Abstract
Background
We have previously shown evidence that polymorphisms within genes controlling leukotriene B4 (LTB4) production (ALOX5AP and LTA4H) are associated with asthma susceptibility in children. Evidence also suggests a potential role of LTB4 in COPD disease mechanisms including recruitment of neutrophils to the lung. The aim of the current study was to see if these SNPs and those spanning the receptor genes for LTB4 (LTB4R1 and LTB4R2) influence baseline lung function and COPD susceptibility/severity in smokers.
Methods
Eight ALOX5AP, six LTA4H and six LTB4R single nucleotide polymorphisms (SNPs) were genotyped in a UK Smoking Cohort (n = 992). Association with baseline lung function (FEV1 and FEV1/FVC ratio) was determined by linear regression. Logistic regression was used to compare smoking controls (n = 176) with spirometry-defined COPD cases (n = 599) and to more severe COPD cases (GOLD stage 3 and 4, n = 389).
Results
No association with ALOX5AP, LTA4H or LTB4R survived correction for multiple testing. However, we showed modest association with LTA4H rs1978331C (intron 11) with increased FEV1 (p = 0.029) and with increased FEV1/FVC ratio (p = 0.020).
Conclusions
These data suggest that polymorphisms spanning ALOX5AP, LTA4H and the LTB4R locus are not major determinants of baseline lung function in smokers, but provide tentative evidence for LTA4H rs1978331C (intron 11) in determining baseline FEV1 and FEV1/FVC ratio in Caucasian Smokers in addition to our previously identified role in asthma susceptibility.
Background
Chronic obstructive pulmonary disease (COPD) is a complex respiratory disease with genetic and environmental contributors to pathophysiology [1,2]. Evidence suggests the dihydroxy leukotriene, leukotriene B4 (LTB4), plays a role in this disease as its production is elevated in the airways of COPD subjects [3,4]. The altered inflammatory response of the airways of COPD sufferers is a result of lymphocytes and neutrophils, suggested in part to be the result of cigarette smoke inhalation [5]. Importantly, LTB4 has been shown to have chemotactic activity recruiting inflammatory cells to the lung [6,7]. LTB4 is implicated in the neutrophillic inflammation of COPD and has been suggested as the major chemotactic agent in more severe forms of this disease [8]. It has been established that the cysteinyl leukotrienes (CysLTs; LTC4, LTD4 and LTE4) play a significant role in bronchoconstriction and airway inflammation in asthma [9] but their role in COPD is less clear.
Studies have suggested that polymorphisms spanning leukotriene pathway genes may be important in determining leukotriene production and susceptibility to allergic disorders, such as asthma [10]. LTB4 and the CysLTs are produced from arachidonic acid in a multi-enzyme pathway called the 5-lipoxygenase (5-LO) pathway. Single nucleotide polymorphisms (SNPs) in two 5-LO pathway genes; 5-lipoxygenase activating protein (ALOX5AP) and leukotriene A4 hydrolase (LTA4H) have shown an association with LTB4 overproduction from ionomycin-stimulated neutrophils and with myocardial infarction (MI) susceptibility [11,12]. 5-lipoxygenase activating protein (FLAP) with 5-LO is involved in the synthesis of LTA4 which can be conjugated with glutathione by LTC4 synthase to form LTC4 and subsequent CysLTs or converted to LTB4 by the enzyme LTA4 hydrolase (LTA4H) [13]. FLAP is involved in the production of all leukotrienes; however LTA4H is specifically involved in LTB4 production.
A recent study has suggested that LTA4H contains an aminopeptidase activity as well as having a role in LTB4 production [14]. This aminopeptidase activity cleaves the neutrophil chemoattractant proline-glycine-proline (PGP), a COPD biomarker, responsible for the influx of neutrophils into the lung - contributing to chronic inflammation. Cigarette smoke was found to inhibit this aminopeptidase activity thereby leading to accumulation of PGP and neutrophils [14]. This dual role could have important consequences for the design of therapeutics targeting LTA4H.
We have recently reported evidence that SNPs spanning ALOX5AP and LTA4H are asthma susceptibility markers [15]. SG13S114A, SG13S89A and SG13S41G (ALOX5AP) and rs1978331C (LTA4H) were associated with asthma and asthma related phenotypes (atopy, FEV1, bronchial hyperresponsiveness) in a family based association study using 341 asthma families with two affected siblings [15]. Several haplotype associations were also observed [15]. To date, no study has investigated the role of these SNPs with respect to COPD or baseline lung function in smokers. Smoking is associated with decline in lung function and is a major risk factor for the development of COPD; we therefore investigated the role of ALOX5AP, LTA4H and LTB4R SNPs in smokers.
The aim of the current study was to determine whether polymorphisms spanning ALOX5AP, LTA4H and the LTB4R locus influence baseline lung function (FEV1 and FEV1/FVC ratio) in smokers and whether they contribute to susceptibility to develop COPD or a more severe form of COPD in smokers. We genotyped twenty SNPs spanning these three loci in a cohort recruited for COPD or smoking history (n = 992 subjects) and completed a series of association analyses.
Methods
Subjects and baseline characteristics
Subjects were recruited from five UK centres for smoking history and/or COPD diagnosis (n = 992) [16]. Subjects collected from Nottingham (n = 537) were Caucasian, > 40 years and had > 10 pack-year smoking history. Subjects collected from other UK centres (n = 455) were recruited for physician and spirometry defined COPD, Caucasian, > 40 years, smokers with > 10 pack-year history. The combined subjects (n = 992) recruited for smoking history or COPD diagnosis was stratified into 'healthy' smokers (smoking controls) (n = 176, post-bronchodilator (BD) salbutamol FEV1 > 80% predicted and postBD FEV1/FVC > 0.7) and COPD cases (n = 599, postBD FEV1 < 80% predicted and postBD FEV1/FVC ratio < 0.7). Subjects not meeting these criteria (or with missing data) were excluded from the case control analyses (n = 217). To investigate whether SNPs determined severity of COPD in the smokers we compared smoking controls with postBD spirometry, i.e. the GOLD classifications [17]. Ethical approval was obtained from the relevant ethics committees (Nottingham, Sheffield, Manchester, Leicester and Oxford) and informed consent from all subjects was obtained.
Selection of SNPs and genotyping
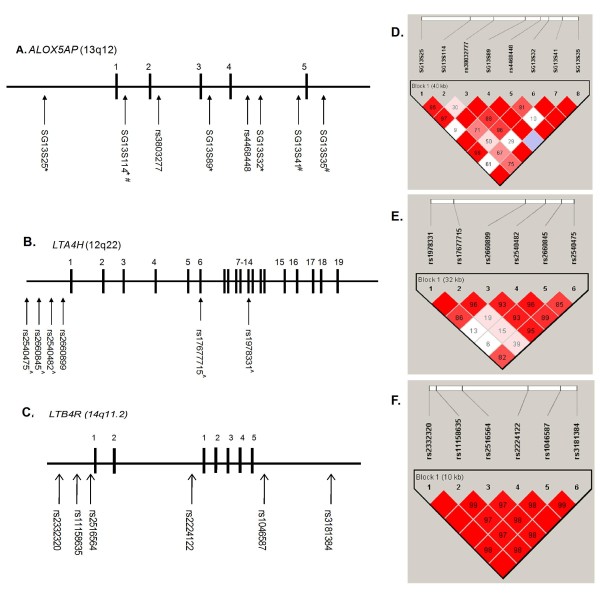
Twenty SNPs were genotyped across ALOX5AP (eight), LTA4H (six) and LTB4R (six) (Figure 1A-C). SNPs spanning ALOX5AP and LTA4H have previously been shown to tag haplotypes associated with myocardial infarction and LTB4 production [11,12] and with asthma susceptibility in our recent study [15]. Six LTB4R SNPs were chosen for their ability to tag linkage disequilibrium (LD) blocks or for inferred function, once the region had been sequenced in Caucasian individuals (22 SNPs validated in 35 Caucasian subjects, data not shown). Genotyping was completed by Kbioscience (Hertfordshire, UK) using KASPar technology. Hardy-Weinberg equilibrium was assessed in all subjects using Haploview software [18].
Figure 1.
Location of SNPs genotyped in ALOX5AP, LTA4H and LTB4R in the smokers and linkage disequilibrium (LD) plot. The LD plot shows the LD displayed as D'/LOD in Haploview software. Numerical values shown correspond to D'. A-C. represents the physical location of the SNPs genotyped in ALOX5AP, LTA4H and LTB4R, respectively. Black boxes represent exons and the spaces between represent introns. D. E. and F. represents the LD plot in the smokers (n = 992) for each loci respectively..* represents SNPs that define the haplotype HapA, #which define HapB and ^ which define the haplotype (HapK) previously associated with MI [11,12].
Statistical analyses
Linear regression (SPSS V15, SPSS Inc., Chicago, IL) was used to determine the contribution of each SNP to baseline FEV1 (litres) or FEV1/FVC ratio using the additive model (e.g. AA vs. AC vs. CC) including age, gender, height and smoking pack-years as covariates. The COPD susceptibility analyses were completed using logistic regression in the additive model in two ways. Firstly, the smoking controls (n = 176) vs. all COPD subjects (n = 599) and then the smoking controls (n = 176) vs. GOLD 3 and 4 subjects (n = 389). Both analyses included age, gender and pack-years as covariates (Table 1). Based on the 80 tests completed (analysis of 20 SNPs with 4 outcomes), a conservative Bonferroni correction suggested a p < 10-4 when reporting results as significant. With respect to power (based on lowest and highest minor allele frequency), there was between 77-99% power to detect a 50 ml difference in FEV1 and between 58-99% power to detect a 5% difference in FEV1/FVC ratio. Analyses of COPD susceptibility were relatively underpowered, with between 28-91% power with an odds ratio of 1.5 and 68-99% power with an odds ratio of 2.0. All analyses considered an error rate of 5%.
Table 1.
Baseline characteristics of the study populations
| UK Smoking Cohort | Smoking Controls |
COPD Cases |
GOLD stage 3 and 4 | Comparison (smoking controls vs. COPD cases | Comparison (smoking controls vs. GOLD 3/4 | |
|---|---|---|---|---|---|---|
| Age | 63.33 ± 10.29 | 54.38 ± 9.52 | 65.96 ± 9.01 | 67.16 ± 8.56 | p < 0.0001 | p < 0.0001 |
| Female (%) | 43.8 | 56.3 | 39.9 | 38.8 | p = 0.037 | p = ns |
| Baseline FEV1 % predicted | 56.05 ± 28.16 | 96.03 ± 12.15 | 40.31 ± 15.63 | 31.46 ± 8.69 | p < 0.0001 | p < 0.0001 |
| FEV1/FVC Ratio | 55.34 ± 17.43 | 77.30 ± 5.90 | 46.3 ± 12.5 | 41.57 ± 11.20 | p < 0.0001 | p < 0.0001 |
| Post BD FEV1 % predicted | 59.08 ± 27.14 | 99.48 ± 11.72 | 44.65 ± 15.52 | 35.28 ± 8.90 | p = 0.049 | p < 0.0001 |
| PostBD FEV1/FVC Ratio | 55.58 ± 17.71 | 79.10 ± 5.05 | 46.2 ± 12.00 | 41.27 ± 10.46 | p < 0.0001 | p < 0.0001 |
| Pack Years | 43.54 ± 26.05 | 32.74 ± 20.04 | 47.61 ± 27.01 | 47.96 ± 27.85 | p < 0.0001 | p < 0.0001 |
| GOLD Stage (%): | ||||||
| • Stage 1 | 6.9 | 0.0 | 0.0 | 0.0 | ||
| • Stage 2 | 32.6 | 0.0 | 34.8 | 0.0 | ||
| • Stage 3 | 42.4 | 0.0 | 45.5 | 69.9 | ||
| • Stage 4 | 18.2 | 0.0 | 19.7 | 30.1 | ||
| Number | 992 | 176 | 599 | 389 | ||
FEV1, Forced expiratory volume in one second; FVC, forced vital capacity; BD, bronchodilator. Control subjects were defined as having postBD (salbutamol) FEV1 > 80% and postBD FEV1/FVC > 0.7. Subjects with COPD were defined as having postBD FEV1 < 80% and FEV1/FVC < 0.7. Individuals who did not meet these criteria were excluded from analyses. Continual variables between groups were compared by Independent T-Test, categorical variables by Pearson chi square.
Results
Clinical characteristics and genotyping
Subject characteristics for the smoking controls (n = 176) and COPD subjects (n = 599) and the entire cohort (n = 992) are shown (Table 1). Comparison of the smoking controls (n = 176) and COPD sufferers (n = 599) show differences in baseline lung function (percent predicted FEV1 96.03% compared to 40.31%) as anticipated (p < 0.0001). These subjects also showed differences for age, gender and pack-years so these variables were included as covariates in analyses. Genotyping completion rates were > 96% for all twenty SNPs genotyped and did not show deviation from Hardy-Weinberg equilibrium (p > 0.05). Minor allele frequencies for ALOX5AP and LTA4H SNPs were similar to those observed in our previous study [15].
Haplotype structure
Figure 1 panels A-C show the location of the SNPs genotyped across ALOX5AP, LTA4H and the LTB4R locus respectively. Panel D shows the linkage disequilibrium (LD) pattern of SNPs genotyped across the ALOX5AP gene on chromosome 13q12. Within ALOX5AP regions of high LD (measured by D') include between SG13S25 (5'UTR) and SG13S114 (intron 1) and of low LD include between SG13S114 (intron 1) and rs38032777 (intron 2). SNPs defining the region SG13S25 (5'UTR) and SG13S35 (3'UTR) show relatively high LD with all other SNPs. Panel E shows the genotyped SNPs for the LTA4H gene on chromosome 12q22. For LTA4H there are regions of high LD between rs17677715 (intron 6) and rs2660899 (5'UTR) and rs2540482 (5'UTR) and rs2660845 (5'UTR). The two distal SNPs defining the extended region are not in strong LD with others. Panel C shows the SNPs genotyped across the LTB4R1 and LTB4R2 genes. There was high LD between all SNPs genotyped in the LTB4R locus indicating there was some redundancy in genotyping (Panel F).
Polymorphisms spanning ALOX5AP, LTA4H and LTB4R are not associated with baseline FEV1 and FEV1/FVC in smokers
To assess whether SNPs in ALOX5AP, LTA4H and LTB4R influence baseline lung function in smokers we determined their role in baseline FEV1 and FEV1/FVC in the entire population (n = 992) using linear regression in the additive model (Additional File 1 Table S1). The FEV1 analyses did not identify any significant association with ALOX5AP, LTA4H or LTB4R SNPs. LTA4H rs1978331C (intron 11) (p = 0.029, mean FEV1 values: TT 1.468 ± 0.039L, TC 1.599 ± 0.034L and CC 1.594 ± 0.057L) and rs2660899 (5'UTR) (p = 0.024; GG 1.580 ± 0.030L, GT 1.504 ± 0.044L and TT 1.192 ± 0.158L) were associated with increased FEV1, although this did not meet Bonferroni correction. Analysis with FEV1/FVC ratio again did not show any significant association. However, the same LTA4H rs1978331C (intron 11) showed p = 0.020 with the same direction of effect, increased FEV1/FVC ratio (mean ratio ranging from 53.8% in the major homozygotes to 57.4% in the minor homozygotes).
Polymorphisms spanning ALOX5AP, LTA4H and LTB4R do not determine COPD susceptibility
To determine whether SNPs spanning ALOX5AP, LTA4H and LTB4R act as determinants of COPD susceptibility in smokers, we completed case-control association analyses comparing the smoking controls (n = 176) with the COPD subjects (n = 599), defined by post-bronchodilator spirometry (Table 2). No significant associations were observed. This analysis was corrected for age, gender and pack-years as these traits were significantly different between the study groups (Table 1).
Table 2.
ALOX5AP, LTA4H and LTB4R SNPs and COPD susceptibility in smokers.
| SNP | Location | Controls (n = 176) | COPD (n = 599) | Additive | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | MAF | 0 | 1 | 2 | MAF | p-value | Odds ratio | 95%CI | ||
| ALOX5AP | ||||||||||||
| SG13S25 (G/A) | 5'UTR | 138 | 35 | 2 | 0.11 | 491 | 100 | 5 | 0.09 | 0.556 | 0.87 | 0.55-1.37 |
| SG13S114 (T/A) | Intron 1 | 92 | 63 | 20 | 0.29 | 248 | 282 | 58 | 0.34 | 0.198 | 1.22 | 0.90-1.65 |
| rs3803277 (C/A) | Intron 2 | 54 | 80 | 42 | 0.47 | 184 | 303 | 103 | 0.43 | 0.318 | 0.87 | 0.65-1.15 |
| SG13S89 (G/A) | Intron 3 | 162 | 13 | 0 | 0.04 | 551 | 45 | 1 | 0.04 | 0.771 | 0.90 | 0.43-1.88 |
| rs4468448 (C/T) | Intron 4 | 100 | 65 | 11 | 0.25 | 333 | 226 | 34 | 0.25 | 0.807 | 1.04 | 0.76-1.45 |
| SG13S32 (C/A) | Intron 4 | 42 | 88 | 43 | 0.49 | 162 | 302 | 130 | 0.47 | 0.351 | 0.87 | 0.66-1.16 |
| SG13S41 (A/G) | Intron 4 | 149 | 21 | 0 | 0.06 | 524 | 62 | 6 | 0.06 | 0.542 | 0.84 | 0.47-1.48 |
| SG13S35 (G/A) | 3'UTR | 146 | 26 | 0 | 0.06 | 479 | 97 | 3 | 0.09 | 0.970 | 1.01 | 0.60-1.71 |
| LTA4H | ||||||||||||
| rs1978331 (T/C) | Intron 11 | 66 | 81 | 27 | 0.39 | 223 | 257 | 109 | 0.40 | 0.419 | 0.89 | 0.68-1.18 |
| rs17677715 (T/C) | Intron 6 | 115 | 50 | 6 | 0.18 | 372 | 188 | 26 | 0.20 | 0.899 | 1.02 | 0.72-1.46 |
| rs2660899 (G/T) | 5'UTR | 130 | 45 | 1 | 0.13 | 418 | 158 | 16 | 0.16 | 0.113 | 1.39 | 0.93-2.08 |
| rs2540482 (T/C) | 5'UTR | 107 | 58 | 8 | 0.21 | 358 | 195 | 34 | 0.23 | 0.473 | 1.13 | 0.81-1.58 |
| rs2660845 (A/G) | 5'UTR | 92 | 71 | 12 | 0.26 | 328 | 224 | 40 | 0.26 | 0.483 | 0.89 | 0.65-1.22 |
| rs2540475 (C/T) | 5'UTR | 108 | 55 | 5 | 0.19 | 358 | 186 | 27 | 0.21 | 0.770 | 0.95 | 0.67-1.35 |
| LTB4R2 | ||||||||||||
| rs2332320 (T/C) | 5'UTR | 130 | 34 | 5 | 0.13 | 440 | 124 | 13 | 0.13 | 0.977 | 1.01 | 0.67-1.52 |
| rs11158635 (G/T) | 5'UTR | 100 | 57 | 9 | 0.23 | 367 | 193 | 26 | 0.21 | 0.165 | 0.78 | 0.55-1.11 |
| rs2516564 (C/T) | 5'UTR | 104 | 59 | 9 | 0.22 | 372 | 193 | 25 | 0.21 | 0.169 | 0.79 | 0.56-1.11 |
| LTB4R1 | ||||||||||||
| rs2224122 (C/G) | 5'UTR | 102 | 55 | 9 | 0.22 | 363 | 190 | 28 | 0.21 | 0.232 | 0.81 | 0.57-1.15 |
| rs1046587 (G/A) | 3'UTR | 43 | 94 | 36 | 0.48 | 155 | 298 | 128 | 0.48 | 0.566 | 1.09 | 0.82-1.45 |
| rs3181384 (C/T) | 3'UTR | 100 | 58 | 10 | 0.20 | 363 | 191 | 28 | 0.21 | 0.111 | 0.76 | 0.54-1.07 |
Logistic regression was used to compare genotype frequencies between smoking controls (n = 176) and total COPD cases (n = 599) using the additive model with the covariates age, gender and pack-years. OR, odds ratio; 95% CI, 95% confidence interval. 0, 1 and 2 represent the number of major homozygote, heterozygote and minor homozygote genotype frequencies.
Polymorphisms spanning ALOX5AP, LTA4H and LTB4R do not determine susceptibility to develop severe COPD
Subjects (where data available) were stratified according to GOLD classifications using post-bronchodilator lung function (GOLD 1 = 44 individuals, GOLD 2 = 209, GOLD 3 = 273 and GOLD 4 = 117). The phenotypic characteristics of these GOLD stratified COPD patients have been previously reported [16]. We did not observe any significant associations with any of the SNPs tested in the control (n = 176) versus severe COPD (n = 389) analyses. rs3803277 (ALOX5AP, intron 2) showed protective association (OR = 0.72, 95% CI = 0.52-0.99, p = 0.045), but this did not survive correction (Table 3).
Table 3.
ALOX5AP, LTA4H and LTB4R SNPs and severe COPD (defined by GOLD stage 3 and 4) in smokers.
| SNP | Location | Controls (n = 176) | GOLD 3 and 4 (n = 389) | Additive | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | MAF | 0 | 1 | 2 | MAF | p-value | Odds ratio | 95%CI | ||
| ALOX5AP | ||||||||||||
| SG13S25 (G/A) | 5'UTR | 138 | 35 | 2 | 0.11 | 325 | 60 | 2 | 0.09 | 0.231 | 0.72 | 0.42-1.23 |
| SG13S114 (T/A) | Intron 1 | 92 | 63 | 20 | 0.29 | 164 | 180 | 36 | 0.33 | 0.282 | 1.21 | 0.86-1.70 |
| rs3803277 (C/A) | Intron 2 | 54 | 80 | 42 | 0.47 | 122 | 203 | 58 | 0.42 | 0.045 | 0.72 | 0.52-0.99 |
| SG13S89 (G/A) | Intron 3 | 162 | 13 | 0 | 0.04 | 356 | 32 | 0 | 0.04 | 0.701 | 0.85 | 0.36-1.99 |
| rs4468448 (C/T) | Intron 4 | 100 | 65 | 11 | 0.25 | 221 | 146 | 18 | 0.24 | 0.882 | 0.97 | 0.67-1.41 |
| SG13S32 (C/A) | Intron 4 | 42 | 88 | 43 | 0.49 | 110 | 199 | 78 | 0.46 | 0.195 | 0.81 | 0.58-1.12 |
| SG13S41 (A/G) | Intron 4 | 149 | 21 | 0 | 0.06 | 343 | 38 | 4 | 0.06 | 0.293 | 0.71 | 0.37-1.35 |
| SG13S35 (G/A) | 3'UTR | 146 | 26 | 0 | 0.06 | 312 | 61 | 2 | 0.09 | 0.677 | 0.88 | 0.49-1.59 |
| LTA4H | ||||||||||||
| rs1978331 (T/C) | Intron 11 | 66 | 81 | 27 | 0.39 | 153 | 165 | 62 | 0.38 | 0.090 | 0.76 | 0.56-1.04 |
| rs17677715 (T/C) | Intron 6 | 115 | 50 | 6 | 0.18 | 241 | 124 | 15 | 0.21 | 0.808 | 1.05 | 0.71-1.56 |
| rs2660899 (G/T) | 5'UTR | 130 | 45 | 1 | 0.13 | 264 | 109 | 10 | 0.17 | 0.114 | 1.44 | 0.92-2.26 |
| rs2540482 (T/C) | 5'UTR | 107 | 58 | 8 | 0.21 | 231 | 129 | 19 | 0.22 | 0.677 | 1.08 | 0.74-1.58 |
| rs2660845 (A/G) | 5'UTR | 92 | 71 | 12 | 0.26 | 209 | 154 | 22 | 0.26 | 0.452 | 0.87 | 0.61-1.25 |
| rs2540475 (C/T) | 5'UTR | 108 | 55 | 5 | 0.19 | 236 | 118 | 16 | 0.20 | 0.895 | 0.97 | 0.65-1.45 |
| LTB4R2 | ||||||||||||
| rs2332320 (T/C) | 5'UTR | 130 | 34 | 5 | 0.13 | 287 | 81 | 8 | 0.13 | 0.523 | 1.17 | 0.73-1.86 |
| rs11158635 (G/T) | 5'UTR | 100 | 57 | 9 | 0.23 | 235 | 131 | 16 | 0.21 | 0.195 | 0.77 | 0.52-1.15 |
| rs2516564 (C/T) | 5'UTR | 104 | 59 | 9 | 0.22 | 238 | 130 | 15 | 0.21 | 0.195 | 0.77 | 0.52-1.14 |
| LTB4R1 | ||||||||||||
| rs2224122 (C/G) | 5'UTR | 102 | 55 | 9 | 0.22 | 228 | 130 | 18 | 0.22 | 0.365 | 0.83 | 0.56-1.23 |
| rs1046587 (G/A) | 3'UTR | 43 | 94 | 36 | 0.48 | 95 | 195 | 83 | 0.48 | 0.857 | 1.03 | 0.75-1.42 |
| rs3181384 (C/T) | 3'UTR | 100 | 58 | 10 | 0.20 | 230 | 131 | 18 | 0.22 | 0.183 | 0.77 | 0.52-1.13 |
Logistic regression was used to compare genotype frequencies between smoking controls (n = 176) and GOLD stage 3/4 cases (n = 389) using the additive model with the covariates age, gender and pack-years. OR, odds ratio; 95% CI, 95% confidence interval. 0, 1 and 2 represent the number of major homozygote, heterozygote and minor homozygote genotype frequencies. Black bold indicated (p = ≤ 0.05).
Discussion
This was the first study to investigate polymorphisms spanning genes involved with LTB4 production and activity with lung function and COPD susceptibility in smokers. A UK smoking cohort comprising n = 992 individuals with > 40 years and > 10 pack-years smoking history was used to determine whether SNPs in ALOX5AP, LTA4H and LTB4R influenced baseline lung function and susceptibility to develop COPD in smokers. LTB4 has been shown to be important for the inflammation observed in COPD, with this mediator upregulated in COPD subjects [4]. We hypothesised that polymorphisms in these genes may influence susceptibility to develop airway obstruction in smokers that is in part driven by LTB4. We have found that polymorphisms spanning ALOX5AP, LTA4H and the LTB4R locus are not associated with lung function or COPD susceptibility in smokers as no SNP survived correction for multiple testing. However, we provide tentative evidence for association between LTA4H rs1978331C (intron 11) and lung function measures in these subjects.
We have previously investigated the role of polymorphic variation in the genes of the 5-lipoxygenase pathway e.g. ALOX5, LTC4S, CYSLTR1 in asthma and allergy susceptibility [15,19,20] and as determinants of clinical responses to therapies targeting this pathway [21]. These studies provide accumulating evidence that polymorphic variation in these genes influence disease phenotypes in disorders where leukotrienes play a significant role [10], also confirmed with other non-respiratory diseases e.g. MI [11,12]. To date, no study has specifically looked at genetic determinants of leukotriene production/activity in smokers with or without COPD. While no association survived the Bonferroni correction, additive model analyses with rs1978331C (LTA4H, intron 11) showed a p = 0.029 with an increase in FEV1 and p = 0.020 with FEV1/FVC ratio. The mean FEV1 and FEV1/FVC values for the TC heterozygotes and CC homozygotes were similar, but the presence of the minor C-allele for these genotype groups gave higher trait values when compared to the TT homozygotes, suggesting a dosage effect does not occur. These findings provide tentative evidence suggesting that variants in LTA4H may determine lung function in COPD.
We next sought to investigate whether polymorphisms spanning these genes determine susceptibility to develop COPD. Case-control association analyses were completed with 'healthy' control smokers and smokers with physician diagnosed COPD (including spirometry). No significant associations with polymorphisms spanning ALOX5AP, LTA4H and LTB4R were identified. We also completed another case-control analysis involving COPD sufferers at the severe end of the spectrum. GOLD groups 3 and 4 were chosen as this represented the most severe cases based on spirometry. Again no significant associations were observed.
There is an interesting link for LTA4H with COPD and asthma; in our group's previous study, we showed preliminary association with rs1978331C (LTA4H, intron 11) and asthma susceptibility in 341 families (protection, p = 0.036) [15]. A recent study has shown a similar effect in a different disease; heterozygosity at two LTA4H SNPs, one rs1978331 (intron 11), is significantly associated with protection from tuberculosis infection, lower mortality amongst patients with severe tuberculosis infection and protection from the development of severe leprosy disease [22]. These two studies show the same protective direction of association and provide further support for a functionally significant role of rs1978331 or (another SNP tagged by this) in determining LTA4H expression or activity.
Suggestive association with COPD (p = 0.02 to 0.05) with four LTA4H SNPs within the promoter region (these SNPs were not analysed in our current study) was reported by another group [23]. We have not identified any association with LTA4H SNPs located in the 5'UTR (rs2540482, rs2660845 and rs2540475) with lung function or COPD susceptibility in smokers. Interestingly, this group also tested different ALOX5AP SNPs to our current study and found no association with COPD [23]. These and our own data provide suggestive support for a role of LTA4H SNPs in determining baseline lung function in smokers potentially suggesting a role for genetically determined LTB4 in COPD (LTA4H converts LTA4 to LTB4). This may be a result of neutrophilic inflammation being important in COPD and severe COPD [24]. While this study did not show significant protective association with LTA4H rs1978331C with lung function in smokers, the same direction of effect was observed with asthma susceptibility in our previous study [15] and with the HapK (rs1978331(T/C), rs17677715(T/C), rs2540482(T/C), rs2660845(A/G) and rs2540475(C/T)) haplotype association that conferred a modest risk of myocardial infarction in Icelandic subjects for rs1978331T [12]. As previously mentioned, a protective effect has also been observed with tuberculosis infection [22].
LTA4H has a pro-inflammatory role generating LTB4 through its epoxide-hydrolase activity (intracellular) and an anti-inflammatory role through its amino-peptidase activity to breakdown PGP, facilitating resolution (extracellular). Cigarette smoke selectively inhibits the ability of LTA4H to break-down PGP leading to neutrophil accumulation and contributing to COPD pathogenesis [14]. rs1978331 may affect the levels of transcription of the LTA4H gene. Decreased transcription could lead to decreased protein levels of LTA4H which may contribute to the protective physiological effect, through reduction in the formation of the inflammatory LTB4. However, this mechanism would lead to the accumulation of PGP and so neutrophillic inflammation, counteracting the situation. LTB4 and PGP are both neutrophil chemoattractants [25,26]. rs1978331 may alter splicing efficiency of LTA4H. rs1978331 is in intron 11 of the gene and exon 10 and 11 of LTA4H contains the zinc-binding domain which is required for both the epoxide hydrolase and aminopeptidase activities [27]. The two functional sites are different but overlapping [28]. Altered splicing in this region could affect the ability of LTA4H to generate LTB4 and/or degrade PGP. Presence of the C-allele may cause splicing events that reduce LTB4 formation, but the aminopeptidase activity may remain functional, which could lead to less neutrophil chemotaxis and so less inflammation. Presence of the T-allele may cause splicing events that lead to increased LTB4 production. The T-allele in HapK was functionally associated with LTB4 overproduction from ionmycin stimulated neutrophils in MI patients [12]. Other factors could complicate this potential mechanism, such as the lung environments in asthma and COPD and the presence/absence of cigarette smoke. This information could have important consequences for the design of any therapeutics inhibiting LTA4H. Reducing LTA4H activity will reduce LTB4 production, but neutrophilic inflammation will persist as PGP will no longer be degraded. A more selective inhibitor strategy would be required to block LTB4 production, but leave the aminopeptidase activity intact. This could take advantage of the different substrate specificities of the non-overlapping regions of the 'active site', small molecules which bind to this hydrophobic part of the site can alter the substrate preference of the aminopeptidase activity [28]. Consideration of both LTB4 and PGP and consideration of the SNPs spanning LTA4H will be required when designing therapeutics.
This is the first study investigating lung function in smokers and genetic variants specific to genes involved with LTB4 production and activity. Overall we have identified that polymorphisms spanning ALOX5AP, LTA4H and LTB4R are not major determinants of lung function in smokers. However, these data highlight the potential importance of LTA4H polymorphisms in particular rs1978331C (LTA4H, intron 11). Although the rs1978331 association did not survive correction for multiple testing, the previous associations with; asthma/lung function [15], MI [12] and TB [22] suggest it may be a true association of modest effect size and this SNPs does influence LTA4H expression and/or activity. While no association survived the Bonferroni correction, additive model analyses with rs1978331C (LTA4H, intron 11) showed a p = 0.029 with an increase in FEV1 and p = 0.020 with FEV1/FVC ratio. The mean FEV1 and FEV1/FVC values for the TC heterozygotes and CC homozygotes were similar, but the presence of the minor C-allele for these genotype groups gave higher trait values when compared to the TT homozygotes, suggesting a dosage effect does not occur. For rs1978331 TT versus TC genotype groups there was a 131 ml difference in FEV1 and for TT versus CC a 126 ml difference in FEV1 was observed. The level of FEV1 at a given time depends on 1) the maximum lung function obtained during development, and 2) the rate of decline of lung function with age. Lung function reaches a maximum by age 25-35 years [29]. In smokers the rate of decline in FEV1 is accelerated and has been calculated to be ~38.2 ml/year in males and 23.9 ml/year in females [29] therefore the differences observed between LTA4H rs1978331 genotypes can be considered clinically relevant and equate to > 3 years decline in FEV1. These findings therefore provide tentative evidence suggesting that variants in LTA4H may determine clinically relevant lung function levels in smokers.
It is important to acknowledge the limitations of our study. Other SNPs spanning these large genes could be important. There may also be another functional variant in linkage disequilibrium with rs1978331. We cannot exclude the contribution of polymorphisms spanning other 5-LO pathway genes e.g. ALOX5, although existing data did not support their inclusion [20,30,31]. The magnitude of effect of SNPs are modest but in line with the predicted relative risk attributed to a single SNP in a single gene in complex disorders. Finally, the number of individuals used in this study was modest and we have not completed extensive replication in multiple cohorts and so caution is required in the interpretation of our novel findings. To our knowledge these SNPs did not show association with lung function and/or COPD in recent GWAS studies. We have also completed a comprehensive look up of genes previously associated with lung function including LTA4H and ALOX5AP in 20,288 individuals from the general population (the SpiroMeta consortium) and did not identify these genes as containing major determinants of lung function in this large general population cohort [32].
Conclusions
In conclusion, these data did not confirm the hypothesis that polymorphisms in genes specific to LTB4 production and activity are major determinants of baseline lung function in smokers and do not determine susceptibility to develop COPD. However, rs1978331 (LTA4H, intron 11) may have a modest effect on lung function parameters in smokers. Heterozygosity of this polymorphism has previously been correlated with LTB4 production, asthma and TB. These findings may be important when considering potential approaches to target LTA4H in COPD.
List of Abbreviations
5-LO: (pathway) 5-lipoxygenase (pathway); 95% CI: 95% confidence interval; ALOX5: 5-lipoxygenase; ALOX5AP: 5-lipoxygenase activating protein; COPD:Chronic obstructive pulmonary disease; CysLT: Cysteinyl leukotriene; CYSLTR1: Cysteinyl leukotriene receptor 1; FEV1: Forced expiratory volume in one second: FEV1/FVC: Ratio of FEV1 to FVC; FLAP: 5-lipoxygenase activating protein; FVC: Forced vital capacity; GOLD: Global Initiative for Obstructive Lung Diseases; LD: Linkage disequilibrium; LTA4, B4, C4, D4, E4: Leukotriene A4, B4, C4, D4, E4; LTA4H: Leukotriene A4 hydrolase; LTB4R1/2: Leukotriene B4 receptor 1/2; LTC4S: Leukotriene C4 synthase; MAF: Minor allele frequency; MI: Myocardial infarction; OR: Odds ratio; PGP Proline-glycine-proline; PostBD: Post bronchodilator; SNP: Single nucleotide polymorphism; TB: Tuberculosis; UK: United Kingdom; UTR: Untranslated region.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
IS and AST designed the study and drafted the manuscript. AST completed the statistical analyses. SGP, MFM, AJW and MJC recruited and clinically characterised subjects. All authors contributed to the final version of the manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Baseline lung function (FEV1 and FEV1/FVC ratio) and ALOX5AP, LTA4H and LTB4R SNPs in the smokers (n = 992). This table shows the results of the association analysis between leukotriene pathway SNPs and baseline FEV1 and FEV1/FVC using the additive model. Covariates included in the model were age, gender, height and pack years. Associations with p < 0.05 are shown in bold black.
Contributor Information
Asif S Tulah, Email: astulah@gmail.com.
Stuart G Parker, Email: S.G.Parker@sheffield.ac.uk.
Miriam F Moffatt, Email: m.moffatt@imperial.ac.uk.
Andrew J Wardlaw, Email: aw24@leicester.ac.uk.
Martin J Connolly, Email: martin.connolly@waitematadhb.govt.nz.
Ian Sayers, Email: ian.sayers@nottingham.ac.uk.
Acknowledgements
We acknowledge the contribution of Dr. Charlotte Ruse to the collection and phenotyping of the COPD cohort. This work was funded by the Medical Research Council UK and the University of Nottingham.
References
- Meyers DA, Larj MJ, Lange L. Genetics of asthma and COPD. Similar results for different phenotypes. Chest. 2004;126(2 Suppl):105S–110S. doi: 10.1378/chest.126.2_suppl_1.105S. discussion 159S-161S. [DOI] [PubMed] [Google Scholar]
- Kleeberger SR, Peden D. Gene-environment interactions in asthma and other respiratory diseases. Annu Rev Med. 2005;56:383–400. doi: 10.1146/annurev.med.56.062904.144908. [DOI] [PubMed] [Google Scholar]
- Kostikas K, Gaga M, Papatheodorou G, Karamanis T, Orphanidou D, Loukides S. Leukotriene B4 in exhaled breath condensate and sputum supernatant in patients with COPD and asthma. Chest. 2005;127(5):1553–1559. doi: 10.1378/chest.127.5.1553. [DOI] [PubMed] [Google Scholar]
- Montuschi P, Kharitonov SA, Ciabattoni G, Barnes PJ. Exhaled leukotrienes and prostaglandins in COPD. Thorax. 2003;58(7):585–588. doi: 10.1136/thorax.58.7.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes PJ. New concepts in chronic obstructive pulmonary disease. Annu Rev Med. 2003;54:113–129. doi: 10.1146/annurev.med.54.101601.152209. [DOI] [PubMed] [Google Scholar]
- Ford-Hutchinson AW, Bray MA, Doig MV, Shipley ME, Smith MJ. Leukotriene B, a potent chemokinetic and aggregating substance released from polymorphonuclear leukocytes. Nature. 1980;286(5770):264–265. doi: 10.1038/286264a0. [DOI] [PubMed] [Google Scholar]
- Sehmi R, Wardlaw AJ, Cromwell O, Kurihara K, Waltmann P, Kay AB. Interleukin-5 selectively enhances the chemotactic response of eosinophils obtained from normal but not eosinophilic subjects. Blood. 1992;79(11):2952–2959. [PubMed] [Google Scholar]
- Woolhouse IS, Bayley DL, Stockley RA. Sputum chemotactic activity in chronic obstructive pulmonary disease: effect of alpha(1)-antitrypsin deficiency and the role of leukotriene B(4) and interleukin 8. Thorax. 2002;57(8):709–714. doi: 10.1136/thorax.57.8.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Calhoun WJ. The role of leukotrienes in airway inflammation. J Allergy Clin Immunol. 2006;118(4):789–798. doi: 10.1016/j.jaci.2006.08.009. quiz 799-800. [DOI] [PubMed] [Google Scholar]
- Duroudier NP, Tulah AS, Sayers I. Leukotriene pathway genetics and pharmacogenetics in allergy. Allergy. 2009;64(6):823–839. doi: 10.1111/j.1398-9995.2009.02015.x. [DOI] [PubMed] [Google Scholar]
- Helgadottir A, Manolescu A, Thorleifsson G, Gretarsdottir S, Jonsdottir H, Thorsteinsdottir U, Samani NJ, Gudmundsson G, Grant SF, Thorgeirsson G. et al. The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat Genet. 2004;36(3):233–239. doi: 10.1038/ng1311. [DOI] [PubMed] [Google Scholar]
- Helgadottir A, Manolescu A, Helgason A, Thorleifsson G, Thorsteinsdottir U, Gudbjartsson DF, Gretarsdottir S, Magnusson KP, Gudmundsson G, Hicks A. et al. A variant of the gene encoding leukotriene A4 hydrolase confers ethnicity-specific risk of myocardial infarction. Nat Genet. 2006;38(1):68–74. doi: 10.1038/ng1692. [DOI] [PubMed] [Google Scholar]
- Devillier P, Baccard N, Advenier C. Leukotrienes, leukotriene receptor antagonists and leukotriene synthesis inhibitors in asthma: an update. Part I: synthesis, receptors and role of leukotrienes in asthma. Pharmacol Res. 1999;40(1):3–13. doi: 10.1006/phrs.1998.0458. [DOI] [PubMed] [Google Scholar]
- Snelgrove RJ, Jackson PL, Hardison MT, Noerager BD, Kinloch A, Gaggar A, Shastry S, Rowe SM, Shim YM, Hussell T. et al. A critical role for LTA4H in limiting chronic pulmonary neutrophilic inflammation. Science. 2010;330(6000):90–94. doi: 10.1126/science.1190594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway JW, Barton SJ, Holgate ST, Rose-Zerilli MJ, Sayers I. The role of LTA4H and ALOX5AP polymorphism in asthma and allergy susceptibility. Allergy. 2008;63(8):1046–1053. doi: 10.1111/j.1398-9995.2008.01667.x. [DOI] [PubMed] [Google Scholar]
- Stewart CE, Hall IP, Parker SG, Moffat MF, Wardlaw AJ, Connolly MJ, Ruse C, Sayers I. PLAUR polymorphisms and lung function in UK smokers. BMC Med Genet. 2009;10:112. doi: 10.1186/1471-2350-10-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli BR, MacNee W. Standards for the diagnosis and treatment of patients with COPD: a summary of the ATS/ERS position paper. Eur Respir J. 2004;23(6):932–946. doi: 10.1183/09031936.04.00014304. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Hao L, Sayers I, Cakebread JA, Barton SJ, Beghe B, Holgate ST, Sampson AP, Holloway JW. The cysteinyl-leukotriene type 1 receptor polymorphism 927T/C is associated with atopy severity but not with asthma. Clin Exp Allergy. 2006;36(6):735–741. doi: 10.1111/j.1365-2222.2006.02511.x. [DOI] [PubMed] [Google Scholar]
- Sayers I, Barton S, Rorke S, Beghe B, Hayward B, Van Eerdewegh P, Keith T, Clough JB, Ye S, Holloway JW. et al. Allelic association and functional studies of promoter polymorphism in the leukotriene C4 synthase gene (LTC4S) in asthma. Thorax. 2003;58(5):417–424. doi: 10.1136/thorax.58.5.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson AP, Siddiqui S, Buchanan D, Howarth PH, Holgate ST, Holloway JW, Sayers I. Variant LTC(4) synthase allele modifies cysteinyl leukotriene synthesis in eosinophils and predicts clinical response to zafirlukast. Thorax. 2000;55(Suppl 2):S28–31. doi: 10.1136/thorax.55.suppl_2.S28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin DM, Vary JC Jr, Ray JP, Walsh GS, Dunstan SJ, Bang ND, Hagge DA, Khadge S, King MC, Hawn TR. et al. The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell. 2010;140(5):717–730. doi: 10.1016/j.cell.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims AM, Damholt A, Knutsson M, Carlholm M, Carlsson LG, Lofdahl CG, Postma DS, Semb KF. The American Thoracic Society. New Orleans; 2010. Genes in the 5-Lipoxygenase (5-LO) pathway: association with COPD. [Google Scholar]
- Keatings VM, Collins PD, Scott DM, Barnes PJ. Differences in interleukin-8 and tumor necrosis factor-alpha in induced sputum from patients with chronic obstructive pulmonary disease or asthma. Am J Respir Crit Care Med. 1996;153(2):530–534. doi: 10.1164/ajrccm.153.2.8564092. [DOI] [PubMed] [Google Scholar]
- Sato E, Koyama S, Takamizawa A, Masubuchi T, Kubo K, Robbins RA, Nagai S, Izumi T. Smoke extract stimulates lung fibroblasts to release neutrophil and monocyte chemotactic activities. Am J Physiol. 1999;277(6 Pt 1):L1149–1157. doi: 10.1152/ajplung.1999.277.6.L1149. [DOI] [PubMed] [Google Scholar]
- Weathington NM, van Houwelingen AH, Noerager BD, Jackson PL, Kraneveld AD, Galin FS, Folkerts G, Nijkamp FP, Blalock JE. A novel peptide CXCR ligand derived from extracellular matrix degradation during airway inflammation. Nat Med. 2006;12(3):317–323. doi: 10.1038/nm1361. [DOI] [PubMed] [Google Scholar]
- Mancini JA, Evans JF. Cloning and characterization of the human leukotriene A4 hydrolase gene. Eur J Biochem. 1995;231(1):65–71. doi: 10.1111/j.1432-1033.1995.tb20671.x. [DOI] [PubMed] [Google Scholar]
- Jiang X, Zhou L, Wu Y, Wei D, Sun C, Jia J, Liu Y, Lai L. Modulating the substrate specificity of LTA4H aminopeptidase by using chemical compounds and small-molecule-guided mutagenesis. Chembiochem. 2010;11(8):1120–1128. doi: 10.1002/cbic.200900788. [DOI] [PubMed] [Google Scholar]
- Kohansal R, Martinez-Camblor P, Agusti A, Buist AS, Mannino DM, Soriano JB. The natural history of chronic airflow obstruction revisited: an analysis of the Framingham offspring cohort. Am J Respir Crit Care Med. 2009;180(1):3–10. doi: 10.1164/rccm.200901-0047OC. [DOI] [PubMed] [Google Scholar]
- Sayers I, Barton S, Rorke S, Sawyer J, Peng Q, Beghe B, Ye S, Keith T, Clough JB, Holloway JW. et al. Promoter polymorphism in the 5-lipoxygenase (ALOX5) and 5-lipoxygenase-activating protein (ALOX5AP) genes and asthma susceptibility in a Caucasian population. Clin Exp Allergy. 2003;33(8):1103–1110. doi: 10.1046/j.1365-2222.2003.01733.x. [DOI] [PubMed] [Google Scholar]
- Sayers I, Sampson AP, Ye S, Holgate ST. Promoter polymorphism influences the effect of dexamethasone on transcriptional activation of the LTC4 synthase gene. Eur J Hum Genet. 2003;11(8):619–622. doi: 10.1038/sj.ejhg.5201015. [DOI] [PubMed] [Google Scholar]
- Obeidat M, Wain LV, Shrine N, Kalsheker N, Artigas MS, Repapi E, Burton PR, Johnson T, Ramasamy A, Zhao JH, A comprehensive evaluation of potential lung function associated genes in the SpiroMeta general population sample. PLoS One. p. e19382. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Baseline lung function (FEV1 and FEV1/FVC ratio) and ALOX5AP, LTA4H and LTB4R SNPs in the smokers (n = 992). This table shows the results of the association analysis between leukotriene pathway SNPs and baseline FEV1 and FEV1/FVC using the additive model. Covariates included in the model were age, gender, height and pack years. Associations with p < 0.05 are shown in bold black.