Abstract
A significant challenge in natural product discovery is the initial discrimination of discrete secondary metabolites alongside functionally similar primary metabolic cellular components within complex biological samples. A property that has yet to be fully exploited for natural product identification and characterization is the gas phase collision cross section, or, more generally, the mobility-mass correlation. Peptide natural products possess many of the properties that distinguish natural products as they are frequently characterized by a high degree of intramolecular bonding, and possess extended and compact conformations among other structural modifications. This report describes a rapid structural mass spectrometry technique based on ion mobility-mass spectrometry for the comparison of peptide natural products to their primary metabolic congeners using mobility-mass correlation. This property is empirically determined using ion mobility-mass spectrometry, applied to the analysis of linear versus modified peptides, and used to discriminate peptide natural products in a crude microbial extract. Complementary computational approaches are utilized to understand the structural basis for the separation of primary metabolism derived linear peptides from secondary metabolite cyclic and modified cyclic species. These findings provide a platform for enhancing the identification of secondary metabolic peptides with distinct mobility-mass ratios within complex biological samples.
The diverse activities of peptide natural products are partially a function of their unique structural attributes, which are defined by incorporation of nonproteinogenic amino acids and by extensive ‘post-translational’ modifications including macrocyclization, heterocyclization, and oxidation/elimination reactions. These modifications are responsible for the diversity of reported cyclic and heterocyclic scaffolds. Additionally, conformational restraints introduced through these modifications have been implicated in the biological activity of these secondary metabolites, as the entropic loss is less significant upon active site binding compared to non-constrained analogues.1 Some microbial peptide secondary metabolites with medical relevance include cyclosporin (immunosuppressive), bialaphos (herbicide), vancomycin and penicillin (antibiotic), to name a few, and their activities have inspired the continuing search for new peptide natural products from microbial sources. This search has been reinvigorated in recent years by the discovery of a large reservoir of cryptic peptide gene clusters, both ribosomally encoded and nonribosomally encoded, in microbial genomes, potentially expressing new peptide natural products.2, 3 Correspondingly, the conditions under which these gene clusters are expressed as well as the identity of their products are a matter of significant interest to the natural product biosynthesis and discovery communities, respectively. However, these discovery endeavors are challenged by low secondary metabolite abundance and the peptide-rich background of biological extracts from which they must be isolated. In this report we describe a structural mass spectrometry technique, ion mobility-mass spectrometry (IM-MS), to rapidly distinguish peptide natural products from other species present in complex mixtures. This method of discrimination is then applied to a crude extract for natural product candidate prioritization. Additionally, a simulated annealing computational approach provides a model to understand the structural differences characteristic of linear and modified peptides in the gas phase.
Briefly, IM-MS is a two-dimensional separation technique that first separates ions in a dimension related to structure (charge-to-surface area ratio) based on their collision cross section (CCS), which represents the area of the ion available for collisions with neutral molecules in the gas phase, and in a second dimension by mass-to-charge. Many forms of ion mobility exist: high-field asymmetric waveform ion mobility (FAIMS), differential mobility, traveling wave ion mobility (TWIM) and uniform field ion mobility (IM). However, for CCS measurements uniform field ion mobility is the only of these methods that presently allows for calculation of absolute CCS using the kinetic theory of gases.4 For this reason, we choose to focus on this instrument as the foundation of this study.
Importantly, by using IM-MS, biomolecular classes can be separated in a complex mixture by their structure due to their different gas phase packing efficiencies dictated by the prevailing intramolecular folding forces5–7 The average correlation for each biomolecular class is referred to as a mobility-mass correlation, or trendline, and significant deviation, i.e., > 7% in CCS for linear peptides (as it has been shown that ~94% of linear peptides fall within this range), from this correlation is observed for modifications and conserved secondary structure in the gas phase.6, 7 A previous study found that the cyclic peptide gramicidin S (cyclo-VOLFPVOLFP) adopted a more compact structure than linear analogs.8, 9 To understand the full potential for IM-MS to distinguish cyclic from linear peptides in complex samples, we have evaluated the general utility of structural separations for a suite of microbially produced peptide natural products in comparison to linear peptide congeners. Moreover, we have applied this method to the detection of the known tricyclic peptide siamycin II, expressed in an extract from a recently isolated cave actinomycete, demonstrating the potential of IM-MS for distinguishing modified peptides in crude extracts.
RESULTS AND DISCUSSION
Measurements of Collision Cross Sections of Peptide Secondary Metabolites
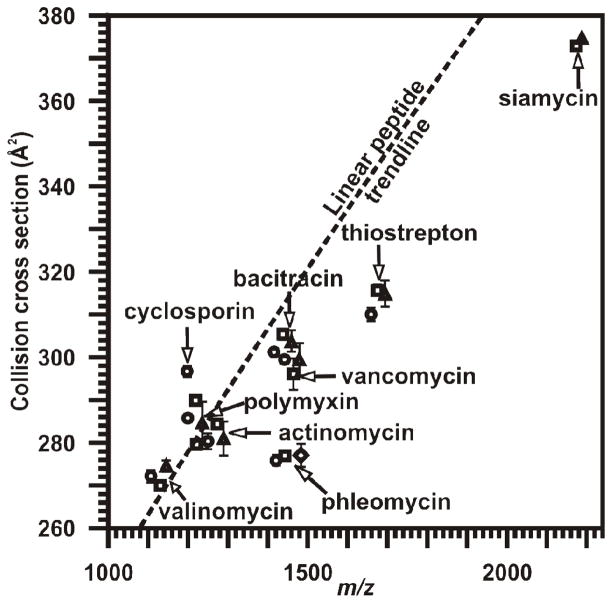
Collision cross sections for a series of peptide natural products including thiostrepton, vancomycin, ampicillin, valinomycin, phleomycin, cyclosporin A, polymyxin B, actinomycin D, bacitracin, and siamycin, were determined and compared to the values for a large suite of collision cross sections for linear peptides. Peptide CCS measurements are reported in Table 1. The peptide natural products were prepared and analyzed using a MALDI-IM-MS instrument which was described previously,10 and further information about the sample preparation and collision cross section calculations can be found in the Experimental Section below. The collision cross sections for all of the quasimolecular ions present (i.e., [M+H]+, [M+Na]+, [M+K]+, and [M+Cu]+) in the IM-MS plot were compared to the mobility-mass correlation for the relevant 280 of 607 reported linear peptides which occur within the m/z range reported (see Table 1, Figure 1).7 A power-fit line of regression was used to best describe the linear peptide data over this region (y=2.8269x0.6475 R2=0.8926). Notably, 76% of peptide natural product signals fall below the peptide trendline consistent with peptide natural product’s compact and modified structures.
Table 1. Peptide Natural Products Analysed in the Present Studies.
Quasi-molecular ion types are shown with their associated CCS values. Ion deviation from expected linear peptide conformation is depicted in Figure 1. Number of measurements for each CCS is shown in parentheses.
| Peptide natural product | Ion Adduct | M/Z (Da) | Collision cross section (Å2) |
|---|---|---|---|
| ampicillin | [M+Na]+ | 372.1 | 110.1±1.3(20) |
| valinomycin | [M+H]+ | 1111.6 | 271.9±1.4(19) |
| valinomycin | [M+Na]+ | 1133.6 | 269.7±0.8(19) |
| valinomycin | [M+K]+ | 1149.6 | 274.5±0.6(19) |
| cyclosporine | [M+H]+ | 1202.8 | 296.6±1.2(19) |
| polymyxin | [M+H]+ | 1203.8 | 285.6±1.0(19) |
| cyclosporine | [M+Na]+ | 1224.8 | 289.9±1.2(19) |
| polymyxin | [M+Na]+ | 1225.7 | 279.4±0.9(19) |
| polymyxin | [M+K]+ | 1241.7 | 284.4±4.9(19) |
| actinomycin | [M+H]+ | 1255.6 | 280.1±1.9(18) |
| actinomycin | [M+Na]+ | 1277.6 | 284.2±0.7(19) |
| actinomycin | [M+K]+ | 1293.6 | 280.8±4.1(17) |
| bacitracin | [M+H]+ | 1422.8 | 301.2±0.8(19) |
| phleomycin | [M+H]+ | 1427.7 | 275.7±1.3(13) |
| bacitracin | [M+Na]+ | 1444.7 | 305.3±1.0(19) |
| phleomycin | [M+Na]+ | 1448.4 | 276.7±1.2(19) |
| vancomycin | [M+H]+ | 1448.4 | 299.4±0.9(20) |
| bacitracin | [M+K]+ | 1460.7 | 303.6±2.3(10) |
| vancomycin | [M+Na]+ | 1470.4 | 296.0±3.7(20) |
| vancomycin | [M+K]+ | 1486.4 | 299.4±3.8(15) |
| phleomycin | [M+Cu]+ | 1489.5 | 276.8±2.7(19) |
| thiostrepton | [M+H]+ | 1664.5 | 310.0±1.6(20) |
| thiostrepton | [M+Na]+ | 1686.5 | 315.5±1.0(20) |
| thiostrepton | [M+K]+ | 1702.5 | 314.8±3.0(10) |
| siamycin I | [M+Na]+ | 2185.8 | 373.2±0.8(15) |
| siamycin I | [M+K]+ | 2201.8 | 375.2±1.7(14) |
Figure 1.
Conformational space plot depicting the relative increased gas-phase density of cyclic peptides when compared to linear peptides. IM-MS plot comparison of the collision cross sections of peptide natural products compared to a trendline best representative of linear peptides for the mass range of 1100–2300 Da (y=2.8269x0.6475 R2=0.8926). The collision cross section values and associated error of measurements can be found in Table 1. Symbols are as follows: ●-[M+H]+; ■-[M+Na]+; ▲-[M+K]+; and ◆-[M+Cu]+.
Computational Modeling of Peptide Natural Products in the Gas Phase
A molecular dynamics model was developed to interpret the theoretical basis of these results. For conformational analysis, a protocol analogous to simulated annealing was used in which snapshots of conformations were taken periodically during the heating procedure and these conformations were subsequently cooled to allow for energy minimization, generating 24,000 unique low-energy conformations. This method was performed for representative peptide natural products based upon their chemical and structural diversity, as illustrated in Figure 2.11 Further details regarding the modeling protocol can be found in the Experimental Section. Valinomycin is a macrocyclic depsipeptide comprised of a mixture of D- and L-Val. Vancomycin is a highly cross-linked tricyclic peptide compacted and rigidified by aromatic C-C and C-O (ether) cross-links. Cyclosporin A is a nonribosomally synthesized macrocyclic molecule containing D-Ala along with a butenyl-N-methyl-L-threonine residue. Finally, polymyxin B is a lariat macrolactam with a hydrophobic tail and hydrophilic macrocyclic core. Shown in Figure 2 are the most representative conformations describing the data discriminated using experimentally derived CCS values, which were extracted from the aforementioned 24,000 unique structures generated for each modeled structure. For structural analyses, larger sets (ca. 18–22 representative conformations) were generated using clustering analyses and interrogated to determine prevalent structural motifs (see Supporting Information).
Figure 2.
Representative structures for peptide natural products. From left to right: sodiated cyclosporin A (24000, 807), valinomycin (24000, 302), polymyxin B (24000, 891), and vancomycin (24000, 2010) conformations which are most representative (top row) of the extracted conformers discriminated based upon experimentally derived CCS values. The number of unique structures initially calculated and the number that correspond with the IM-MS empirical collision cross section are shown in parenthesis, respectively. The percent deviation of each calculated CCS value from predicted CCS values for isobaric linear peptides are 2.69%±0.43%, 0.43%±0.30%, −1.08%±0.32%, −6.88%±1.16%, respectively. Sodium coordination distances are labeled. Carbon atoms are shown in grey, hydrogen white, nitrogen blue, oxygen red, chlorine green, and sodium yellow. Shown below are the associated chemical structures.
These molecular dynamics calculations provide a model for rationalizing conformational differences for cyclic versus linear peptides. The primary features that influence gas phase conformational density are: (1) the loss of degrees of freedom due to macrocyclization, (2) the presence of highly coordinated metal atoms, and (3) the presence of atoms with intrinsically higher density, such as halides.12 While it is expected that torsional freedom is significantly reduced for cyclic peptides compared to linear peptides, this effect was visualized in these models. Notably, the degree of cyclization and the number of residues per ring has a large influence on trendline deviation. For example, valinomycin (a 12 residue macrocycle) and cyclosporin (an 11 residue macrocycle), each adopt CCS values greater than that predicted by the linear peptide trendline, while polymyxin B (a lariat compound with seven residues present in the macrocycle) falls below the trendline. However, vancomycin (a modified tricyclic peptide with three - four amino acids per ring) falls significantly below the linear peptide trendline. Modeling indicates extensive coordination of sodium within the ring of cyclosporin and valinomycin, likely resulting in the less dense conformations, as this extends the ring. This is supported by the increased collision cross section of valinomycin when coordinated to potassium, which likely results in an extended ring conformation, as reported in the literature (as valinomycin is a potassium specific ionophore).13 Sodium coordination distances and proximities in peptide natural products are consistent with those from quantum mechanical calculations previously reported for gas phase linear peptides.14, 15 The observed high degree of coordination to sodium appears to be facilitated by the more constrained orientation of carbonyls, esters, ethers, amines, and other functional groups (e.g., chlorine), but should not be considered an exclusive product of cyclization. It should be noted that post-translational modifications and the addition of atoms such as chlorine increase the density of the compounds, resulting in further deviation from the linear peptide trendline. Therefore, although cyclization provides increased density, glycosylation and halide inclusion can further enhance the deviation from linear peptides in ion mobility based separations.
In individual cases, these models aid in rationalizing deviations. For example, the relatively low collision cross section deviation of polymyxin B is attributed to coordinating sodium by both the ring and tail portions of the lariat peptide. The small number of residues present in the ring allows for increased density due to this constraint as well. The extended butenyl-methyl-L-threonine group of cyclosporin A likely expands its conformation relative to other peptides, while the large number of residues present in the ring structure preclude adopting a dense gas phase conformation. Vancomycin is an example of a molecule possessing both deoxysugars and halides present in addition to three small macrocyclic structures, factors that apparently contribute to the higher gas-phase packing efficiency.
Discriminating Linear Peptides from Peptide Secondary Metabolites
Intuitively, the apparent decreased mobility-mass correlation of modified and structurally constrained peptides relative to linear peptides can potentially aid in discriminating peptide natural products from linear peptide ions. As previously noted, a line of regression best describing the mobility-mass correlation for a large number of linear peptides was generated. This creates a baseline of comparison, in which we explored the utility of analyzing percent deviation from this line of regression as a discriminator for peptide natural products, termed the threshold. Figure 3 displays the percentage of peptide natural products (white fill) versus unmodified linear peptides (black fill) that fall beyond the applied threshold (i.e., have a CCS value smaller than the applied threshold). Using this threshold analysis, the enrichment of modified peptides is evident. For instance, 80% of peptide natural products possess CCS-mass ratios beneath a −3% deviation, whereas only 20% of linear peptides are observed. Using a more stringent analysis, beneath a 6.0% deviation threshold, 95% of linear peptides are excluded from analysis, while 40% of the peptide natural products in the dataset remain. Beyond a −8% deviation threshold, ~99% of linear peptides are removed from analysis, at the expense of excluding about two-thirds of peptide natural products. Therefore, while not an absolute discriminator of cyclic from linear peptides, we propose that the deviation of observed mobility-mass measurements from the linear trendline is a useful metric for enhancing the identification of peptide natural products in extracts containing competitive background levels of linear peptide signals.
Figure 3.
Percentage of analyzed linear (black fill) and secondary metabolic (white fill) peptides that deviate from the linear peptide mobility-mass correlation greater than an applied threshold. As a function of percent deviation from the linear peptide trendline, the percentages of analyzed species that occupy a more dense CCS value are indicated. Notably, with a threshold of −6.0%, 95% of linear peptides are excluded from analysis, while 40% of peptide natural products have more dense CCS values.
Secondary Metabolic Peptide Discrimination in an Extract
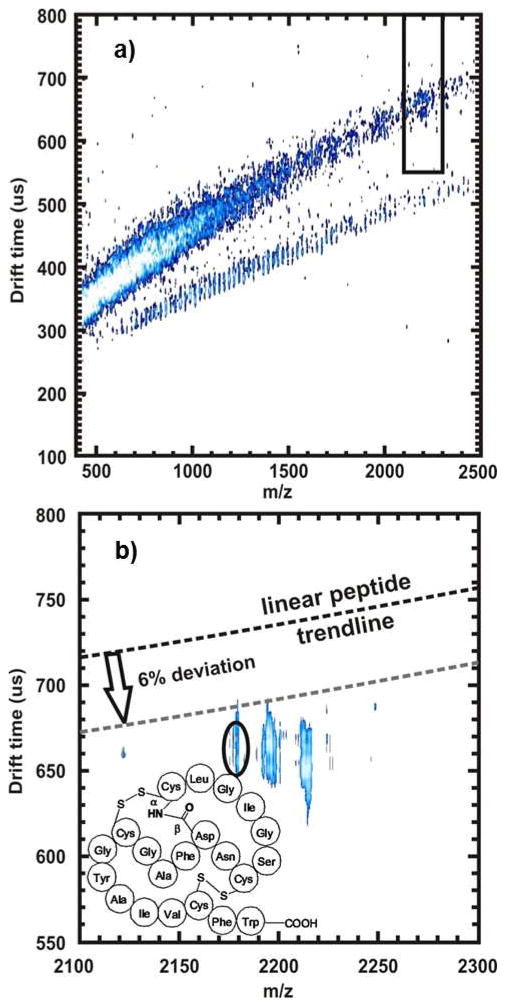
To demonstrate the potential of IM-MS to discriminate peptide natural products within a crude microbial extract, we analyzed an extract derived from actinomycete “BBBLUE19”, an organism recently isolated from a hypogean ecosystem (Blue Springs Cave, Sparta, TN). This organism was fermented and extracted under conditions typical for natural product discovery and a total hydrophobic extract of BBBLUE19 was analyzed using the aforementioned MALDI-IM-MS methodology. Correspondingly, the linear peptide mobility-mass correlation was superimposed on the IM-MS snapshot of the extract (Figure 4a). A threshold of −6.0% was applied to enrich for potential peptide natural products, and peaks were prioritized upon this basis. A series of high abundance peaks with significant trendline deviation were thus selected for further identification, one of which (accurate mass = 2175.88, CCS value 360.8±4.5Å2) possessed > 11% deviation (predicted linear peptide CCS at this mass = 409.7Å2, Figure 4b), which corresponds to a <1% probability of being a linear peptide. The observed mass was most consistent with siamycin II (within 5 ppm mass error), previously isolated from Streptomyces sp. AA3891.16–18 Subsequently, the isolated compound was dereplicatively identified and validated as Siamycin II by comparison to the reported 1H NMR spectrum. As a result, the tricyclic peptide natural product siamycin II was successfully prioritized from a crude extract using the unique CCS value adopted in the gas phase.
Figure 4.
Shown are the analyses of the BBBLUE19 Streptomycete extract using IM-MS methodologies for the determination of peptide natural product presence. (a) 2-dimensional MALDI-IM-MS spectrum of the extract using drift tube mobility separations. The red-boxed in region is expanded in (b). In (b), the red-circled region annotates a peak that, upon initial inspection, has dense gas phase packing efficiency. Collision cross section measurements were performed, resulting in a CCS value of 360.8 ± 4.5 Å2, a deviation of 11.9% below the linear peptide trendline for this m/z (predicted CCS value of 409.7Å2). The sample was then analyzed using ESI-IM-high resolution MS (not shown). This compound was identified to be siamycin II, a tricyclic peptide, using high mass accuracy measurements obtained from an IM-high resolution MS and confirmed by isolation and NMR analyses. The structure is shown as an inset in (b).
Conclusions and Perspectives
Uniform field IM-MS instruments as described herein remain an uncommon instrument in drug discovery laboratories. Commercially available IM-MS instruments typically use an alternative mobility separation paradigm termed a traveling wave, which is distinct from uniform field instruments and not amenable to absolute CCS determination using the kinetic theory of gases.4 As such, the utility of using the mobility-mass correlation as a means for discrimination of natural products using the commercially available traveling wave IM-MS are currently being explored. The primary difficulty in comparing the two separations is the inability to extract CCS values from first principles using traveling wave IM separations, as the electric field gradient experienced by ions is time dependent, although CCS calibrants can be used to obtain relative CCS measurements.19, 20 Ongoing studies in our laboratory seek to describe the degree to which these deflections are observed in traveling wave based instrumentation. We envision the mobility differences resulting from oxidation, cyclization and macrocyclization to be consistent with those reported here for uniform field IM-MS.
EXPERIMENTAL SECTION
MALDI Sample Preparation and Analysis
The modified cyclic peptides, thiostrepton, vancomycin, ampicillin, valinomycin, phleomycin, cyclosporine A, polymyxin B, actinomycin D, and bacitracin, were obtained from Sigma and used without further purification. The extract was reconstituted in 50:50 MeOH:H2O at a concentration of ~5 mg/mL. To determine collision cross sections, MALDI ionization was performed by mixing analytes in a 200:1 molar ratio of saturated 2,5-dihydroxybenzoic acid in 20% MeOH or 25 mg/mL α-cyano-4-hydroxycinnamic acid in 50% MeOH with analyte. Samples were spotted onto a stainless steel plate, and allowed to dry before IM-MS analysis. The MALDI-IM-TOFMS is equipped with a 13.9 cm ion mobility drift cell that is maintained at a pressure of ca. 3 Torr helium and an orthogonal reflectron TOFMS with a 1 m flight path maintained at 5 × 10−8 Torr. Further instrumental details have been published previously.10
Collision Cross Section Calculations
The ion-neutral collision cross section (CCS, Ω) was calculated by determining the drift time (td) of the ions across the cell under low-field conditions. Drift time values were obtained at multiple field strengths of between 69 and 91 Townsend (Td), to ensure that measurements were performed under the low-field limit. Collision cross sections were calculated by the following:
where L is the length of the drift cell, E is the electrostatic-field strength (~90–120 V cm−1), z is the charge state of the ion, e is elementary charge, mi and mn are the masses of the ion and the neutral drift gas, respectively, N0 is the gas number density at standard temperature and pressure, kB is the Boltzmann constant, and p and T are the pressure (~3 Torr) and temperature (~293 K), respectively.4
Computational Modeling Charge Derivation Protocol
All molecules were first built in ChemBioDraw Ultra. Molecules were broken into fragments for charge derivation of feasible size for quantum based energy minimization and charge calculations (i.e. ~120 atoms or less). An initial energy minimization was performed using a B3LYP/HF 6-31G* level of quantum theory in Gaussian03.21 After determining the calculation had converged upon a local minimum, the frequency values were interrogated to ensure the calculation had not rested upon a saddle point. These energy minimized structures were then subjected to further ab initio optimization and electrostatic potential calculations using Gaussian03, mimicking the protocol derived for the general Amber force field, applying a HF/6-31G* basis set and the Merz-Singh-Kollman scheme for charge parameterization.
Computational Modeling Molecular Dynamics Protocol
All molecular dynamics simulations were generated using the Amber 11 Molecular Dynamics Package.22 Charge densities were converted to restrained electrostatic potential (RESP) values and prep files using antechamber.
Intact molecules were assembled and coordinated with sodium in LeAP using prep files. GAFF 23(general AMBER force field) was used for atom type descriptions. To define the sodium cation, AMBER99SB force field was used.24, 25 Topology and coordinate files were generated for molecular dynamic simulations in sander. Initially, restraint files were generated for the sodium cation such that full exploration of the assembled molecule was possible, but if the cation began to leave the molecule, a force would be applied to return the sodium within reasonable distance constraints of sodium-peptide interaction. Upon inspection of trajectory files, this restraint was not necessary, as the cation remained proximal to the molecule, yet was still able to fully explore all regions of the molecule. Prior to simulation, an energy minimization was performed using sander to ensure a stable starting structure. The simulated annealing protocol was carried out in three steps. First, the structure was heated to ~1200–1500K over 10 picoseconds using 0.25 femtosecond time steps. The structure was then held at this temperature for 9 nanoseconds using 0.25 femtosecond time steps, extracting a conformation every 3 picoseconds, resulting in 3000 conformational snapshots. These snapshots were cooled to ~320K over 15 picoseconds to allow the conformation to find a local minima. To generate the desired 24,000 conformations, this protocol was performed 8 times, using unique starting conformations for each to promote enhanced sampling of conformational space. Each conformation was then converted to a collision cross section using MOBCAL.26, 27 The collision cross section and potential energy for each conformation was then extracted and visualized using a scatter plot. Data were then discriminated using the measured collision cross sections, with the sampling window ± 2 sigma of the IM CCS measurement.
Computational Modeling Clustering Protocol
The extracted conformations were then aligned using superpositioning software, suppose, written by Jarrod Smith (Vanderbilt Center for Structural Biology). These structures were then clustered based upon their root mean square distance (RMSD) over all atoms. A cutoff was then determined based upon the RMSD which resulted in approximately 19–21 conformations. This value was then imposed, and the most representative conformation from each branch of the cluster was extracted. This in effect distilled the data from ~400–2000 conformations to 20 which could be interrogated (see Supporting Information). PDB files for each were generated and interrogated using Molecular Operating Environment (MOE).28
Fermentation and Extraction Conditions
BBBLUE19 was cultivated in a 1-L Erlenmeyer flask containing 500 mL of medium EA (5% lactose, 0.5% corn steep solids, 0.5% glucose, 1.5% glycerol, 1% soybean flour, 0.5% bacto-peptone, 0.3% CaCO3, 0.2% (NH4)2 SO4, 0.01% FeSO4 3 7H2O, 0.01% ZnCl2, 0.01% MnCl2, 0.05% MgSO4 3 7H2O) for 7 days at 30 °C on rotary shaker 170 rpm. The 25 g of Diaion HP-20 resin was added to the 500 mL culture and resin/culture mixture was agitated on a rotary shaker at 170 rpm for 1 h. The mixture was separated by centrifugation and resin/cells layer was extracted separately with 300 mL of MeOH and 300 mL of acetone. Combined organic fractions were evaporated to dryness to produce the extract. The closest relative by 16S RNA sequencing was Streptomyces mirabilis NBRC 13450 (99.9 % identity). The 16S RNA sequence for BBBLUE19 can be found in GeneBank with the accession number: BankIt1497245 BBBLUE19 JQ085995.
Supplementary Material
Acknowledgments
We thank the Vanderbilt Center for Structural Biology for computational support. This work was funded by the National Institute of Health (1R01GM092218-01 and RC2DA028981), the U.S. Defense Threat Reduction Agency (HDTRA-09-1-0013), and the Vanderbilt University College of Arts and Sciences, the Vanderbilt Institute of Chemical Biology, and the Vanderbilt Institute for Integrative Biosystems Research and Education. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Structures, clustering trees and representative structures from computational studies are available in Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kessler H. Angew Chem, Int Ed Engl. 1982;21:512–523. [Google Scholar]
- 2.Wilkinson B, Micklefield J. Nat Chem Biol. 2007;3:379–386. doi: 10.1038/nchembio.2007.7. [DOI] [PubMed] [Google Scholar]
- 3.Challis GL. J Med Chem. 2008;51:2618–2628. doi: 10.1021/jm700948z. [DOI] [PubMed] [Google Scholar]
- 4.Mason EA, McDaniel EW. John Wiley & Sons, Inc. New York: 1988. [Google Scholar]
- 5.Fenn L, McLean J. Anal Bioanal Chem. 2008;391:905–909. doi: 10.1007/s00216-008-1951-x. [DOI] [PubMed] [Google Scholar]
- 6.Fenn LS, Kliman M, Mahsut A, Zhao SR, McLean JA. Anal Bioanal Chem. 2009;394:235–244. doi: 10.1007/s00216-009-2666-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao L, McLean JR, McLean JA, Russell DH. J Am Soc Mass Spectrom. 2007;18:1232–1238. doi: 10.1016/j.jasms.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 8.McLean JA, Ruotolo BT, Gillig KJ, Russell DH. Int J Mass Spectrom. 2005;240:301–315. [Google Scholar]
- 9.Ruotolo BT, Tate CC, Russell DH. J Am Soc Mass Spectrom. 2004;15:870–878. doi: 10.1016/j.jasms.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Sundarapandian S, May JC, McLean JA. Anal Chem. 2010;82:3247–3254. doi: 10.1021/ac902980r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyttenbach T, vonHelden G, Bowers MT. J Am Chem Soc. 1996;118:8355–8364. [Google Scholar]
- 12.Valentine SJ, Counterman AE, Hoaglund-Hyzer CS, Clemmer DE. J Phys Chem B. 1999;103:1203–1207. [Google Scholar]
- 13.Forester TR, Smith W, Clarke JHR. J Phys Chem. 1994;98:9422–9430. [Google Scholar]
- 14.Heaton AL, Moision RM, Armentrout PB. J Phys Chem A. 2008;112:3319–3327. doi: 10.1021/jp711649g. [DOI] [PubMed] [Google Scholar]
- 15.Cerda BA, Hoyau S, Ohanessian G, Wesdemiotis C. J Am Chem Soc. 1998;120:2437–2448. [Google Scholar]
- 16.Frechet D, Guitton JD, Herman F, Faucher D, Helynck G, Monegier du Sorbier B, Ridoux JP, James-Surcouf E, Vuilhorgne M. Biochemistry. 1994;33:42–50. doi: 10.1021/bi00167a006. [DOI] [PubMed] [Google Scholar]
- 17.Constantine KL, Friedrichs MS, Detlefsen D, Nishio M, Tsunakawa M, Furumai T, Ohkuma H, Oki T, Hill S, Bruccoleri RE, Lin PF, Mueller L. J Biomol NMR. 1995;5:271–286. doi: 10.1007/BF00211754. [DOI] [PubMed] [Google Scholar]
- 18.Detlefsen DJ, Hill SE, Volk KJ, Klohr SE, Tsunakawa M, Furumai T, Lin PF, Nishio M, Kawano K, Oki T, Lee MS. J Antibiot. 1995;48 doi: 10.7164/antibiotics.48.1515. [DOI] [PubMed] [Google Scholar]
- 19.Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH. Int J Mass Spectrom. 2007;261:1–12. [Google Scholar]
- 20.Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Anal Chem. 2010;82:9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- 21.Frisch M, Trucks G, Schlegel H, Scuseria G, Robb M, Cheeseman J, Montgomery JJ, Vreven T, Kudin K, Burant J, Millam J, Iyengar S, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson G, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox J, Hratchian H, Cross J, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann R, Yazyev O, Austin A, Cammi R, Pomelli C, Ochterski J, Ayala P, Morokuma K, Voth G, Salvador P, Dannenberg J, Zakrzewski V, Dapprich S, Daniels A, Strain M, Farkas O, Malick D, Rabuck A, Raghavachari K, Foresman J, Ortiz J, Cui Q, Baboul A, Clifford S, Cioslowski J, Stefanov B, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin R, Fox D, Keith T, Al-Laham M, Peng C, Nanayakkara A, Challacombe M, Gill P, Johnson B, Chen W, Wong M, Gonzalez C, Pople J. Wallingford CT. 2004 [Google Scholar]
- 22.Case DA, Darden TA, Cheatham ITE, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvai I, Wong KF, Paesani F, Vanicek J, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. 2010 [Google Scholar]
- 23.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 24.Hornak V, Okur A, Rizzo RC, Simmerling C. 2006;103:915–920. doi: 10.1073/pnas.0508452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Proteins: Struct, Funct, Bioinf. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. J Phys Chem. 1996;100:16082–16086. [Google Scholar]
- 27.Shvartsburg AA, Jarrold MF. Chem Phys Lett. 1996;261:86–91. [Google Scholar]
- 28.Chemical Computing Group. Amber, version 10. 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.