Abstract
The malachite green aptamer binds two closely related ligands, malachite green (MG) and tetramethylrosamine (TMR), with near equal affinity. The MG ligand consists of three phenyl rings emanating from a central carbon, while TMR has two of the three rings connected by an ether linkage. The binding pockets for MG and TMR in the aptamer, known from high-resolution structure, differ only in the conformation of a few nucleotides. Herein, we applied isothermal titration calorimetry (ITC) to compare the thermodynamics for binding of MG and TMR to the aptamer. Binding heat capacities were obtained from ITC titrations over the temperature range of 15 to 60 °C. Two temperature regimes were found for MG binding: one from 15 to 45 °C where MG bound with a large negative heat capacity and an apparent stoichiometry (n) of ~0.4, and another from 50 to 60 °C where MG bound with positive heat capacity and n~1.1. The binding of TMR, on the other hand, revealed only one temperature regime for binding, with a more modest negative heat capacity and n~1.2. The large difference in heat capacity between the two ligands suggests that significantly more conformational rearrangement occurs upon the binding of MG than TMR, which is consistent with differences in solvent accessible surface area calculated for available ligand-bound structures. Lastly, we note that binding stoichiometry of MG was improved not only by raising the temperature, but also by lowering the concentration of Mg2+ or increasing the time between ITC injections. These studies suggest that binding of a dynamical ligand to a functional RNA requires the RNA itself to have significant dynamics.
Keywords: RNA dynamics, heat capacity, thermodynamics, aptamer
Introduction
Aptamers are RNA molecules obtained via in vitro selection that bind a small molecule or protein.1-3 RNA aptamers typically have high affinity and specificity for their targets, which has led to them being employed in numerous applications.4-8 The malachite green aptamer is a 38 nt RNA that was selected to bind the dye malachite green (MG) (Figure 1 A, B).9-11 This aptamer has the unusual feature of binding equally well to its cognate ligand, MG, and a related but non-cognate ligand, tetramethylrosamine (TMR)10,12 that possesses an ether linkage between the two dimethylamino-containing phenyl rings (Figure 1C). Rigidity conferred by this connection has been suggested to lower the conformational entropy penalty for binding.11,12
Figure 1.
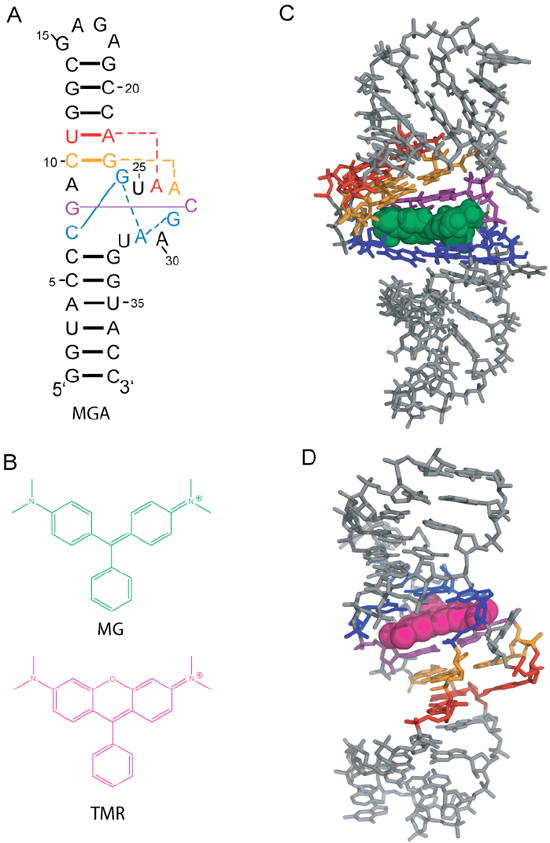
Secondary and tertiary structures of the malachite green aptamer free and bound to MG and TMR. A.) Secondary structure of the aptamer.11 Key base interactions involved in the binding pocket are colored and correspond with panels C and D. B.) Structures of MG and TMR, which differ only in the presence of an ether linkage in TMR. C.) NMR solution structure of the MG-bound aptamer (1Q8N).11 MG is green spacefilling. D.) X-ray crystallography structure of the TMR-bound aptamer (1F1T).10 TMR is magneta spacefilling. Structures were rendered with PyMOL (Schrodinger, Cambridge MA).
There are two high-resolution structures available for the MG aptamer: an x-ray crystal structure bound to TMR at 2.8 Å resolution (Figure 1D),10 and an NMR solution structure bound to MG (Figure 1C).11 Both complexes have similar binding pockets wherein the ligand stacks between the G8:C28 base pair and the C7:G24:A31:G29 base quadruple, with C10:G23:A27 and U11:A22:A26 base triples providing key structural portions of the binding cavity’s shape. These structures also have differences in the binding pocket architecture, especially with regards to syn bases (Figure S1).13 The TMR-bound complex has two syn bases in the binding pocket, G24 and A30, while the MG-bound complex has three syn bases in the binding pocket, G24, G29, and A31, with A30 in the standard anti conformation. Moreover, both of the syn bases in the TMR complex are in the full syn conformation, while the three syn bases in the MG complex are intermediate syn.13 These differences in binding pocket architecture argue that the RNA structure likely plays a major role in aptamer selectivity.
The binding heat capacity (ΔCp), obtained by measuring binding enthalpy (ΔH) as a function of temperature, provides a measure of the conformational changes associated with binding and can be expressed in terms of bond formation (ΔH) or conformational rearrangement (ΔS),14
| (1) |
Equation 1 suggests that if binding involves compaction of the RNA that increases in magnitude with temperature then binding should have a large negative ΔCp.15 A large negative heat capacity has been associated with formation of RNA structure16 and with specificity of binding in protein-nucleic acid17,18 and small molecule-RNA interactions.19
In this study, we investigated binding of MG and TMR to the MG aptamer using ITC. We find that the MG aptamer exhibits a negative ΔCp for interaction with both ligands (for temperatures up to ~45 °C), but that the ΔCp is significantly larger in magnitude for MG. This observation suggests that larger conformational rearrangement of the RNA occurs upon binding of MG. In addition, the apparent stoichiometry for binding of MG is found to be dependent on temperature, Mg2+ concentration, and time, suggesting that the unbound state of the aptamer consists of a heterogeneous population of folds that require significant dynamics in order to bind the more dynamical ligand.
Materials and Methods
RNA Preparation
MG aptamer was produced by in vitro transcription from a hemi-duplex template 20 constructed from the following sequences:
T7 promotor + G: 5’ TAA TAC GAC TCA CTA TAG
Aptamer template: 5’ GGT ACC ATT CGT TAC CTG GCT CTC GCC AGT CGG GTA CCT ATA GTG AGT CGT ATT A
Following PAGE purification, the band containing the MG aptamer was crushed and soaked in 1X TEN250, and the RNA pellet was obtained via ethanol precipitation. The pellet was dissolved in ITC buffer [10 mM sodium cacodylate (pH 5.8), 10 mM KCl] and dialyzed against the same buffer for 15-18 h at 4 °C using MWCO 1000 membranes (SpectraPor 7, SpectraPor Inc). This window of time was chosen because shorter times (e.g. 10 h) revealed larger injection heats (data not shown), while longer times might lead to RNA degradation. Buffer conditions for ITC were chosen to be similar to crystallization solving conditions.10 Prior to ITC, the aptamer was diluted as needed in the dialysis flow-through buffer and renatured by heating at 90 °C for 2 min, cooling at room temperature for 1 min, adding MgCl2 from a stock made from MgCl2 solid dissolved in 1×ITC buffer, and incubating at 55 °C for 10 min. Typical final concentrations of Mg2+ were 10 mM, although this was lowered in select experiments. RNA concentrations were measured at 90 °C using ε(260 nm) = 376,300 M-1 cm-1, calculated from the RNAcalc program.21
Ligand Preparation
Malachite green oxalate (Sigma Aldrich) and tetramethylrosamine chloride (Invitrogen) were dissolved in the MG aptamer dialysis flow-through buffer, with MgCl2 added from MgCl2 stocks dissolved in 1×ITC buffer prepared as described above. Concentrations of dyes were determined spectrophotometrically using values of ε617 nm = 147,800 M-1 cm-1 and ε552 nm = 52,430 M-1 cm-1 for MG and TMR, respectively, derived from massing a quantity, dissolving it in ITC buffer, and taking an OD reading at the maximum observed wavelength. As a test of purity, MG was re-crystallized from the original oxalate salt to a chloride salt using the procedure of Swain and Hedberg.22 No differences were observed in the calorimetric experiments, indicating that any MG impurities did not affect the data. Care was taken to keep the solutions from light as much as possible, although no photosensitivity was observed.
Isothermal Titration Calorimetry
All ITC experiments were performed on a VP-ITC (MicroCal, Inc. Northampton MA). Concentrations of MG aptamer were in the range of 2 to 4 μM, with the initial ligand concentrations being 12-15 times that of the initial RNA concentration. Samples were degassed at 35 °C for 10 min prior to being loaded into the instrument. The MG or TMR solutions were injected into the sample cell in 7 or 10 μL increments, with 5 min spacing between injections (unless otherwise noted), which was sufficient for the signal to return to baseline. The final 5 points of each titration were averaged and subtracted from each point in the titration to account for ligand dilution. This method of baseline correction was justified because such injection heats were small and consistent; moreover, the small value of these injection heats confirmed that the RNA and ligand, prepared as described above, were well matched in their buffers. Titration data were integrated and analyzed using Origin software (OriginLab Corporation, Northampton MA). Errors are reported as the standard error of the mean of multiple trials, typically 3 or more (see Tables for actual number of trials). Heat capacities were obtained by fitting temperature-dependent enthalpy values to the following equation, using ΔH25 as a reference enthalpy,
| (2) |
Fits were by linear least squares with weighting to the standard error of the mean for the enthalpy.
Solvent Accessible Surface Area (ASA) Calculation
The solvent accessible surface area (ASA) of the MG-bound and the TMR-bound structures was calculated using the web-based program GETAREA23 with a probe size of 1.4 Å, which approximates the radius of a water molecule.24 Each RNA-ligand structure contained the same number of nucleotides allowing direct comparison of ΔASA for the two structures. Water and metal ions were removed prior to calculation, but ligands were left so as to properly represent the final bound state.
Thermal Denaturation Monitored by UV Absorption
All thermal denaturation experiments were carried out in a Gilford Response spectrophotometer using 0.1, 0.5 or 1 cm pathlength quartz cuvettes, which allowed different concentrations of RNA to be used. Samples were prepared identically to ITC samples, with the exclusion of the 55 °C renaturation step for samples studied in the absence of divalent ions. Absorbance at 260 nm was monitored from 20 to 95 °C, with data collected every 0.5 °C. Solutions containing Mg2+ were melted once, while solutions without Mg2+ were melted multiple times and gave superimposable heating and cooling curves. Melting temperatures are determined from the maximum of a first derivative plot with five point smoothing. Data are the average of three melting curves.
UV Crosslinking of 4-thioU-Containing RNA
Experiments generally followed the protocol of McGraw and co-workers.25 Briefly, 4-thiouridine triphosphate (TriLink Biotechnolgies) in a 1:1 mixture with UTP was incorporated into the aptamer via in vitro transcription, which also contained α32P-labeled GTP. The transcription reaction was purified via Sephadex G20 spin column, and mixed with 10X ITC buffer and water to reach the same concentrations as in ITC. Samples were renatured according to the ITC sample procedure, and ligands were added afterwards. While resting on an ice-chilled aluminum block acting as a heat sink, 20 μL drops of sample were irradiated with 312 nm light from a UV-B bulb for 30 min. Samples were then mixed with a 2X EDTA formamide loading buffer and fractionated on 10% PAGE under denaturing conditions.
Results
Ligand Binding to the MG Aptamer is Enthalpically Driven, with an Observed Stoichiometry Less than 1
We begin by considering ligand binding at our reference temperature of 25 °C. As described below, this temperature is centered in the low temperature binding regime for MG. At 25 °C, MG and TMR bound the MG aptamer with nearly equal affinity (apparent Kd of 50 ± 20 nM and 60 ± 30 nM, respectively, Tables 1 and 2), but with very different thermodynamic origins (Figure 2). At this temperature, MG bound with a large favorable enthalpy (-53 ± 5 kcal/mol) and a large penalizing entropy (-140 ± 10 eu), while TMR bound with a modest enthalpy (-15 ± 2 kcal/mol) and a less penalizing entropy (-16 ± 8 eu). These trends in ΔH° and ΔS° are consistent with general expectations of enthalpy-entropy compensation for ligand binding to a macromolecule.26
Table 1.
ITC data for MG binding
| Temperature (°C) | Number of Trials | Apparent Stoichiometry | Apparent Kd (nM) | ΔH° (kcal/mol) | ΔS° (eu) |
|---|---|---|---|---|---|
| 15 | 3 | 0.41 ± 0.03 | 30 ± 10 | -38 ± 4 | -100 ± 10 |
| 20 | 3 | 0.43 ± 0.01 | 30 ± 10 | -42 ± 3 | -109 ± 6 |
| 25 | 3 | 0.40 ± 0.02 | 50 ± 20 | -53 ± 5 | -140 ± 10 |
| 30 | 3 | 0.41 ± 0.02 | 100 ± 20 | -55 ± 4 | -150 ± 10 |
| 35 | 2 | 0.389 ± 0.004 | 180 ± 70 | -57 ± 5 | -160 ± 10 |
| 40 | 2 | 0.345 ± 0.007 | 300 ± 100 | -59 ± 7 | -160 ± 20 |
| 45 | 2 | 0.36 ± 0.07 | 600 ± 300 | -74 ± 6 | -200 ± 20 |
| 50 | 2 | 0.56 ± 0.03 | 1000 ± 400 | -47 ± 3 | -120 ± 10 |
| 55 | 3 | 0.9 ± 0.1 | 1100 ± 200 | -27 ± 4 | -60 ± 10 |
| 60 | 1 | 1.07 | 2300 | -14 | -18 |
All experiments were performed in 10 mM sodium cacodylate (pH 5.8) with 10 mM KCl and 10 mM MgCl2. Errors are the standard error of the mean. Errors in Kd were typically higher than in apparent stoichiometry or enthalpy due to increased sensitivity to scatter in the transition region of the ITC curve.
Table 2.
ITC data for TMR binding
| Temperature (°C) | Number of Trials | Apparent Stoichiometry | Apparent Kd (nM) | ΔH° (kcal/mol) | ΔS° (eu) |
|---|---|---|---|---|---|
| 15 | 3 | 1.1 ± 0.2 | 29 ± 6 | -11.9 ± 0.5 | -7 ± 2 |
| 20 | 2 | 1.4 ± 0.2 | 40 ± 20 | -13 ± 2 | -9 ± 6 |
| 25 | 2 | 1.4 ± 0.2 | 60 ± 30 | -15 ± 2 | -16 ± 8 |
| 30 | 8 | 1.2 ± 0.1 | 70 ± 20 | -19 ± 2 | -29 ± 6 |
| 35 | 4 | 1.2 ± 0.1 | 90 ± 10 | -19 ± 2 | -30 ± 6 |
| 40 | 2 | 1.0 ± 0.3 | 400 ± 200 | -23.2 ± 0.2 | -44 ± 2 |
| 45 | 3 | 1.0 ± 0.2 | 500 ± 200 | -20 ± 2 | -35 ± 5 |
| 50 | 3 | 1.1 ± 0.2 | 700 ± 200 | -25 ± 5 | -50 ± 20 |
| 55 | 3 | 1.3 ± 0.3 | 600 ± 300 | -21 ± 6 | -40 ± 20 |
| 60 | 2 | 1.26 ± 0.06 | 1300 ± 100 | -23 ± 3 | -43 ± 9 |
All experiments were performed in 10 mM sodium cacodylate (pH 5.8) with 10 mM KCl and 10 mM MgCl2. Errors are the standard errors of the mean. Errors in Kd were typically higher than in stoichiometry or enthalpy due to increased sensitivity to scatter in the transition region of the ITC curve.
Figure 2.
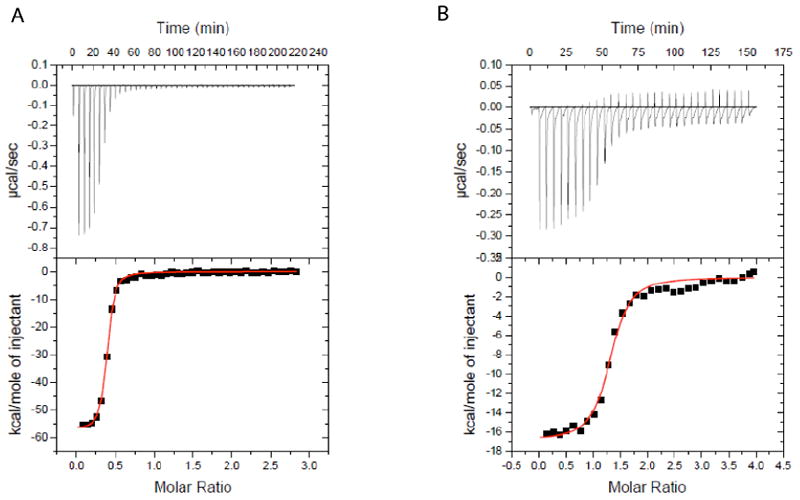
ITC data for ligand binding to the MG aptamer. A.) 53.9 μM MG titrated into 4.36 μM MG aptamer at 25°C. B) 56.6 μM TMR titrated into 3.29 μM MG aptamer at 25°C. Note the greater dynamic range of heat for MG, which reflect its binding with much greater exothermicity. The x-axis is the molar ratio of ligand added to total RNA. Binding thermodynamic parameters are provided in Tables 1 and 2.
The observed binding stoichiometry differs significantly between the two ligands. At 25 °C, TMR binds with an apparent stoichiometry near 1:1 (n=1.4 ± 0.2) while MG binds with a significantly smaller apparent stoichiometry (n=0.40 ± 0.02). Moreover, these values were consistent across multiple independent trials (Tables 1 and 2). It should be noted that the observed stoichiometry does not affect the enthalpy values: ΔH° is calculated as kcal per mol ligand injected, all of which has plenty of RNA to bind to in the initial injections. Indeed, the integrated heat for the early injections (Figure 2A, bottom panels) is approximately equal to the ΔH calculated from model fitting. In addition, the value of n does not affect the apparent Kd value, as the dissociation constant is dependent only upon the slope and curvature of the transition region.
Temperature-Dependent Enthalpy Changes for MG Binding Reveal Two Temperature Regimes
We next examined ligand binding at different temperatures. In order to elucidate the heat capacity for ligand binding, ITC measurements were made over a wide temperature range, 15 to 60 °C. Tables 1 and 2 give the data for these measurements, while Figure 3 provides plots of ΔH° versus temperature. The heat capacity plot in Figure 3 reveals that MG binding has two distinct temperature regimes: one at the lower temperatures of 15 to 45 °C, wherein binding becomes increasingly exothermic with temperature (i.e. negative ΔCp value), and the other in the range of 45 to 60 °C, wherein binding becomes decreasingly exothermic with temperature (i.e. positive ΔCp value). The TMR ligand, on the other hand, shows essentially one temperature regime, which extends to at least 50 °C, which has a smaller, negative ΔCp.
Figure 3.
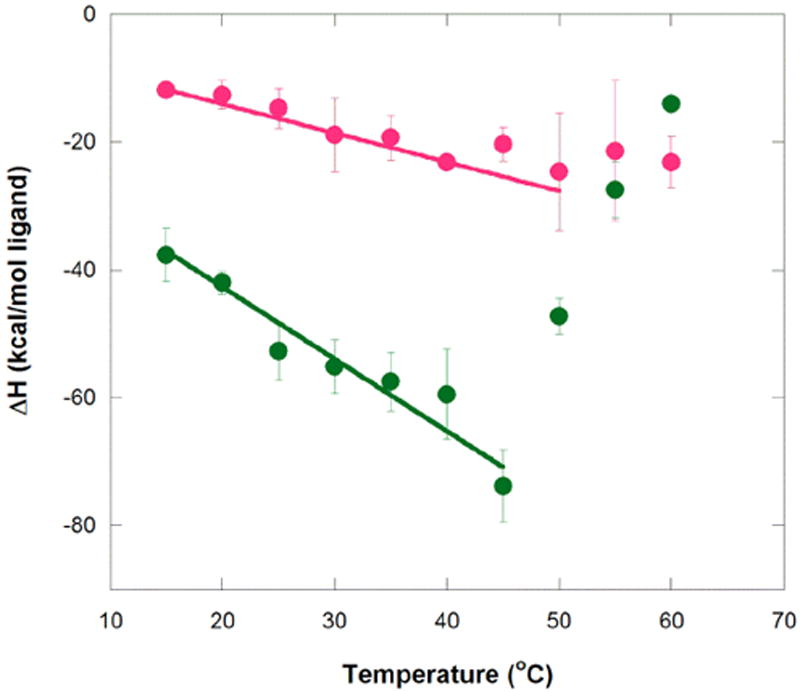
Temperature dependence of enthalpy for aptamer binding to MG and TMR. Circles represent average ΔH° values at a given temperature for MG (green) and TMR (magenta). Error bars are the standard error of the mean. Weighted linear regression of MG binding over the temperature range of 15 to 45 °C gives ΔH(T) = −1.13 kcal/mol°C (T − 25 °C) − 48.3 kcal/mol (R2 = 0.95), while that for TMR binding over the range 15 to 50 °C gives ΔH(T) = −0.45 kcal/mol°C (T − 25 °C) − 16.3 kcal/mol (R2 = 0.97). These plots provide ΔCp values for MG and TMR of −1.13+/−0.17 and −0.45+/−0.035 kcal K−1 mol−1, respectively. Note that the values of the final term in these fits are similar to the actual enthalpies for MG and TMR measured at 25 °C (provided in Tables 1 and 2), as expected. Buffer conditions were 10 mM sodium cacodylate (pH 5.8), 10 mM KCl, and 10 mM MgCl2.
Values of the binding ΔCp’s for aptamer interaction with MG and TMR were calculated from the linear fit of ΔH° versus temperature data in the 15 to 45 °C regime (fits provided in Figure 3).1 As mentioned, both ligands exhibit negative heat capacity in this temperature range, but the ΔCp for MG binding is ~2.5-fold larger in magnitude than for TMR binding, −1.13 and −0.45 kcal mol-1 K-1, respectively. These values suggest that binding of MG involves a larger conformational rearrangement of the ligand, the aptamer, or both relative to the binding of TMR. Comparison of apparent Kd values for MG and TMR (Tables 1 and 2, respectively) shows that the binding affinities for the two ligands are nonetheless approximately equal at the low temperature end of the data, with a slight preference for the non-cognate TMR as temperature increases, which is as expected given the more exothermic ΔH for binding of MG.
Next, we consider the binding entropy. Tables 1 and 2 give the data for these measurements, while Figure 4 provides the TΔS° versus temperature plots for both ligands. Like the enthalpy plots, the entropy plots reveal two temperature regimes. The binding entropy for MG becomes increasingly unfavorable with temperature until 50 °C, where it rapidly becomes less unfavorable with temperature (Figure 4). The binding entropy for TMR (Table 2) does not follow this trend, rather it levels off at high temperature, akin to the binding enthalpy.
Figure 4.
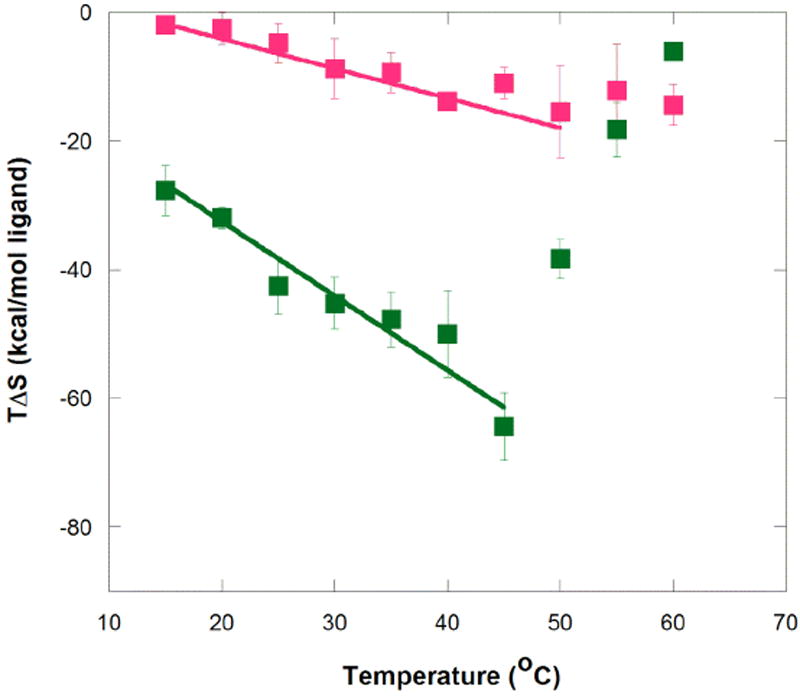
Temperature dependence of entropy for aptamer binding to MG and TMR. Symbols represent average TΔS° values at each temperature for MG (green) and TMR (magenta). Error bars are from the standard error of the mean. Buffer conditions were 10 mM sodium cacodylate (pH 5.8), 10 mM KCl, and 10 mM MgCl2. Slope of the fits here is equal to ΔCp + ΔS, which is ~ΔCp because ΔS is relatively small. The slope of this plot is not interpreted in detail herein because ΔCp is directly obtainable in Figure 3 and ΔS is available in Tables 1 and 2. Nonetheless, this plot provides the trend of TΔS° with temperature.
Since heat capacity for biopolymers is often correlated with ΔASA,17,27,28 we next calculated ΔASA for the TMR-bound structure relative to the MG-bound structure.23 Total ASA for the TMR complex is 5432 Å2, while that of the MG complex is 5183 Å2, providing a ΔASA ~250 Å2. This difference is in line with approximately 1.5 nt less surface area in the MG complex.29 It should be noted that the two structures were solved under different solution conditions. Specifically the MG-bound solution structure was solved in the absence of divalent ions, although the authors pointed out that divalent ions had no effect on the NMR spectra.11,12 With the assumption that both binding interactions in our experiments start with the same initial unbound state (or states), which is reasonable given that the MG aptamer was prepared identically for both ligands, this result indicates that there is greater compaction of the RNA aptamer in the course of binding MG. This ΔASA thus provides a structural basis for observation of larger binding heat capacity for MG than TMR.
Binding Stoichiometry for MG is Near Unity in the High-Temperature Regime
Temperature affects not only the binding enthalpy and entropy, but also the apparent stoichiometry of binding. The binding stoichiometry for MG increases to ~1 at 55 and 60 °C, while the stoichiometry for TMR remains constant at ~1.2 across all temperatures (Figure 5).
Figure 5.
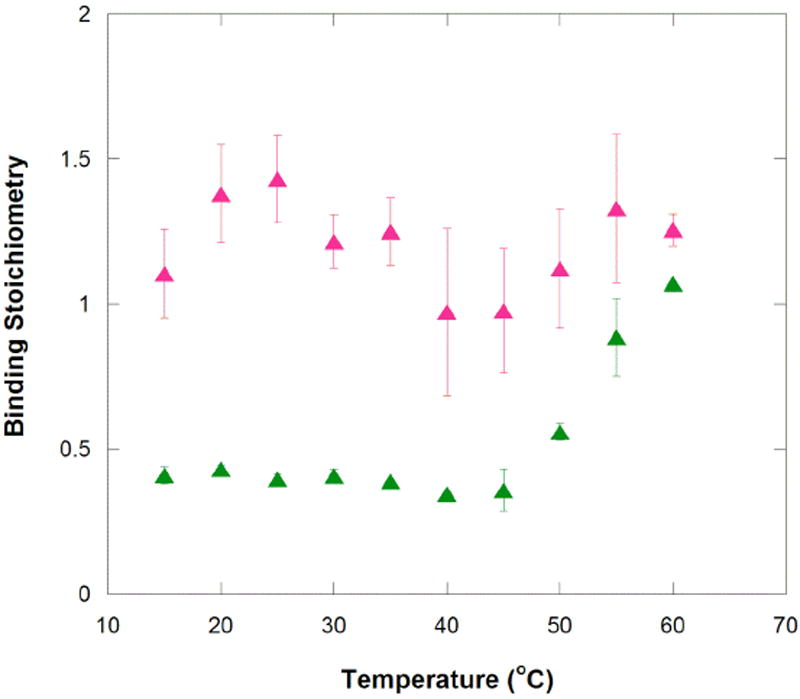
Temperature dependence of apparent stoichiometry for aptamer binding to MG and TMR. Binding stoichiometries for the MG (green) and TMR (magenta) binding to MG aptamer were measured by ITC and are listed in Tables 1 and 2.
As mentioned, the ratio of ligand to macromolecule (n) for the MG interaction in the low-temperature binding regime is ~0.4. This apparent stoichiometry suggests that for every ~5 aptamers, only ~2 MG ligands bind, or in other words ~60% of all aptamers do not readily accommodate an MG ligand. An alternative case was considered as well, in which MG could bind to a dimer of the aptamer strands, given the near 1:2 observed stoichiometry value. Thermal denaturation monitored by UV spectroscopy and UV-crosslinking experiments were performed to assess this possibility. Melts over a ~20-fold range in aptamer concentration are provided in Figure S2 and summarized in Table 3. These experiments, both in the absence and presence of tertiary structure-promoting divalent metal ions, showed no increase in the melting temperature (Tm) of the aptamer with increasing RNA concentration, which supports absence of MG dimerization.
Table 3.
Summary of Tm for the MG aptamer
| [MG aptamer] (μM) | Tm (°C) | [MG aptamer] (μM) | Tm (°C) |
|---|---|---|---|
| in 0 mM Mg2+ | in 10 mM Mg2+ | ||
| 1.3 | 59.7 ± 0.3 | ||
| 2.5 | 58.5 ± 0.5 | 2.5 | 85.2 ± 0.3 |
| 5 | 57 ± 1 | 5 | 85.3 ± 0.8 |
| 10 | 58 ± 1 | 10 | 85 ± 1 |
| 20 | 57 ± 1 | 18 | 84.7 ± 0.3 |
All experiments are an average of 3 scans.
As a second way of assessing the possibility of aptamer dimerization, the photoactivatable crosslinker 4-thioUTP was incorporated into the aptamer, followed by exposure to 312 nm light and denaturing PAGE. This experiment was performed in an effort to crosslink any dimer species (Figure S3). However, no slow mobility bands were observed during gel electrophoresis, further supporting absence of dimer formation. Lastly, we note that if the aptamer were dimerizing it would likely have caused TMR’s stoichiometry to be fractional as well because the same aptamer was used for both ligands.
We also noted that TMR’s binding stoichiometry is slightly higher than unity, with an average value of 1.21 ± 0.05. The possibility that the more planar TMR interacts non-specifically with RNA, perhaps by intercalating into the base pairs30 was tested. We titrated TMR into tRNAphe (Sigma-Aldrich) under conditions identical to the MG aptamer ITC experiments. We chose tRNA because it has a varied combination of secondary and tertiary structure and sequence. As shown in Figure S4, injection heats with tRNA were consistently an order of magnitude smaller than with MG aptamer, and were similar to background injection heats. There is thus no evidence that TMR has non-specific interactions with RNA.
Binding Stoichiometry for MG is Also Improved by Lower Concentrations of Magnesium and Longer Experimental Timescales
In order to explore if the improved stoichiometry of the MG aptamer at high temperatures was due to weakened tertiary structure, ITC experiments were repeated in the presence of lower magnesium ion concentrations, which generally weakens RNA tertiary folding.31 As the divalent metal ion concentration is decreased from 10 to 0.1 mM, the apparent stoichiometry for MG binding improves. At 30 °C, n is 0.41 at 10 mM Mg2+, but 0.55 at 0.5 and 1 mM Mg2+ and 0.59 to 0.60 at 0 and 0.1 mM Mg2+, respectively (Table 4). In addition, the binding enthalpy gets less exothermic as stoichiometry increases, although the binding affinity is not significantly impaired until all divalent ions are removed from the system. We note that a similar trend was found with higher temperature: binding enthalpy became significantly less exothermic as stoichiometry increased in going from 50 to 60 °C (Table 1). We note that increasing the monovalent concentration from 10 to 100 mM KCl or changing the pH from 5.8 to 6.5 did not affect the n= 0.4 result for MG-aptamer interaction (data not shown). The association of apparent stoichiometry improvement with lowering of Mg2+ concentration but not ionic strength in general is suggestive of a role of the unbound state’s tertiary structure in affecting the ability of ligand to bind. Indeed, Dieckmann and co-workers pointed out that the structure of the unbound MG aptamer in the absence of divalent ions has almost no base pairing in the binding pocket, consistent with lack of unbound tertiary structure under these conditions.11 These observations suggest the presence of enhanced intramolecular dynamics in the absence of divalent ions.
Table 4.
ITC data for MG binding as a function of MgCl2 concentration.
| [MgCl2] (mM) | Apparent Stoichiometry | Apparent Kd (nM) | ΔH° (kcal/mol) |
|---|---|---|---|
| 0 | 0.59 | 1600 | -26 |
| 0.1 | 0.60 | 130 | -40. |
| 0.5 | 0.55 | 93 | -42 |
| 1 | 0.55 | 150 | -48 |
| 10 | 0.41 ± 0.02 | 100 ± 20 | -55 ± 4 |
All experiments were performed in 10 mM sodium cacodylate (pH 5.8) with 10 mM KCl. Experiments were at 30 °C, which was chosen because it is in the center of the lower temperature regime.
Observation that both higher temperature and lower Mg2+ concentration improved the stoichiometry of MG binding, suggested that RNA tertiary structure may need to melt prior to MG binding. We reasoned that such a structural transition may be under kinetic control as well. With all titrations for MG and TMR, the injection peaks returned to baseline within 3 min, well within the 5 min injection spacing parameter for the titration. Despite this, we tested the effect of lengthening the time between injections.
Remarkably, increasing the time between the injections improved the MG-aptamer interaction stoichiometry, with the most significant effects occurring in the high temperature binding regime. Increasing injection spacings from 5 to 25 min had a slight effect on the stoichiometry at 25 °C, improving n from 0.40 to 0.48. However, increasing the injection spacings from 5 to 25 min at 50 °C increased the apparent stoichiometry from 0.56 to 0.85. This result is in line with 50 °C being the transition temperature between the two binding regimes (Figure 3), and thus with longer times improving access to a buried active site.
Notably, this improvement in apparent stoichiometry did not lead to any observable signal on the ITC (Figure S5). In fact, we switched the feedback mode on the VP-ITC from high feedback (i.e. standard mode) to either low feedback or passive mode (i.e. no feedback), as recommended by the manufacturer for detecting any heats associated with slow thermal processes; however, no differences were found, indicating that any net heat associated with this very slow process is below the detection level of the instrument (Figure S5). The molecular basis for absence of observable heat during the longer injection times is unclear, but could reflect the relatively slow rate of binding during this time, or could be reflective of a thermoneutral process in which bond breaking and making are roughly balanced.
Discussion
The malachite green aptamer has a complex thermodynamic profile with regards to ligand binding. It binds two related ligands, MG and TMR, with similar affinities but different enthalpy and entropy profiles. For both ligands, binding is enthalpically driven with unfavorable binding entropy, but the relative magnitude of these factors differs dramatically. The aptamer binds MG with ~3-fold greater magnitude in enthalpy than TMR, but also with 4 to 10 fold-greater penalty in entropy. Comparison of ligand structures suggests that at least some of this difference is due to the order imposed on the aromatic rings of MG within the confines of the binding pocket, which does not occur in the case of TMR since it is already fixed in a largely co-planar conformation (see Figure 1).
Comparison of the binding pocket architecture of the two complexes indicates that the MG complex has a larger ΔASA (i.e. more compact) than the TMR complex, by ~250 Å2. With the assumption that both complexes start from the same initial unbound state and that the different solution conditions of the ligand-bound PDB structures do not affect the interactions between the ligand and the binding pocket (see Results), these structural differences help explain thermodynamic differences. In particular, binding of MG is characterized by a ~2.5-fold larger binding heat capacity (-1.13 kcal/molK) than TMR (-0.45 kcal/molK) (Figure 3). Magnitude of heat capacity changes have been correlated to conformational changes in the ligand-macromolecule system for many binding systems,27 including riboswitch-metabolite and DNA-protein interactions.17-19
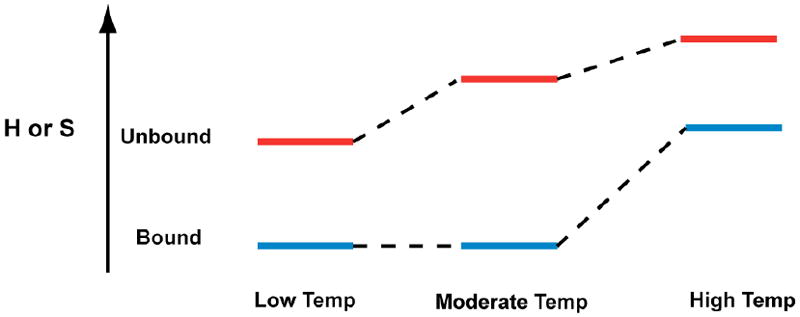
As we noted there are two regimes to the temperature dependence of enthalpy and entropy (Figures 3 and 4). To help account for this, we provide a model in Scheme 1. Over the low temperature regime of 15 to 45 °C, the data suggest that the unfolded state becomes more disordered with temperature (negative ΔCp), while over the high temperature regime of 45 to 60 °C, the data suggest that the MG-bound folded state becomes even more disordered with temperature (positive ΔCp). It is noteworthy that TMR does not show such a steep apparent positive ΔCp in the high temperature regime, which may be reflective of TMR not requiring a highly ordered folded state, perhaps because the initial selections were not for TMR.
Scheme 1.
Heterogeneous Unbound State in the MG Aptamer
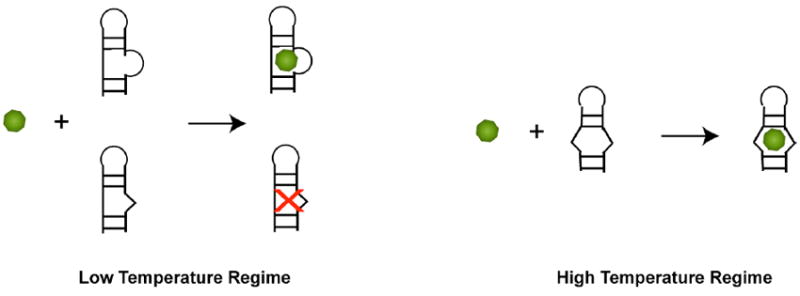
The different binding stoichiometry for MG and TMR, along with the dependence of binding stoichiometry for MG on temperature, divalent ion, and time provides evidence for a heterogeneous unbound state of the aptamer. Several lines of evidence suggest that the effect of these parameters is on the RNA rather than the ligand: RNA is known to be denatured by higher temperature and lower divalent ion concentration, whereas a ligand is not expected to be strongly sensitive to divalent ion concentration. In addition, RNA can get trapped in stable misfolds, whereas this is unlikely for a small molecule ligand. The observed stoichiometry of n~0.4 for MG binding suggests that the unbound aptamer binding pocket has two or more folds (Scheme 2). The two temperature binding regimes observed for MG may be due to partial melting of the pre-bound binding pocket. In the low temperature regime, MG can bind only a portion of the aptamer population, and the conformational rearrangement that the RNA and ligand must undergo is extensive, judging by the value of the binding heat capacity. In contrast, TMR can likely access the entirety of pre-bound MG aptamer population in this temperature range with less structural changes. As temperature increases (or magnesium concentration decreases) to where the pre-bound state’s tertiary structure has been denatured to a certain extent (Scheme 2B), all of the MG aptamer molecules are accessible to MG and it binds 1:1. At the same time, TMR continues to access all of the MG aptamer population at these higher temperatures. We note that a heterogeneous free state has been suggested in the case of the theophylline-RNA complex as well, which was revealed primarily through the Mg2+-dependence of binding kinetics.32
Scheme 2.
The MG Aptamer is Not Selective for MG or TMR
Affinities of the MG aptamer for the two ligands, MG and TMR, are approximately equal in the low temperature regime, and the affinity is only slightly lower for MG in the high temperature regime. The specificity of the MG aptamer for its cognate ligand is therefore quite poor, at least with respect to TMR. This is especially striking when compared to the theophylline aptamer, which can distinguish caffeine from theophylline by a single methyl group.33 The nonselective nature of the MG aptamer for a closely related ligand is not surprising, however, as there was no counter-selection against TMR,9 as there was against caffeine in the theophylline aptamer.33
A recent thermodynamic study on the MG aptamer explored the specificity of the aptamer as a function of monovalent and divalent metal ion concentration using ITC.12 Their results showed that the selectivity for MG over TMR can be tuned via metal ion concentration, with MG binding slightly tighter at low salt conditions (1 mM NaCl), and equivalently at conditions comparable to our study (10 mM MgCl2). These results indicate that tertiary structure stability is a key factor in the function of the MG aptamer, which is in line with our findings with the dependence of binding stoichiometry on temperature and magnesium.
In closing, the MG aptamer reveals that dynamics in the RNA, promoted by higher temperature or lower Mg2+ concentration and captured by waiting long times, are important for binding a dynamic ligand MG, but not a rigid one TMR. These results suggest that that the binding pocket structure of the MG aptamer consists of a heterogeneous population of folds, some of which could be intrinsically disordered, that exclude a dynamic ligand but not a more static one. The ability of the MG aptamer to bind its cognate ligand can be improved by adopting conditions that weaken tertiary structure of the unbound aptamer. These findings have general ramifications for the ability of functional RNAs to interact with dynamic substrates.
Supplementary Material
Acknowledgments
We would like to thank Olke Uhlenbeck and George Makhatadze for helpful discussions. This work was funded by a National Science Foundation Grant (MCB-0527102) and a NIH Predoctoral fellowship (1F31NS054492-01) to J. Sokoloski.
Abbreviations
- MG
Malachite Green
- TMR
Tetramethylrosamine
- ITC
Isothermal Titration Calorimetry
- ASA
Accessible Surface Area
Footnotes
We did not calculate ΔCp for the higher temperature regime because that region likely couples ligand binding with global folding of the aptamer (see Discussion).
Supporting Information Overlay of the TMR and MG binding pockets; UV melting of the MG aptamer; 4-thioUTP crosslinking of the MG aptamer; TMR titrated into tRNA measured by ITC; ITC injection peak morphology. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ellington AD. RNA selection. Aptamers achieve the desired recognition. Curr Biol. 1994;4:427–429. doi: 10.1016/s0960-9822(00)00093-2. [DOI] [PubMed] [Google Scholar]
- 2.Joyce GF. In vitro evolution of nucleic acids. Curr Opin Struct Biol. 1994;4:331–336. doi: 10.1016/s0959-440x(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 3.Mayer G. The chemical biology of aptamers. Angew Chem Int Ed Engl. 2009;48:2672–2689. doi: 10.1002/anie.200804643. [DOI] [PubMed] [Google Scholar]
- 4.Giovannoli C, Baggiani C, Anfossi L, Giraudi G. Aptamers and molecularly imprinted polymers as artificial biomimetic receptors in affinity capillary electrophoresis and electrochromatography. Electrophoresis. 2008;29:3349–3365. doi: 10.1002/elps.200800004. [DOI] [PubMed] [Google Scholar]
- 5.Cho EJ, Lee JW, Ellington AD. Applications of aptamers as sensors. Annu Rev Anal Chem. 2009;2:241–264. doi: 10.1146/annurev.anchem.1.031207.112851. [DOI] [PubMed] [Google Scholar]
- 6.Barbas AS, Mi J, Clary BM, White RR. Aptamer applications for targeted cancer therapy. Future Oncol. 2010;6:1117–1126. doi: 10.2217/fon.10.67. [DOI] [PubMed] [Google Scholar]
- 7.Becker RC, Povsic T, Cohen MG, Rusconi CP, Sullenger B. Nucleic acid aptamers as antithrombotic agents: Opportunities in extracellular therapeutics. Thromb Haemost. 2010;103:586–595. doi: 10.1160/TH09-10-0716. [DOI] [PubMed] [Google Scholar]
- 8.Khati M. The future of aptamers in medicine. J Clin Pathol. 2010;63:480–487. doi: 10.1136/jcp.2008.062786. [DOI] [PubMed] [Google Scholar]
- 9.Grate D, Wilson C. Laser-mediated, site-specific inactivation of RNA transcripts. Proc Natl Acad Sci U S A. 1999;96:6131–6136. doi: 10.1073/pnas.96.11.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baugh C, Grate D, Wilson C. 2.8 A crystal structure of the malachite green aptamer. J Mol Biol. 2000;301:117–128. doi: 10.1006/jmbi.2000.3951. [DOI] [PubMed] [Google Scholar]
- 11.Flinders J, DeFina SC, Brackett DM, Baugh C, Wilson C, Dieckmann T. Recognition of planar and nonplanar ligands in the malachite green-RNA aptamer complex. Chembiochem. 2004;5:62–72. doi: 10.1002/cbic.200300701. [DOI] [PubMed] [Google Scholar]
- 12.Bernard Da Costa J, Dieckmann T. Entropy and Mg(2+) control ligand affinity and specificity in the malachite green binding RNA aptamer. Mol Biosyst. 2011;7:2156–2163. doi: 10.1039/c1mb05075c. [DOI] [PubMed] [Google Scholar]
- 13.Sokoloski JE, Godfrey SA, Dombrowski SE, Bevilacqua PC. Prevalence of syn nucleobases in the active sites of functional RNAs. RNA. 2011;17:1775–1787. doi: 10.1261/rna.2759911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callen HB. Thermodynamics and an Introduction to Thermostatistics. Second. John Wiley and Sons, Inc.; New York: 1985. [Google Scholar]
- 15.Prabhu NV, Sharp KA. Heat capacity in proteins. Annu Rev Phys Chem. 2005;56:521–548. doi: 10.1146/annurev.physchem.56.092503.141202. [DOI] [PubMed] [Google Scholar]
- 16.Mikulecky PJ, Feig AL. Heat capacity changes associated with DNA duplex formation: salt- and sequence-dependent effects. Biochemistry. 2006;45:604–616. doi: 10.1021/bi0517178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 18.Jen-Jacobson L, E LE, Ames JT, Kurpiewski MR, Grigorescu A. Thermodynamic Parameters of Specific and Nonspecific Protein-DNA Binding. Supramol Chem. 2000;12:143–160. [Google Scholar]
- 19.Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J Mol Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Bevilacqua PCB, T S, Chadalavada D, Parente AD, Yajima R. In: Kinetic Analysis of Ribozyme Cleavage. Johnson KA, editor. Oxford University Press; Oxford: 2003. pp. 49–74. [Google Scholar]
- 21.McDowell JA. 1995 [Google Scholar]
- 22.Swain CG, Hedberg K. The Mechanism of Oxidation of Leuco Malachite Green by Ceric Sulfate. J Am Chem Soc. 1950;72:3373–3375. [Google Scholar]
- 23.Rychkov G, Petukhov M. Joint neighbors approximation of macromolecular solvent accessible surface area. J Comput Chem. 2007;28:1974–1989. doi: 10.1002/jcc.20550. [DOI] [PubMed] [Google Scholar]
- 24.Shrake A, Rupley JA. Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J Mol Biol. 1973;79:351–371. doi: 10.1016/0022-2836(73)90011-9. [DOI] [PubMed] [Google Scholar]
- 25.McGraw AP, Mokdad A, Major F, Bevilacqua PC, Babitzke P. Molecular basis of TRAP-5’SL RNA interaction in the Bacillus subtilis trp operon transcription attenuation mechanism. RNA. 2009;15:55–66. doi: 10.1261/rna.1314409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsson TS, Ladbury JE, Pitt WR, Williams MA. Extent of enthalpy-entropy compensation in protein-ligand interactions. Protein Sci. 2011;20:1607–1618. doi: 10.1002/pro.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shelton VM, Sosnick TR, Pan T. Applicability of urea in the thermodynamic analysis of secondary and tertiary RNA folding. Biochemistry. 1999;38:16831–16839. doi: 10.1021/bi991699s. [DOI] [PubMed] [Google Scholar]
- 29.Singh YH, Andrabi M, Kahali B, Ghosh TC, Mizuguchi K, Kochetov AV, Ahmad S. On nucleotide solvent accessibility in RNA structure. Gene. 2010;463:41–48. doi: 10.1016/j.gene.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Saucier JM, Festy B, Le Pecq JB. The change of the torsion of the DNA helix caused by intercalation. II. Measurement of the relative change of torsion induced by various intercalating drugs. Biochimie. 1971;53:973–980. doi: 10.1016/s0300-9084(71)80065-2. [DOI] [PubMed] [Google Scholar]
- 31.Misra VK, Draper DE. On the role of magnesium ions in RNA stability. Biopolymers. 1998;48:113–135. doi: 10.1002/(SICI)1097-0282(1998)48:2<113::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 32.Jucker FM, Phillips RM, McCallum SA, Pardi A. Role of a heterogeneous free state in the formation of a specific RNA-theophylline complex. Biochemistry. 2003;42:2560–2567. doi: 10.1021/bi027103+. [DOI] [PubMed] [Google Scholar]
- 33.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


