yang P, Yin X, Rutishauser U. Intercellular space is affected by the polysialic acid content of NCAM. J Cell Biol. 1992;116:1487–1496. doi: 10.1083/jcb.116.6.1487. doi:10.1083/jcb.116.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young WW, Jr, Johnson HS, Tamura Y, Karlsson KA, Larson G, Parker JM, Khare DP, Spohr U, Baker DA, Hindsgaul O, et al. Characterization of monoclonal antibodies specific for the Lewis a human blood group determinant. J Biol Chem. 1983;258:4890–4894. [PubMed] [Google Scholar]
- Yuasa N, Zhang W, Goto T, Sakaue H, Matsumoto-Takasaki A, Kimura M, Ohshima H, Tsuchida Y, Koizumi T, Sakai K, et al. Production of anti-carbohydrate antibodies by phage display technologies: Potential impairment of cell growth as a result of endogenous expression. J Biol Chem. 2010;285:30587–30597. doi: 10.1074/jbc.M110.107284. doi:10.1074/jbc.M110.107284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Matsumoto-Takasaki A, Kusada Y, Sakaue H, Sakai K, Nakata M, Fujita-Yamaguchi Y. Isolation and characterization of phage-displayed single chain antibodies recognizing nonreducing terminal mannose residues. 2. Expression, purification, and characterization of recombinant single chain antibodies. Biochemistry. 2007;46:263–270. doi: 10.1021/bi0618767. doi:10.1021/bi0618767. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nakayama J, Ohyama C, Suzuki M, Suzuki A, Fukuda M, Fukuda MN. Sialyl Lewis X-dependent lung colonization of B16 melanoma cells through a selectin-like endothelial receptor distinct from E- or P-selectin. Cancer Res. 2002;62:4194–4198. [PubMed] [Google Scholar]