Abstract
Laing distal myopathy (MPD1) is a genetically dominant myopathy characterized by early and selective weakness of the distal muscles. Mutations in the MYH7 gene encoding for the β-myosin heavy chain are the underlying genetic cause of MPD1. However, their pathogenic mechanisms are currently unknown. Here we measure the biological effects of the R1500P and L1706P MPD1 mutations in different cellular systems. We show that, while the two mutations inhibit myosin self-assembly in non-muscle cells, they do not prevent incorporation of the mutant myosin into sarcomeres. Nevertheless, we find that the L1706P mutation affects proper anti-parallel myosin association by accumulating in the bare zone of the sarcomere. Furthermore, bimolecular fluorescence complementation assay (BiFC) shows that the alpha-helix containing the R1500P mutation folds into homodimeric (mutant/mutant) and heterodimeric (mutant/WT) myosin molecules that are competent for sarcomere incorporation. Both mutations also form aggregates consisting of cytoplasmic vacuoles surrounding paracrystalline arrays, and amorphous rod-like inclusions that sequester WT myosin. Myosin aggregates were also detected in transgenic nematodes expressing the R1500P mutation. By showing that the two MPD1 mutations can have dominant effects on distinct components of the contractile apparatus, our data provide the first insights into the pathogenesis of the disease.
Keywords: Laing, Myopathy, Myosin, Aggregation, BiFC
Introduction
Laing distal myopathy (MPD1) is an autosomal dominant disease characterized by initial weakness of the lower leg anterior compartment that affects ankle and great toe dorsiflexion.1,2 Although distal weakness is often noted in the first five years of life and with time progresses to the proximal muscles without impacting life expectancy, the onset of disease can range from birth to adulthood.3 Histological findings are often variable and include a change in muscle fiber size with type I hypotrophy, co-expression of slow and fast myosins, core/minicore structures, mitochondrial abnormalities, and mild necrosis and regeneration.3,4 Recently, a study on a large cluster of MPD1 patients has shown that the severity of the disease has also wide variance: patients can be asymptomatic, mildly disabled or wheelchair-confined.3
All the mutations associated with MPD1 have been mapped to the MYH7 gene that encodes the muscle motor β-myosin heavy chain (MyHC), the primary myosin isoform expressed in the human heart and in type I skeletal muscle fibers. Like other members of the sarcomeric MyHC family, β-myosin is a hexameric molecule consisting of a pair of heavy chains and two pairs of non-identical light chains (MLC). Whereas dimerization of two heavy chains into a parallel coiled-coil rod promotes myosin assembly into thick filaments, interaction between actin filaments and the two N-terminal motor domains of the molecule generates force and movement.5 Ten MPD1 mutations occur in the light meromyosin (LMM) domain corresponding to the C-terminal third of the myosin rod;6-9 two mutations showing MPD1-like clinical and biopsy findings were also identified in the myosin motor domain.10,11 Surprisingly, despite the high level of β-myosin expression in the heart, only a minority of patients with MPD1 rod mutations has detectable signs of cardiomyopathy.3,8,10-12
The mutations in the rod mainly consist of codon deletions or missense mutations that introduce proline residues,6-9 but more recently, charge reversals (Glu→Lys) have also been identified.8 The first two genetic defects are predicted to negatively impact the proper folding and assembly of myosin coiled-coil structure. Amino acid deletions potentially affect folding and stability of coiled-coils by changing the configuration of apolar and charged residues along of the heptad repeat, the well defined 7-residue periodic pattern (abcdefg)n that characterizes the typical coiled-coil structure.13 The presence of proline residues in the middle of a coiled-coil generally introduces a kink of the α-helix axis that results in structural deformations.14
We have recently begun the biochemical characterization of several rod mutations associated with cardiomyopathy as well as distal myopathies and demonstrated that the MPD1 amino acid substitution R1500P alters the thermodynamic stability and filament forming properties of LMM in vitro.15 In an effort to broaden our insights into the mechanisms of MPD1 pathogenesis, we have expressed myosin proteins bearing the R1500P and L1706P missense mutations6 in cells and nematode-based systems, and evaluated their molecular effects on myosin assembly and muscle function. For phenotypic comparison, we have also analyzed the R1500W mutation that causes dilated cardiomyophaty (DCM) without distal myopathy.16 The data presented here indicate that muscle activity could be impaired by the incorporation of homo and heterodimeric mutant myosin molecules into the sarcomere, and by their mislocalization in the bare zone of the thick filament. Additionally, the detection of mutant cytoplasmic aggregates suggests a potential toxic effect on muscle cell physiology.
Results
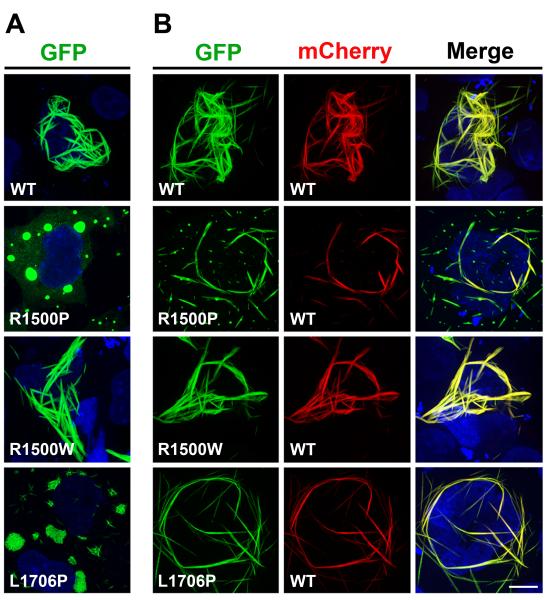
We and others have previously shown that sarcomeric MyHC expressed in non-muscle cells can self-assemble into spindle-shaped periodic structures consisting of bundles of myosin filaments.17,18 We have also shown that MyHC mutations linked to familial hypertrophic cardiomyopathy (HCM) can interfere with the formation of these spindles.19 Since the absence of other sarcomeric proteins in non-muscle cells allows us to ascribe mutation phenotypes directly to MyHC assembly properties, we first analyzed the effects of MPD1 MyHC rod mutations in COS-7 cells. We expressed N-terminally tagged GFP or mCherry wild type (WT) and mutant MyHC rods, which do not require specific muscle chaperones but can fold spontaneously.20 After transient transfection, the morphology and cytoplasmic distribution of the fluorescently-tagged rods were examined by confocal microscopy. While the WT rod forms long filaments distributed through the cytoplasm (Figure 1A, WT), the mutant R1500P rod, carrying the mutation in the f position of the heptad repeat, fails to form organized assemblies, and localizes in discrete foci where it forms dense aggregates (Figure 1A, R1500P). This is a new cellular phenotype not previously observed in truncated rod mutants showing instead a diffuse pattern of distribution.21
Figure 1.
The R1500P and L1706P mutations alter the formation of myosin ordered structures in COS-7 cells. COS-7 cells were transfected with GFP-tagged myosin rod constructs as indicated. 12 h later, cells were imaged by confocal microscopy. While micrographs of cells transfected with GFP-tagged constructs alone (WT, R1500P, R1500W, L1706) are shown in Panel A, co-transfections with mCherry-tagged WT myosin rod are shown in Panel B. Bar, 10 μm.
The R1500W mutation that causes only dilated cardiomyopathy (DCM) and shows distinct thermodynamic and structural properties compared to the R1500P mutation,15 does not appreciably affect myosin filament formation (Figure 1A, R1500W). The other MPD1 proline mutation (L1706P), located in the a position of the heptad repeat, also forms foci as observed for the R1500P mutant. However, these myosin aggregates contain small filaments not detected with the R1500P mutation (Figure 1A, L1706P). These two proline substitutions analyzed are dominant heterozygous mutations; therefore, the mutated myosin interferes, directly or indirectly, with the function of the WT allele. To dissect these molecular events, we co-expressed the mCherry-tagged WT myosin with the GFP mutant constructs, and asked whether the WT would co-aggregate with the mutant or improve its phenotype. Virtually all the cells transfected with the WT constructs show good co-localization of the two tagged myosins in the cytoplasmic assemblies (Figure 1B, WT, Merge panels). Although aggregates containing both tagged proteins were detected in the majority of cells co-expressing GFP-R1500P and mCherry-WT, filamentous-like structures consisting of mutant and WT myosins were also present in a small percentage of cells (Figure 1B, R1500P, Merge panels). While the R1500W DCM mutation does not impair co-assembly of the mutant with the WT myosin, (Figure 1B, R1500W, WT, Merge panels), the presence of the mCherry-WT myosin substantially improves the cellular phenotype linked to the other MPD1 mutation (Figure 1B, L1706P, Merge panels).
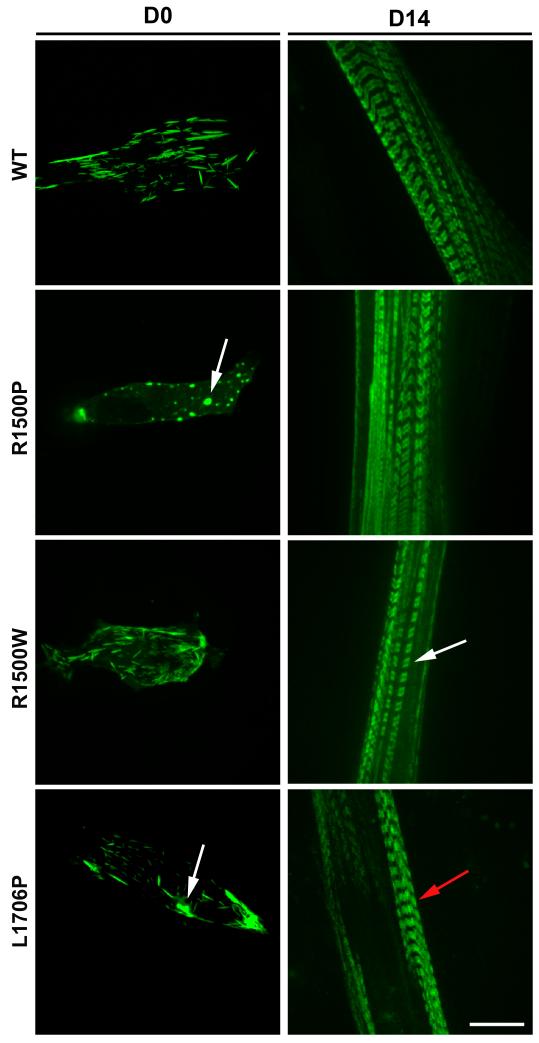
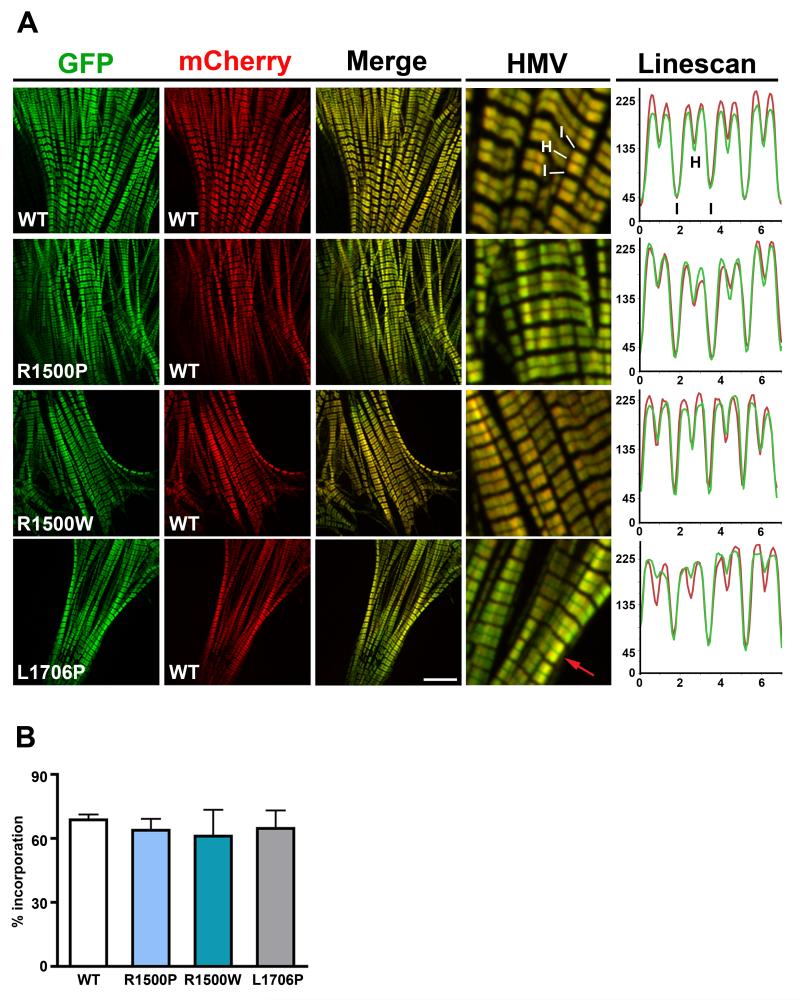
We next examined the behavior of the mutants in a more physiologically relevant context by transfecting the C2C12 mouse muscle cell line. Consistent with the phenotype observed in COS-7 cells, we found that the R1500P and L1706P mutations induce myosin aggregates in myoblasts (Figure 2, D0). However, 14 days after differentiation, both mutants were efficiently incorporated into myotube thick filaments (Figure 2, D14). Intriguingly, the bare zone, corresponding to the central part of the thick filament where myosin molecules are arranged in a bipolar array, was not clearly visible in cells expressing the L1706P myosin, indicating potential accumulation/mislocalization of the mutant in proximity of the M band (Figure 2, D14, red arrow). In order to examine muscle sarcomeres at higher resolution,22 we transfected the WT and mutant tagged constructs in neonatal rat ventricular myocytes (NRVMs) which have more highly ordered myofibrils. This approach confirmed and extended the results obtained in C2C12. Proper and homogeneous incorporation of both GFP and mCherry-tagged WT myosin rods in the ~ 1.6 μm long thick filaments (A-bands) was observed with good discrimination of the thin actin filaments (I-band) and the bare zone (H-band) (Figure 3A, WT panels). Furthermore, exclusion of fluorescence from the H-band indicates that the N-terminal tags are spatially located close to the endogenous myosin heads and out of the bare zone. The two WT proteins also fully co-localize as judged by the merged images (Figure 3A, Merge, and HMV (High Magnification View) panels) and by the quantitative Linescan analysis plotting the intensity profile of each fluorophore along the sarcomere topology (Figure 3A, Linescan panel). An evaluation of the three GFP-myosin mutants reveals appropriate distribution along the A-band and good co-localization with the mCherry-WT myosin for all constructs (Figure 3A, R1500P, R1500W, L1706P, Merge, HMV, and Linescan panels). However, as previously observed in C2C12, the fluorescence signal of the L1706P myosin was detected in the center of the bare zone with no effect on the proper distribution of the mCherry-tagged WT myosin (Figure 3A, L1706P, Merge, HMV and Linescan panels). Although scoring of NRVMs transfected with WT and mutant myosins did not unveil any disruptive effect of the mutants on thick filament formation and organization (Figure 3B) we found protein aggregates in a small percentage (< than 10%) of cells transfected with either of the two proline substitutions. Figure 4A depicts the aggregate phenotype observed for the R1500P construct. The mutant myosin is incorporated into the sarcomeres, but we found also cytoplasmic aggregates that partially sequester the mCherry-WT myosin. Significantly, aggregates were never observed with the WT or the R1500W tagged myosins in either C2C12 or NRVMs.
Figure 2.
Analysis of mutants in C2C12 cells. Cells transfected with the WT and mutant GFP-tagged myosin rod constructs as depicted, were imaged 24 h (D0) or 14 days (D14) after transfection. Pictures shown in D0 and D14 do not correspond to the same cell followed over time. Bar, 10 μm. D0 arrows indicate myosin aggregates; D14 white arrow indicates sarcomere bare zone, red arrow indicates mutant accumulation in the bare zone.
Figure 3.
Analysis of mutants in NRVMs. (A) NRVMs were electroporated with the WT/mutant GFP-tagged and WT mCherry-tagged myosin rod constructs as indicated. Cells were imaged by confocal microscopy 96 h later. HMV: High Magnification View of the merged images; I, I-Band; H, H-zone (bare zone); the red arrow in the L1706P HMV panel indicates the presence of fluorescence in the sarcomere bare zone. Bar, 10 μm. Linescan panels show a typical graphical representation of the fluorescence intensity values of myosin constructs expressing GFP-tagged WT, R1500P, R1500W, L1706P myosin (green line) and mCherry-tagged WT myosin (red line) measured along 4 sarcomeres; x-axis, distance (μm); y-axis, gray levels (avg). (B) The percentage of transfected NRVMs showing organized sarcomeres containing GFP-myosin was scored 96 h after electroporation. Data were obtained from 3 independent transfections and a total of ~ 4000 cells/construct scored blind.
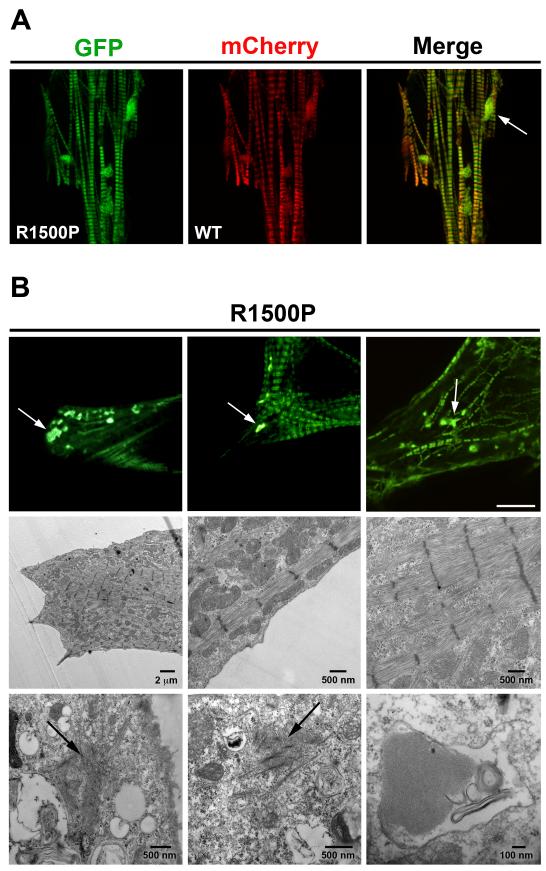
Figure 4.
The R1500P mutation causes myosin cytoplasmic accumulation. (A) A representative cardiomyocyte co-transfected with the GFP-tagged R1500P and WT-mCherry myosin constructs showing myosin aggregates 96 h after electroporation. (B) Top panels: NRVMs were plated on gridded coverslips and 3 individual cells containing myosin cytoplasmic accumulations were identified and then retrieved for electron microscopy analysis. Bar, 10 μm. Middle and bottom panels show respectively the sarcomere organization and myosin cytoplasmic accumulations of the first cell shown in the top left panel analyzed by electron microscopy. Bars, as indicated. White arrows indicate myosin aggregates, black arrows indicate amorphous matrix with disorganized rod shaped structures.
To investigate the ultrastructural nature of the aggregates, we performed correlative light-electron microscopy on single transfected cardiomyocytes. Transfected NRVMs were plated on gridded coverslips and cells showing fluorescent myosin aggregates were located (Figure 4B, top panels) and subjected to electron microscopy (EM) high-resolution analysis. In agreement with the fluorescence light microscopy data, EM imaging shows good sarcomeric organization with no signs of myofibrillar disarray in every cell examined (Figure 4B, middle panels). However, this analysis also reveals two distinct sarcoplasmic aggregate structures: amorphous matrix containing disorganized rod shaped formations, and crystalline arrays of protein inside vacuoles (Figure 4B, bottom panels). Consistently, the biopsy of a patient carrying the L1706P mutation also showed sarcoplasmic inclusion bodies, rod bands, and autophagic vacuoles.4
To study in more detail the molecular basis of the disease, we assessed by bimolecular fluorescence complementation assay (BiFC)23 whether the R1500P mutant rod can form homodimers (mut/mut) and/or heterodimers (mut/WT) in vivo, and how these coiled-coil combinations affect assembly and incorporation of myosin into the sarcomere. GFP was split into two non-fluorescent fragments and each of them was fused through 2-4 nm long linkers to the N-terminal end of WT and mutant MyHC rods (since the axial spacing between adjacent levels of myosin heads in thick filaments is 14.3 nm, these linkers are too short to promote BiFC between neighboring coiled-coils). Formation of homo or heterodimeric coiled-coil myosin molecules that promote reconstitution of fluorescence by bringing the two GFP fragments into proximity was then monitored by confocal microscopy.
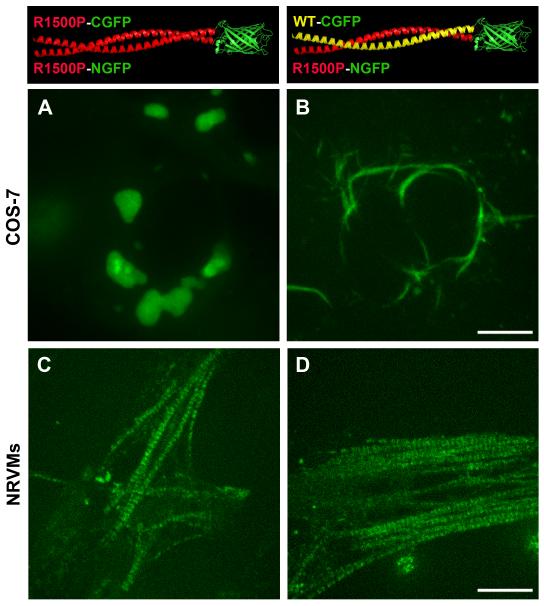
Transfection of different combinations of split GFP constructs into COS-7 cells reveals that the R1500P mutation does not prevent the coiling of homo and hetero α-helices; although the homodimers do not assemble into spindle-like structures, the heterodimers assemble relatively well (Figure 5, COS-7, A and B panels). Thus as previously shown (Figure 1B, R1500P, Merge), in non-muscle cells the presence of the WT myosin molecule improves the R1500P phenotype by assisting the incorporation of the mutant in heterodimeric coiled-coils (Figure 5, COS-7, B panel). In contrast, the BiFC assay carried out in cardiomyocytes shows that the R1500P mutant can be found in the sarcomeres as homodimers as well as heterodimers (Figure 5, NRVMs, C and D panels).
Figure 5.
Bimolecular fluorescence complementation (BiFC) assay detects R1500P homo/heterodimeric myosin molecules in live cells. COS-7 cells (Panels A and B) and NRVMs (Panels C and D) were co-transfected with the combinations of myosin rods fused to the amino (NGFP, 1-157) or carboxyl (CGFP158-230) portions of GFP as depicted in the top panels. A, and C: formation of R1500P homodimers in COS-7 and NRVMs cells respectively; B and D formation of R1500P/WT heterodimers in COS-7 and NRVMs cells respectively. Cells were imaged by confocal microscopy 24 h (COS-7) and 48 h (NRVMs) after tranfection. Bars, 10 μm.
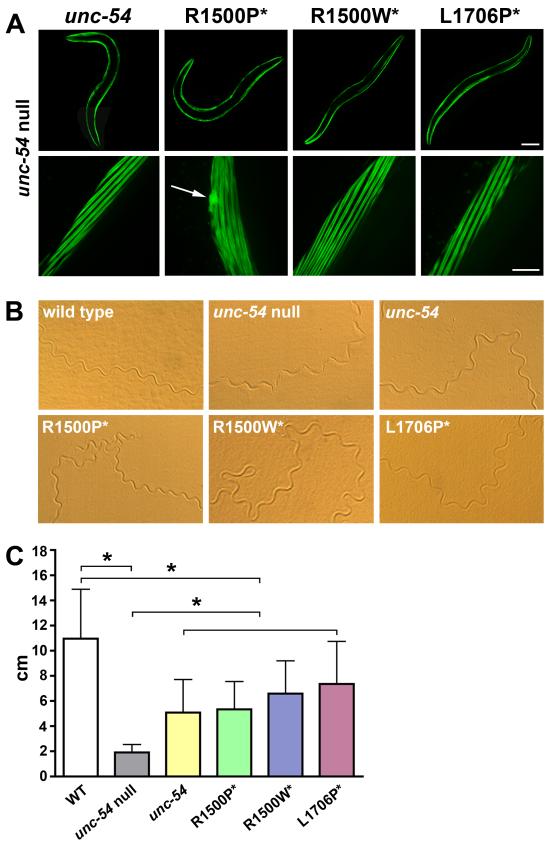
To further examine and quantify the effects of proline mutations on sarcomere mechanics, we carried out a set of genetic analyses on Caenorhabditis elegans, a platform successfully used to study the influence of mutations located in the myosin motor domain,24 to model Muscular Dystrophy,25 and Spinal Muscular Atrophy.26 The body wall muscle cells of C. elegans contain two myosin isoforms: MHC A assembles in an antiparallel organization with paramyosin and several other proteins to form the bare zone; MHC B is the major myosin isoform, and fills the arms of the thick filament (A-band) by packing in a parallel manner.27 MHC B null animals (lacking expression of the unc-54 gene) have disorganized body wall musculature, reduced number of thick filaments28 and display a slow phenotype as larvae, progressing to severe paralysis as they become adults. Since microinjection of the unc-54 gene into the null worm rescues muscle function,29 the pathological effects of mutations on the contractile apparatus can be quantified by monitoring sarcomere morphology and nematode locomotion. After finding that the rod elements predicted to be important for myosin assembly (amino acid sequence, ~28 residues periodicity, and charge profile) share high levels of conservation between vertebrate and MHC B myosins (Figure S1), we generated transgenic nematodes by microinjecting in the null unc-54 background the WT and mutant MHC B expressing the proline/tryptophan mutations in the corresponding residues of the unc-54 rod. To insure proper level of expression and prevent tissue mislocalization, transgene transcription was controlled by the endogeous unc-54 promoter. To evaluate muscle cell distribution and homogenous myosin expression, rescuing proteins were tagged at the N-terminus with GFP, a configuration that does not affect myosin motor activity or filament formation.22,30 Although fluorescent images of transgenic nematodes showed correct localization of WT and all mutant myosins into the A-band of the myofibrils of body wall muscles, nematodes expressing the R1500P mutations sometimes also exhibit, as previously detected in the cellular assays, myosin aggregates (Figure 6A). The effect of mutations on worm motility was then assessed by analyzing the level of rescue of the compromised sinusoidal movement observed in the unc-54 null (Figure 6B, wild type and unc-54 null). Phenotypic comparison of the tracks imprinted on agar plates by transgenic nematodes expressing WT and mutant MHC B, show equivalent rescue of the alternate contraction of the dorsal and ventral body muscles that is associated with normal body movement (Figure 6B, unc-54, R1500P*, R1500W*, L1706P*). We furthermore quantified the effects of proline mutations on locomotion by measuring the distance traveled by the nematodes over a 20 minute time-frame (Figure 6C). Deficiency of MHC B distinctly compromises the motility of the unc-54 null worms that averaged only 18% of the distance covered by the WT animals (2 versus 11 cm). Partial rescue of the paralyzed phenotype, comparable to the functional levels previously reported,24,29,31 was obtained after microinjection and expression of the unc-54 gene, as well as of the myosin mutant constructs carrying both proline and tryptophan mutations. Moreover, we did not observe a negative impact of the mutations on sarcomere function during physiological aging (worm strains with extended lifespan were not analyzed, since longevity mutations activate the expression of chaperones that delay the onset of diseases characterized by protein aggregation32).
Figure 6.
Analysis of mutations in transgenic C. elegans. The motor domain of C. elegans myosin heavy chain B (MHC B) was tagged with GFP and human mutations* were introduced in the corresponding positions of the rod: R1500P*/W* →R1512P/W; L1706P* →A1718P. Constructs were injected in the unc-54 null worms lacking MHC B (CB 190, unc-54 (e190)). (A) Fluorescence images of body wall muscles of transgenic worms injected with the indicated constructs. Nematodes expressing the R1500P mutation displayed normal sarcomere organization; a representative picture of the R1500P mutant aggregation (indicated by arrow) is shown. Top bar, 100 μm, bottom bar, 10 μm. (B) Sinusoidal movement of WT, unc-54 null, unc-54 null rescued with the unc-54 gene and the mutants as indicated (R1500P*, R1500W*, and L1706P*). (C) Histogram showing the distance covered by the WT, unc-54 null, and rescued transgenic worms. Error bars indicate the standard deviations of the means (n=20 for each group). * P <0.01, one-way analysis of variance (ANOVA) with Tukey post hoc analysis.
Thus, in the nematode null system, parallel thick filament formation and muscle force generation are not affected by the presence of the MD1 mutations or, as in the case of the R1500P, by the presence of myosin aggregates.
Discussion
It has been proposed that the R1500P and L1706P mutations could affect the incorporation of myosin in the thick filament by hindering the formation of the coiled-coil domain of the molecule.6 In fact, proline residues are known to destabilize and distort α-helical strands by their inability to form hydrogen bonds and by their steric hindrance.33 Moreover, proline substitutions have been predicted to cause short and long-range disruptive changes in the secondary structure of keratin.34-35 Although the proper folding of the rod is probably affected by the two proline mutations studied, the data presented in Figure 3B showing the percentage of transfected cardiomyocytes having organized sarcomeres containing GFP-myosin, indicate that all the mutants studied are incorporated into thick filaments as efficiently as the wild type myosin. However, it is likely that thick filaments containing the MPD1 mutants could be subjected to pathological instability and turn over. In fact, we have previously shown that paracrystals formed by the R1500P mutant are less stable than those formed with WT protein.15 Additionally, detection of sarcomeric R1500P homodimers, predicted to cause a more severe structural and biochemical phenotype, and heterodimers perturbing myosin mechanical performance, suggests that their relative percentage in the thick filaments could also modulate the degree of mechanical impairment.
Our sarcomere analysis suggests that the N-terminal GFP tag of the L1706P mutant could be mislocalized in the sarcomere bare zone. This finding indicates that this mutation perturbs myosin anti-parallel-more than parallel-association. In COS-7 cells both R1500P and L1706P mutant aggregate and fail to self-organize into ordered myosin filaments that require the presence of antiparallel- and parallel-packed rods. Their phenotype improves however, in the presence of the WT myosin, and is suppressed in C. elegans unc-54 null, where the bare zone is formed by paramyosin and MHC A, and the MHC B mutants assemble only in a parallel fashion. Sequence analysis of the myosin hydrophobic core residues of the rod domain, shows the presence of amino acid clusters (3 or more residues) that can both stabilize or destabilize the coiled-coil structure;36 as expected, leucine, that has ideal hydrophobic and packing properties, is the predominant residue found in the stabilizing clusters. Interestingly, the L1706P mutation produces a new cluster of destabilizing amino acids (thr, arg, ala, pro, thr) that could alter myosin assembly by modifying the structure and flexibility of the rod as well as its ability to undergo conformational changes. Based on their localization, the R1500P and L1706P mutations should not directly interfere with the mapped interactions between the myosin rod and M-band proteins (myosin residues 1505-1673),37 myosin-binding protein-C (myosin residues 1554-1581),38 and titin (myosin residues 1815-1831),39 as previously proposed.6 The L1706P mutation could however affect the interaction between myosin and Myomasp/LRRC39, a novel sarcomeric component predicted to anchor myosin to the M band.40 Thus, by disrupting the protein scaffold ensuring proper myosin M-band localization,41 the L1706P mutation could cause myosin rod misalignment.
There are no well-defined morphological hallmarks for MPD1 that is mainly characterized by variation in muscle fiber diameter, a likely switch from type I to type II fiber and mild muscular necrosis. Our data provide evidence that both R1500P and L1706P mutations induce myosin cytoplasmic aggregation. By sequestering wild type and mutant proteins in the cytoplasm, the mutations could cause disruption of the sarcomere structure and/or cause haploinsufficiency over time. Accordingly, detailed ultrastructural analysis of another MDP1 mutation (K1729del) has shown multiple areas of myofibrillar disorganization with Z-band streaming, and sarcomere disruption.3
Considering that β-myosin is the main molecular motor in the human heart, it remains unclear why MPD1 mutations are rarely associated with a cardiomyopathy. We have observed that the percentage of cells with aggregates is much higher in C2C12 cells than in NRVMs (data not shown). This finding may simply be caused by expression of the mutants in myoblasts before activation of muscle differentiation, or it may indicate that the skeletal muscle milieu is less tolerant of mutant MyHC misfolding or accelerated turn over. It would be interesting to determine whether the levels of chaperones that control myosin incorporation into thick filaments, or autophagy activity that is responsible for myosin degradation,42 are lower in skeletal muscle. The absence of a functional phenotype in the C. elegans motily assay, may suggest that the rhythmic contractile activity of the nematode muscle cells is more similar to the cardiomyocytes muscle properties. Interestingly, mathematical modeling of the nematode sinusoid motion pattern suggests the existence of a pacemaker or Central Pattern Generator (CPG) controlling the rhythmic activity of the body muscle cells.43
Failure to detect sarcomere disarray suggests that our assays have timing limitations and can expose only the initial stages of the disease characterized by the absence of a manifest muscular impairment. In fact, the two patients carrying the R1500P and L1706P mutations developed weakness of ankle dorsiflexion, and bilateral foot drop 4 and 5 years after birth respectively.6 Lack of aggregates in cells or nematodes expressing the R1500W mutation, identified in a DCM patient showing the first cardiac symptoms at the age of 55,16 also suggests that myosin cytoplasmic accumulation could be accountable for the early onset seen in patients carrying the R1500P and L1706P mutations.
The recent characterization of rod mutations that introduce the charge reversal Glu → Lys causing both cardiomyopathy and distal myopathy,8 indicates that the clinical outputs of MPD1 are modulated by the kind of amino acid replacements and the different cellular environments and physiological demands of the two muscle compartments. Thus, each group of MPD1 mutations probably acts through distinctive and complex pathological mechanisms. Nevertheless, our findings provide for the first time, novel insights into the role of a subset of MPD1 mutations affecting muscle function.
Materials and Methods
DNA constructs
EGFP and mCherry myosin rod constructs were generated by fusing each reporter gene at amino acid 841 of the rat α-cardiac myosin (accession number: X15938). The myosin rod was amplified from the pMT21α High-Fidelity DNA Polymerase (Bio-Rad) and cloned into plasmid pEGFP-C2 (Clontech) as EGFP carboxy-terminal fusion. The mCherry tagged myosin construct was obtained by replacing the EGFP gene. The myosin EGFP mutant constructs R1500P, R1500W and L1706P were generated by inverse PCR. C. elegans constructs used for microinjection were obtained by fusing EGFP to the amino terminus of the unc-54 gene cloned in the expression vector pPD5-41 (gift from Andrew Fire). The human rod mutations were introduced in the corresponding C. elegans residues (R1500P/W=R1512P/W; L1706P=A1718P) by inverse PCR. All cloning strategies and oligonucleotide sequences are reported in the Supplemental Material.
Cell culture and transfection
COS-7 cells (ATCC # CRL-1651) grown in DMEM (Dulbecco’s Modified Eagle’s Medium) High Glucose (Gibco) supplemented with 10% Fetal Bovine Serum were plated on glass coverslips, or on 35 mm glass bottom dishes (MatTek) for the BiFC assay. Cells were transfected with 1 μg of DNA using TransIT-LT1 reagent (Mirrus) according to the manufacturer’s instructions. Coverslips were fixed in 4% paraformaldehyde for 15 minutes at room temperature, washed three times in PBS, then mounted with Vectashield (Vector Laboratories). Neonatal rat ventricular myocytes (NRVMs) were prepared from Sprague-Dawley neonatal rat hearts as previously described.44 Isolated cells (~2.5×106) were transfected with Rat Cardiomyocyte-Neonatal Nucleofector Kit (Lonza) with 2 μg of Endotoxin Free DNA according to the manufacturer’s protocol, and plated on 1% gelatin coated glass coverslips, or on 35 mm glass bottom dishes (MatTek) for the BiFC assay. 18 h post-transfection cells were washed with PBS and incubated in MEM TI-BSA supplemented with 15 μM L-Phenylephrine. After 48 h, glass coverslips were washed, fixed and mounted as previously described. Fixed and live cells were then analyzed by confocal microscopy. Scoring of transfected NRVMs was carried out 96 h after transfection: cells lacking the classical organized sarcomeric pattern were counted as non-striated while cells containing well defined myofibrils (either aligned or misaligned) were counted as striated. For this analysis, coverslips were directly applied onto the cells after plates were washed with PBS. Three independent transfection experiments were performed and about 1300 cells were scored per constructs in each experiment.
Bimolecular Fluorescence Complementation (BiFC) assay
Sequences corresponding to EGFP residues 1-157 (NGFP) and 158-230 (CGFP) were fused to the WT and R1500P mutant myosin rod (residue 841) using the GGSGSGSS linker for connecting the NGFP fragment and the SEASGTSSGTSSTSSGI linker for connecting the CGFP fragment. Cells were transfected with different construct combinations and imaged by confocal microscopy 24 h (COS-7) or 48 h (NRVMs) later.
General C. elegans methods
The wild-type strain N2 and the MHC B null strain CB190, which carries the unc-54 (e190) mutation, were maintained as described.45 Transgenic worms expressing the unc-54 gene and the relative mutants were generated by standard procedures.46 For rescue experiments GFP-tagged myosin constructs were microinjected at 40 μg/ml in the distal gonad arms. Transgenic animals were identified by GFP expression in the body wall muscles using the Leica MZ 16 F fluorescence stereomicroscope.
C. elegans motility assay
60 mm NGM plates were equilibrated at room temperature, layered with 1 ml of an overnight culture of E.coli strain OP50, dried at room temperature, and used within 3 h 47. Single young adult worms (average length = .85 mm; 20 worms for each construct) were transferred on the center of the assay plates and allowed to crawl for 20 min at 25° C. After worm removal, plates were photographed with a Moticam 2000 CCD camera (Motic Instruments) connected to a Leica MZ6 stereo microscope and the total distance covered by each worm was computed with ImageJ, after individual images were merged into a single panorama using Photoshop.
Confocal microscopy
Cells and C. elegans body wall muscles were analyzed with a Nikon Eclipse TE 2000-U microscope coupled with an electron-multiplying charge-coupled device camera (Cascade II; Photometrics) and a Yokogawa spinning disc confocal system (CSU-Xm2; Nikon). Images were taken with a 60x Nikon Plan Apo VC NA 1.4 and 100x Nikon Plan Apo VC NA 1.4 oil objectives. MetaMorph software was used for image acquisition and image analysis.
Correlative light-electron microscopy
Correlative light-electron microscopy was carried out as previously described.48
Supplementary Material
Acknowledgments
We thank R. Thompson and N. Laing for discussion, A. Robinson for NRVM preparations; J. Blanchette and the M. Han laboratory for advice on C. elegans microinjections; D. Galati, C. English and the Woeltz laboratory for help with confocal imaging. We are gratefully indebted to T. Giddings for EM excellent technical assistance. This work was supported by National Institutes of Health grants RO1GM29090 and RO1HL85573.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Laing NG, Laing BA, Meredith C, Wilton SD, Robbins P, Honeyman K, Dorosz S, Kozman H, Mastaglia FL, Kakulas BA. Autosomal dominant distal myopathy: linkage to chromosome 14. Am J Hum Genet. 1995;56:422–7. [PMC free article] [PubMed] [Google Scholar]
- 2.Mastaglia FL, Phillips BA, Cala LA, Meredith C, Egli S, Akkari PA, Laing NG. Early onset chromosome 14-linked distal myopathy (Laing) Neuromuscul Disord. 2002;12:350–7. doi: 10.1016/s0960-8966(01)00287-5. [DOI] [PubMed] [Google Scholar]
- 3.Muelas N, Hackman P, Luque H, Garces-Sanchez M, Azorin I, Suominen T, Sevilla T, Mayordomo F, Gomez L, Marti P, Millan J. Maria, Udd B, Vilchez JJ. MYH7 gene tail mutation causing myopathic profiles beyond Laing distal myopathy. Neurology. 2010;75:732–41. doi: 10.1212/WNL.0b013e3181eee4d5. [DOI] [PubMed] [Google Scholar]
- 4.Lamont PJ, Udd B, Mastaglia FL, de Visser M, Hedera P, Voit T, Bridges LR, Fabian V, Rozemuller A, Laing NG. Laing early onset distal myopathy: slow myosin defect with variable abnormalities on muscle biopsy. J Neurol Neurosurg Psychiatry. 2006;77:208–15. doi: 10.1136/jnnp.2005.073825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craig R, Woodhead JL. Structure and function of myosin filaments. Curr Opin Struct Biol. 2006;16:204–12. doi: 10.1016/j.sbi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Meredith C, Herrmann R, Parry C, Liyanage K, Dye DE, Durling HJ, Duff RM, Beckman K, de Visser M, van der Graaff MM, Hedera P, Fink JK, Petty EM, Lamont P, Fabian V, Bridges L, Voit T, Mastaglia FL, Laing NG. Mutations in the slow skeletal muscle fiber myosin heavy chain gene (MYH7) cause laing early-onset distal myopathy (MPD1) Am J Hum Genet. 2004;75:703–8. doi: 10.1086/424760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamont PJ, Wallefeld W, Junckerstorff R, Laing NG. Multicore myopathy caused by a mutation in MYH7. Neuromuscul Disord. 2009;19:590–1. [Google Scholar]
- 8.Udd B. 165th ENMC International Workshop: distal myopathies 6-8th February 2009 Naarden, The Netherlands. Neuromuscul Disord. 2009;19:429–38. doi: 10.1016/j.nmd.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Dubourg O, Maisonobe T, Behin A, Suominen T, Raheem O, Penttila S, Parton M, Eymard B, Dahl A, Udd B. A novel MYH7 mutation occurring independently in French and Norwegian Laing distal myopathy families and de novo in one Finnish patient. J Neurol. 2011 doi: 10.1007/s00415-011-5900-9. [DOI] [PubMed] [Google Scholar]
- 10.Darin N, Tajsharghi H, Ostman-Smith I, Gilljam T, Oldfors A. New skeletal myopathy and cardiomyopathy associated with a missense mutation in MYH7. Neurology. 2007;68:2041–2. doi: 10.1212/01.wnl.0000264430.55233.72. [DOI] [PubMed] [Google Scholar]
- 11.Homayoun H, Khavandgar S, Hoover JM, Mohsen AW, Vockley J, Lacomis D, Clemens PR. Novel mutation in MYH7 gene associated with distal myopathy and cardiomyopathy. Neuromuscul Disord. 2011;21:219–22. doi: 10.1016/j.nmd.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Hedera P, Petty EM, Bui MR, Blaivas M, Fink JK. The second kindred with autosomal dominant distal myopathy linked to chromosome 14q: genetic and clinical analysis. Arch Neurol. 2003;60:1321–5. doi: 10.1001/archneur.60.9.1321. [DOI] [PubMed] [Google Scholar]
- 13.Parry DA, Fraser RD, Squire JM. Fifty years of coiled-coils and alpha-helical bundles: a close relationship between sequence and structure. J Struct Biol. 2008;163:258–69. doi: 10.1016/j.jsb.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 14.Barlow DJ, Thornton JM. Helix geometry in proteins. J Mol Biol. 1988;201:601–19. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]
- 15.Armel TZ, Leinwand LA. Mutations at the same amino acid in myosin that cause either skeletal or cardiac myopathy have distinct molecular phenotypes. J Mol Cell Cardiol. 2010;48:1007–13. doi: 10.1016/j.yjmcc.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karkkainen S, Helio T, Jaaskelainen P, Miettinen R, Tuomainen P, Ylitalo K, Kaartinen M, Reissell E, Toivonen L, Nieminen MS, Kuusisto J, Laakso M, Peuhkurinen K. Two novel mutations in the beta-myosin heavy chain gene associated with dilated cardiomyopathy. Eur J Heart Fail. 2004;6:861–8. doi: 10.1016/j.ejheart.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 17.Vikstrom KL, Rovner AS, Saez CG, Bravo-Zehnder M, Straceski AJ, Leinwand LA. Sarcomeric myosin heavy chain expressed in nonmuscle cells forms thick filaments in the presence of substoichiometric amounts of light chains. Cell Motil Cytoskeleton. 1993;26:192–204. doi: 10.1002/cm.970260303. [DOI] [PubMed] [Google Scholar]
- 18.Moncman CL, Rindt H, Winkelmann DA. Segregated assembly of muscle myosin expressed in nonmuscle cells. Mol Biol Cell. 1993;4:1051–67. doi: 10.1091/mbc.4.10.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Straceski AJ, Geisterfer-Lowrance A, Seidman CE, Seidman JG, Leinwand LA. Functional analysis of myosin missense mutations in familial hypertrophic cardiomyopathy. Proc Natl Acad Sci U S A. 1994;91:589–93. doi: 10.1073/pnas.91.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srikakulam R, Winkelmann DA. Myosin II folding is mediated by a molecular chaperonin. J Biol Chem. 1999;274:27265–73. doi: 10.1074/jbc.274.38.27265. [DOI] [PubMed] [Google Scholar]
- 21.Sohn RL, Vikstrom KL, Strauss M, Cohen C, Szent-Gyorgyi AG, Leinwand LA. A 29 residue region of the sarcomeric myosin rod is necessary for filament formation. J Mol Biol. 1997;266:317–30. doi: 10.1006/jmbi.1996.0790. [DOI] [PubMed] [Google Scholar]
- 22.Wang Q, Moncman CL, Winkelmann DA. Mutations in the motor domain modulate myosin activity and myofibril organization. J Cell Sci. 2003;116:4227–38. doi: 10.1242/jcs.00709. [DOI] [PubMed] [Google Scholar]
- 23.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu Rev Biophys. 2008;37:465–87. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tajsharghi H, Pilon M, Oldfors A. A Caenorhabditis elegans model of the myosin heavy chain IIa E706K [corrected] mutation. Ann Neurol. 2005;58:442–8. doi: 10.1002/ana.20594. [DOI] [PubMed] [Google Scholar]
- 25.Gieseler K, Grisoni K, Segalat L. Genetic suppression of phenotypes arising from mutations in dystrophin-related genes in Caenorhabditis elegans. Curr Biol. 2000;10:1092–7. doi: 10.1016/s0960-9822(00)00691-6. [DOI] [PubMed] [Google Scholar]
- 26.Briese M, Esmaeili B, Fraboulet S, Burt EC, Christodoulou S, Towers PR, Davies KE, Sattelle DB. Deletion of smn-1, the Caenorhabditis elegans ortholog of the spinal muscular atrophy gene, results in locomotor dysfunction and reduced lifespan. Hum Mol Genet. 2009;18:97–104. doi: 10.1093/hmg/ddn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller DM, 3rd, Ortiz I, Berliner GC, Epstein HF. Differential localization of two myosins within nematode thick filaments. Cell. 1983;34:477–90. doi: 10.1016/0092-8674(83)90381-1. [DOI] [PubMed] [Google Scholar]
- 28.Epstein HF, Waterston RH, Brenner S. A mutant affecting the heavy chain of myosin in Caenorhabditis elegans. J Mol Biol. 1974;90:291–300. doi: 10.1016/0022-2836(74)90374-x. [DOI] [PubMed] [Google Scholar]
- 29.Fire A, Waterston RH. Proper expression of myosin genes in transgenic nematodes. EMBO J. 1989;8:3419–28. doi: 10.1002/j.1460-2075.1989.tb08506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Resnicow DI, Hooft AM, Harrison BC, Baker JE, Leinwand LA. GFP fails to inhibit actin-myosin interactions in vitro. Nat Methods. 2008;5:212–3. doi: 10.1038/nmeth0308-212. author reply 213-4. [DOI] [PubMed] [Google Scholar]
- 31.Sassi HE, Renihan S, Spence AM, Cooperstock RL. Gene CATCHR--gene cloning and tagging for Caenorhabditis elegans using yeast homologous recombination: a novel approach for the analysis of gene expression. Nucleic Acids Res. 2005;33:e163. doi: 10.1093/nar/gni164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–5. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 33.MacArthur MW, Thornton JM. Influence of proline residues on protein conformation. J Mol Biol. 1991;218:397–412. doi: 10.1016/0022-2836(91)90721-h. [DOI] [PubMed] [Google Scholar]
- 34.Smith TA, Steinert PM, Parry DA. Modeling effects of mutations in coiled-coil structures: case study using epidermolysis bullosa simplex mutations in segment 1a of K5/K14 intermediate filaments. Proteins. 2004;55:1043–52. doi: 10.1002/prot.20089. [DOI] [PubMed] [Google Scholar]
- 35.Natsuga K, Nishie W, Smith BJ, Shinkuma S, Smith TA, Parry DA, Oiso N, Kawada A, Yoneda K, Akiyama M, Shimizu H. Consequences of Two Different Amino-Acid Substitutions at the Same Codon in KRT14 Indicate Definitive Roles of Structural Distortion in Epidermolysis Bullosa Simplex Pathogenesis. J Invest Dermatol. 2011;131:1869–76. doi: 10.1038/jid.2011.143. [DOI] [PubMed] [Google Scholar]
- 36.Kwok SC, Hodges RS. Stabilizing and destabilizing clusters in the hydrophobic core of long two-stranded alpha-helical coiled-coils. J Biol Chem. 2004;279:21576–88. doi: 10.1074/jbc.M401074200. [DOI] [PubMed] [Google Scholar]
- 37.Obermann WM, Gautel M, Weber K, Furst DO. Molecular structure of the sarcomeric M band: mapping of titin and myosin binding domains in myomesin and the identification of a potential regulatory phosphorylation site in myomesin. EMBO J. 1997;16:211–20. doi: 10.1093/emboj/16.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flashman E, Watkins H, Redwood C. Localization of the binding site of the C-terminal domain of cardiac myosin-binding protein-C on the myosin rod. Biochem J. 2007;401:97–102. doi: 10.1042/BJ20060500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Houmeida A, Holt J, Tskhovrebova L, Trinick J. Studies of the interaction between titin and myosin. J Cell Biol. 1995;131:1471–81. doi: 10.1083/jcb.131.6.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Will RD, Eden M, Just S, Hansen A, Eder A, Frank D, Kuhn C, Seeger TS, Oehl U, Wiemann S, Korn B, Koegl M, Rottbauer W, Eschenhagen T, Katus HA, Frey N. Myomasp/LRRC39, a heart- and muscle-specific protein, is a novel component of the sarcomeric M-band and is involved in stretch sensing. Circ Res. 2010;107:1253–64. doi: 10.1161/CIRCRESAHA.110.222372. [DOI] [PubMed] [Google Scholar]
- 41.Gautel M. The sarcomeric cytoskeleton: who picks up the strain? Curr Opin Cell Biol. 2011;23:39–46. doi: 10.1016/j.ceb.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 42.Willis MS, Schisler JC, Portbury AL, Patterson C. Build it up-Tear it down: protein quality control in the cardiac sarcomere. Cardiovasc Res. 2009;81:439–48. doi: 10.1093/cvr/cvn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karbowski J, Schindelman G, Cronin CJ, Seah A, Sternberg PW. Systems level circuit model of C. elegans undulatory locomotion: mathematical modeling and molecular genetics. J Comput Neurosci. 2008;24:253–76. doi: 10.1007/s10827-007-0054-6. [DOI] [PubMed] [Google Scholar]
- 44.Maass AH, Buvoli M. Cardiomyocyte preparation, culture, and gene transfer. Methods Mol Biol. 2007;366:321–30. doi: 10.1007/978-1-59745-030-0_18. [DOI] [PubMed] [Google Scholar]
- 45.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–70. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cronin CJ, Mendel JE, Mukhtar S, Kim YM, Stirbl RC, Bruck J, Sternberg PW. An automated system for measuring parameters of nematode sinusoidal movement. BMC Genet. 2005;6:5. doi: 10.1186/1471-2156-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rieder CL, Cassels G. Correlative light and electron microscopy of mitotic cells in monolayer cultures. Methods Cell Biol. 1999;61:297–315. doi: 10.1016/s0091-679x(08)61987-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.