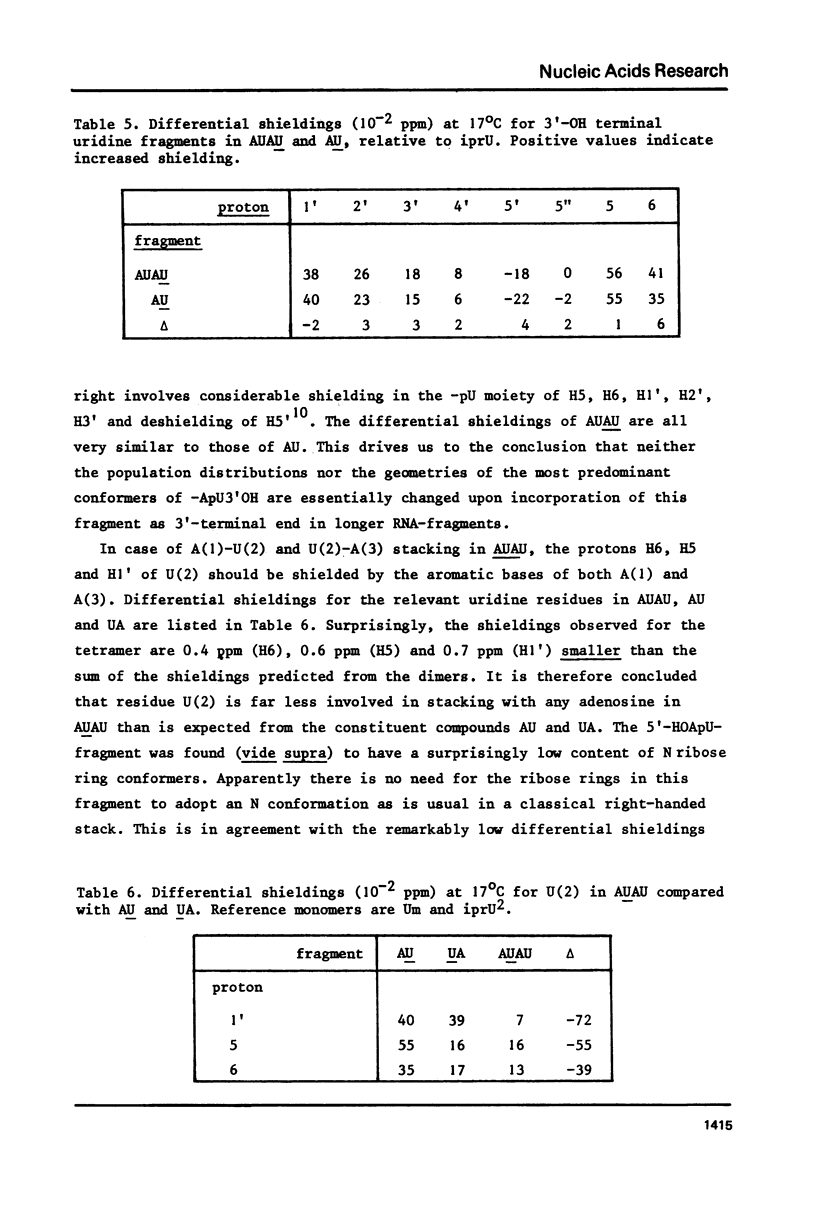
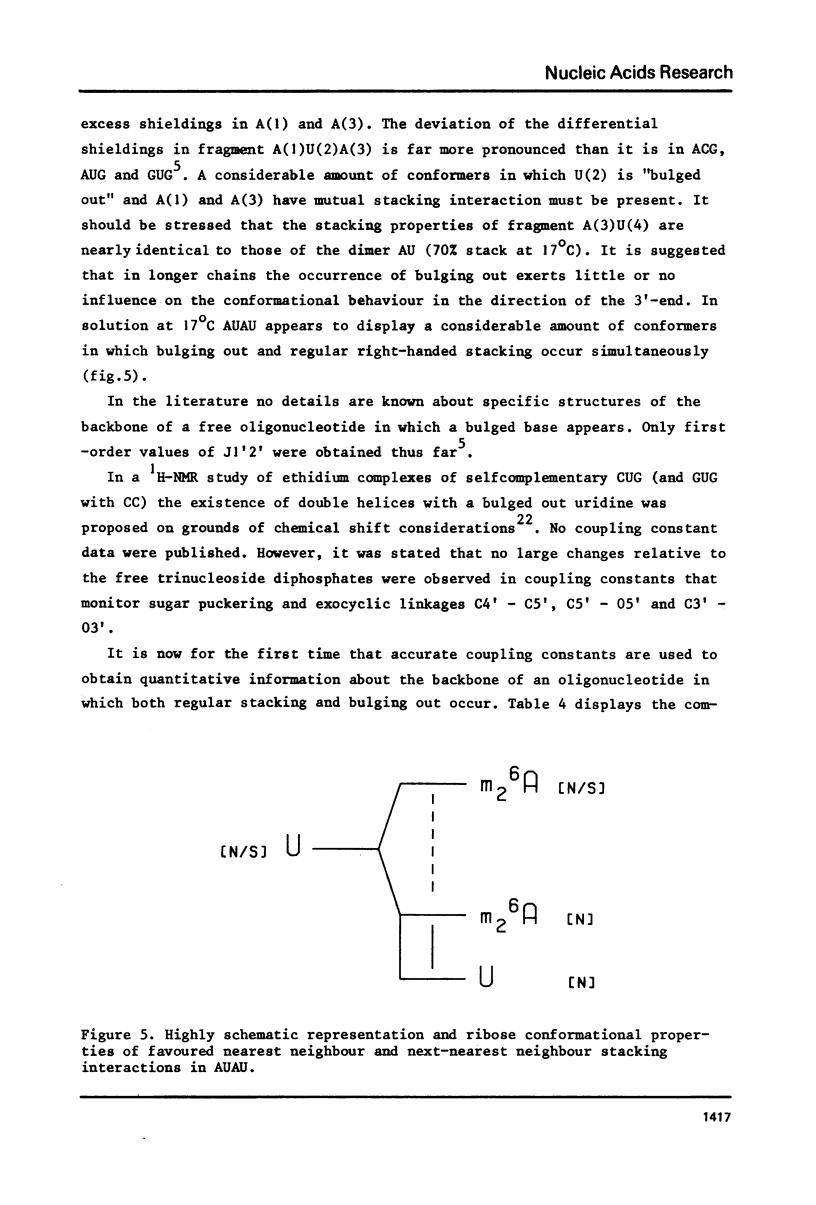
Abstract
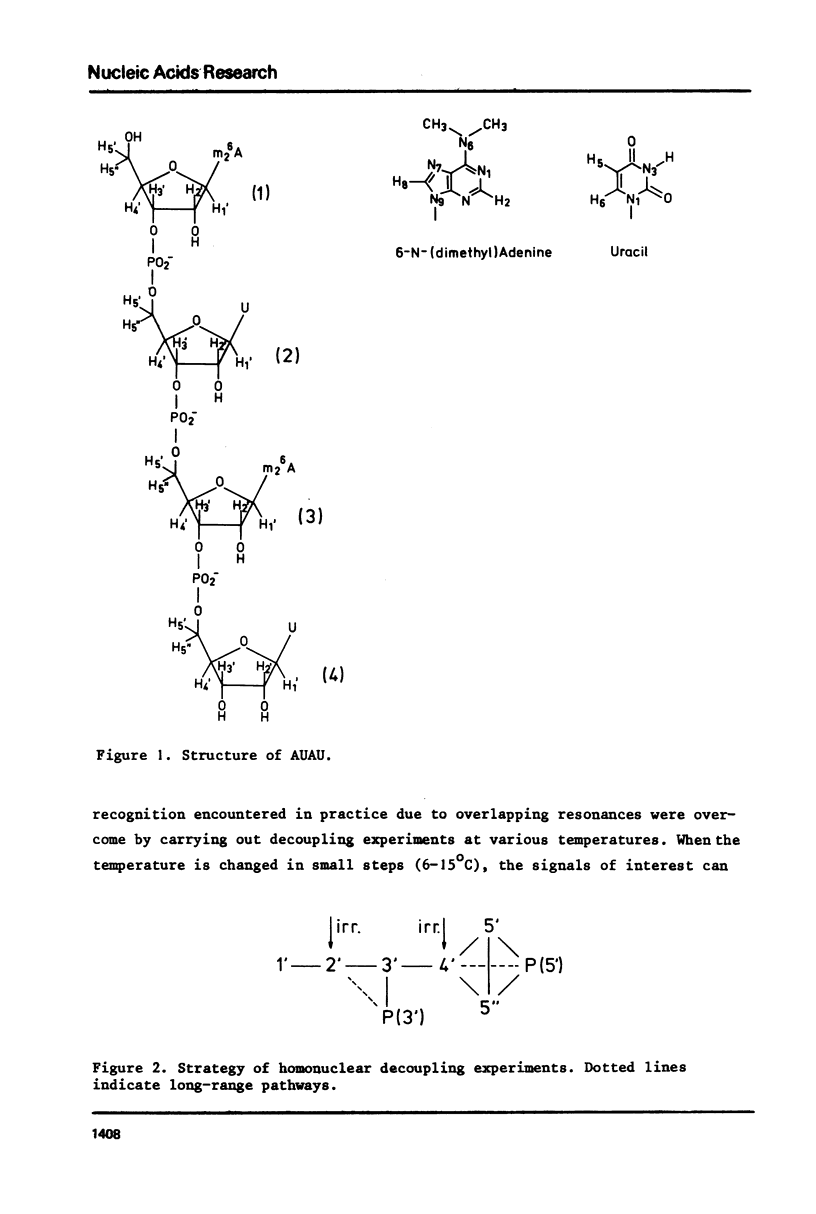
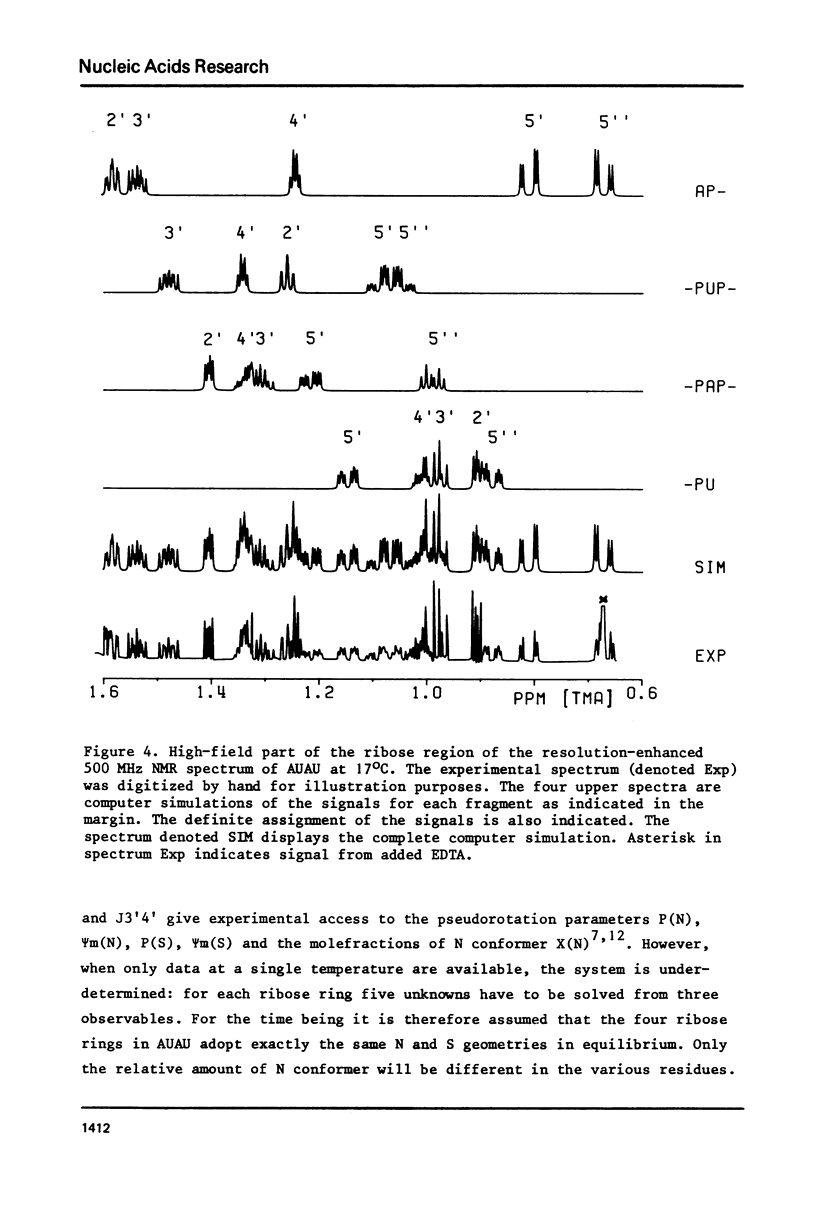
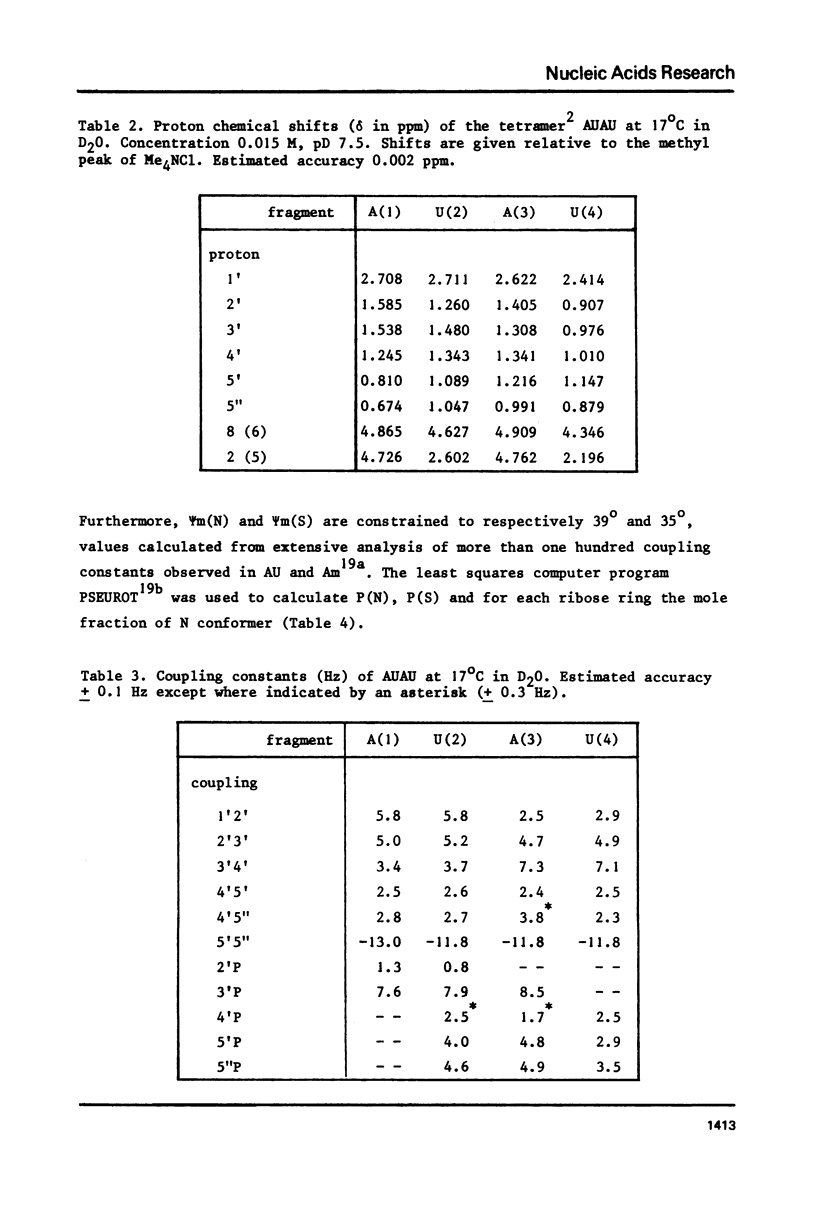
The complete and unequivocal assignment of the 24 ribose proton signals of m6(2)A(1)-U(2)-m6(2)(3)-U(4) by means of 500 MHz NMR spectroscopy at 17 degrees C is given. this assignment is based on scrupulous decoupling experiments carries out at various temperatures. Analysis of the observed chemical shifts and coupling constants of the tetramer shows that the two fragments -m6(2)A(3)-U(4) comprising the 3'-end occur mainly in the classical right-handed stack conformation, whereas the 5'-end the -U(2)- residue appears bulged out in favour of a less well-defined stacking interaction between the bases m6(2)A(1)-and -m6(2)A(3)-. Conformational populations about each of the torsional degrees of freedom along the backbone are discussed. A modernized version of pseudorotation analysis is used to delineate the conformational behaviour of the four ribose rings.
Full text
PDF












Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altona C., Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J Am Chem Soc. 1972 Nov 15;94(23):8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- Altona C., Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. Improved method for the interpretation of proton magnetic resonance coupling constants. J Am Chem Soc. 1973 Apr 4;95(7):2333–2344. doi: 10.1021/ja00788a038. [DOI] [PubMed] [Google Scholar]
- Altona C., Van Boom J. H., Haasnoot C. A. Conformational analysis of a DNA triplet in aqueous solution. Thymidylyl-(3'-5')-thymidylyl-(3'-5')-2'-deoxyadenosine, d(T-T-A), studied by 1H nuclear magnetic resonance at 360 MHz. Eur J Biochem. 1976 Dec 11;71(2):557–562. doi: 10.1111/j.1432-1033.1976.tb11145.x. [DOI] [PubMed] [Google Scholar]
- Cheng D. M., Danyluk S. S., Dhingra M. M., Ezra F. S., MacCoss M., Mitra C. K., Sarma R. H. Conformational flexibility of the 3' acceptor end of transfer ribonucleic acid. Biochemistry. 1980 May 27;19(11):2491–2497. doi: 10.1021/bi00552a030. [DOI] [PubMed] [Google Scholar]
- Evans F. E., Sarma R. H. Nucleotide rigidity. Nature. 1976 Oct 14;263(5578):567–572. doi: 10.1038/263567a0. [DOI] [PubMed] [Google Scholar]
- Gray D. M., Tinoco I., Jr, Chamberlin M. J. The circular dichroism of synthetic ribonucleic acids and the influence of uracil on conformation. Biopolymers. 1972;11(6):1235–1258. doi: 10.1002/bip.1972.360110609. [DOI] [PubMed] [Google Scholar]
- Lee C. H., Sarma R. H. Aqueous solution conformation of rigid nucleosides and nucleotides. J Am Chem Soc. 1976 Jun 9;98(12):3541–3548. doi: 10.1021/ja00428a026. [DOI] [PubMed] [Google Scholar]
- Lee C. H., Tinoco I., Jr Conformation studies of 13 trinucleoside diphosphates by 360 MHz PMR spectroscopy. A bulged base conformation. I. Base protons and H1' protons. Biophys Chem. 1980 Apr;11(2):283–294. doi: 10.1016/0301-4622(80)80031-7. [DOI] [PubMed] [Google Scholar]
- Lee C. H., Tinoco I., Jr Mutagen--oligonucleotide complexes with a bulged base as models for frameshift mutation. Nature. 1978 Aug 10;274(5671):609–610. doi: 10.1038/274609a0. [DOI] [PubMed] [Google Scholar]
- Olsthoorn C. S., Haasnoot C. A., Altona C. Circular dichroism studies of 6-N-methylated adenylyladenosine and adenylyluridine and their parent compounds. Thermodynamics of stacking. Eur J Biochem. 1980 May;106(1):85–95. doi: 10.1111/j.1432-1033.1980.tb05999.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg J. M., Seeman N. C., Day R. O., Rich A. RNA double-helical fragments at atomic resolution. II. The crystal structure of sodium guanylyl-3',5'-cytidine nonahydrate. J Mol Biol. 1976 Jun 14;104(1):145–167. doi: 10.1016/0022-2836(76)90006-1. [DOI] [PubMed] [Google Scholar]



