Abstract
Viral RNA helicases of the NS3/NPH-II group unwind RNA duplexes by processive, directional translocation on one of the duplex strands. The translocation is preceded by a poorly understood unwinding initiation phase. For NPH-II from vaccinia virus, unwinding initiation is rate limiting for the overall unwinding reaction. To develop a mechanistic understanding of the unwinding initiation, we studied kinetic and thermodynamic aspects of this reaction phase for NPH-II in vitro, using biochemical and single molecule fluorescence approaches. Our data show that NPH-II functions as a monomer and that different stages of the ATP-hydrolysis cycle dictate distinct binding preferences of NPH-II for duplex vs. single stranded RNA. We further find that the NPH-II-RNA complex does not adopt a single but at least two distinct conformations in each of the analyzed stages of ATP hydrolysis. These conformations interconvert with rate constants that depend on the stage of the ATP hydrolysis cycle. Our data establish a basic mechanistic framework for unwinding initiation by NPH-II and suggest that the various stages of the ATP hydrolysis cycle do not induce single, stage-specific conformations in the NPH-II-RNA complex, but primarily control transitions between multiple states.
Keywords: Fluorescence, FRET, Translocation, SF2, Single Molecule
INTRODUCTION
Most aspects of RNA metabolism in eukaryotes, bacteria and many viruses involve RNA helicases of the helicase superfamily 2, ubiquitous enzymes that use ATP to bind and remodel RNA and RNA protein complexes1; 2,3. SF2 helicases are further classified into distinct families, based on structural, sequence and mechanistic characteristics1. SF2 helicases of the NS3/NPH-II family, named after NS3 from hepatitis C virus and NPH-II from vaccinia virus, are encoded by diverse viruses, where these proteins are essential for viral replication4; 5. HCV NS3 functions in conjunction with the viral polymerase and several other viral proteins in complexes that replicate the HCV genome5. NPH-II has been implicated in transcription termination in vaccinia virus and in the export of viral RNAs out of the virion4.
The biological functions of NS3/NPH-II helicases correlate with NTP-dependent unwinding activities6,7, but the connections between helicase activities and physiological functions are not well understood. Nonetheless, HCV NS3 and NPH-II have become attractive model systems for the mechanistic analysis of RNA helicase activity8. Both, HCV NS3 and NPH-II unwind RNA and DNA duplexes, in a unidirectional, stepwise, and processive fashion6,8–12. This unwinding mode resembles canonical DNA helicases13.
In vitro, RNA duplex unwinding by NPH-II and NS3 requires substrates with a single stranded overhang 3' to the duplex 6,9,14,15. The helicases bind to the overhang prior to unwinding, and are thereby oriented for subsequent translocation 3' to 5' along this "loading" strand 16–18. These binding and orientation events collectively constitute the unwinding initiation process.
In the NS3/NPH-II helicases for which this has been tested, unwinding initiation is markedly slower than the subsequent unwinding steps6,10. In NPH-II, unwinding initiation is at least two orders of magnitudes slower than subsequent strand separation steps6. Initiation is thus rate limiting for overall unwinding under usual reaction conditions6. A further important characteristic of the initiation process by NPH-II is the continuous hydrolysis of ATP before the start of the actual unwinding6. ATPase measurements suggest that the enzyme turns over hundreds of ATP before starting unwinding19. These findings indicate that unwinding initiation is itself a complex process.
Despite the significance of initiation for the overall unwinding reaction by NS3/NPH-II helicases, it is unclear which processes occur during this reaction phase. Here, we studied kinetic and thermodynamic aspects of this reaction phase for NPH-II in vitro. We examined specific stages of the ATPase cycle using non-hydrolyzable ATP analogs and a combination of biochemical and single molecule fluorescence approaches. Our data show that NPH-II functions as a monomer in which different stages of the ATP-hydrolysis cycle dictate distinct binding preferences for duplex vs. single stranded RNA. We also found that the NPH-II-RNA complex does not adopt a single but at least two distinct conformations in each of the analyzed stages of ATP hydrolysis. These conformations interconvert with rate constants that depend on the stage of the ATP hydrolysis cycle. Our data establish a basic mechanistic framework for the unwinding initiation process by NPH-II and suggest that the various stages of the ATP hydrolysis cycle do not induce single, stage-specific conformations in the NPH-II-RNA complex, but primarily control interconversion between multiple conformational states.
RESULTS
Different stages of the ATP hydrolysis cycle dictate distinct binding preferences of NPH-II for ssRNA and duplex RNA
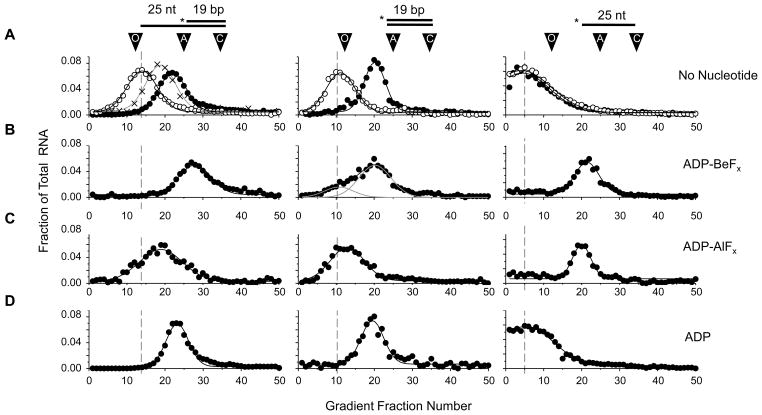
To characterize unwinding initiation by NPH-II, we first measured RNA binding at the main stages of the ATP hydrolysis cycle. We performed density gradient centrifugation with NPH-II and radiolabeled RNA without nucleotide, with ADP, and with the non-hydrolyzable ATP analogs ADP-beryllium fluoride (ADP-BeFx, ground state analog20) and ADP-aluminum fluoride (ADP-AlFx, transition state analog 20)(Fig.1). ADP-BeFx and ADP-AlFx have been widely employed in mechanistic and structural studies of other RNA helicases, including HCV NS321–25.
Figure 1. Binding of NPH-II to RNA measured by sucrose gradient centrifugation.
(A) Binding of NPH-II to three RNA substrates without nucleotide. Cartoons above the plots show the substrates; asterisks mark the radiolabel. For RNA sequences see Materials and Methods. Radiolabeled RNA in gradient fractions was quantified by scintillation counting. Plots show the fraction of total RNA (or NPH-II) in each gradient fraction (numbered at the x-axis). All experiments were repeated 3 to 5 times, and identical results were obtained. Representative data are shown. Open circles: RNA alone; filled circles: RNA in the presence of 100 nM NPH-II; crosses: free NPH-II, as quantified by the relative ATPase activity in the gradient fractions. Arrows indicate the average peak migration in the gradient of the three size standards, ovalbumin (49 kD, fraction 13), aldolase (150 kD, fraction 25), and catalase (240 kD, fraction 34). The dashed lines indicate the migration of each of the three RNAs. The apparent molecular weight of the complexes was calculated based on the sedimentation of the size standards, assuming a linear distribution of molecular weight for primarily globular protein or protein-RNA complexes. Errors represent one standard deviation from the Gaussian fit of the gradient pattern. The migration of RNA alone was slightly lower than expected based solely on molecular weight; most likely due to the higher density of free RNA and deviations from globular shape. Apparent molecular weights were for NPH-II alone: Mw = 82.7 ± 15.8 kD; bipartite RNA, Mw = 27.7 ± 13.4 kD; NPH-II + bipartite RNA, Mw = 113.1 ± 25.2 kD; blunt-end duplex RNA alone, Mw = 14.7 ± 9.8 kD; NPH-II + blunt-end duplex, Mw = 106.0 ± 7.5 kD. The single-strand RNA migrated lower than the predicted molecular weight of Mw = 8 kD. (B) Binding of NPH-II to RNA with ADP-BeFx. ADP-BeFx was at 3.5 mM. Apparent molecular weights were for NPH-II + bipartite RNA + ADP-BeFx , Mw = 180.0 ± 8.7 kD; NPH-II + blunt-end duplex + ADP-BeFx, Mw = 114.7 ± 15.1 kD; NPH-II + single-strand RNA + ADP-BeFx, Mw = 106.5 ± 6.4 kD. (C) Binding of NPH-II to RNA with ADP-AlFx. ADP-AlFx was at 3.5 mM. Apparent molecular weights were for NPH-II + bipartite RNA + ADP-AlFx , Mw = 84.3 ± 17.3 kD. NPH-II + blunt-end duplex + ADP-AlFx , Mw = 22.6 ± 12.3 kD; NPH-II + single-strand RNA + ADP-AlFx, Mw = 104.9 ± 11.4 kD. (D) Binding of NPH-II to RNA with ADP. ADP was at 3.5 mM. Apparent molecular weights were for NPH-II + bipartite RNA + ADP, Mw = 119.9 ± 15.0 kD; NPH-II+ blunt-end duplex + ADP, Mw = 105.7 ± 12.7 kD.
We determined how many protomers of NPH-II bound a duplex RNA with a 3’ ssRNA overhang. This RNA substrate (calculated molecular weight MW = 21 kD) had been previously used in unwinding studies of NPH-II6,16.Without nucleotide, NPH-II (MW = 77kD) shifted the RNA to an apparent molecular weight consistent with a single protomer of NPH-II bound to the RNA (Mw(C) = 113 ± 25 kD, Fig.1A, left panel). Next, we examined binding of NPH-II to RNAs containing only the duplex or the single stranded part of the substrate (Fig.1A, middle and right panels). We observed binding of a single NPH-II protomer to the duplex, but no association to the ssRNA (Fig.1A, middle and right panels).
In the presence of the ATP ground state analog ADP-BeFx, NPH-II shifted the RNA substrate to an apparent molecular weight consistent with binding of two protomers (Fig.1B, left panel). Binding of NPH-II to both, duplex and ssRNA was detected, but only a single protomer appeared to bind in each case (Fig.1B, middle and right panels). With ADP-AlFx, NPH-II shifted the RNA to an apparent molecular weight most consistent with binding of a single protomer (Fig.1C). The broadening of the peak, compared to the peak without nucleotide (Fig.1A) suggests a distinct conformation of the NPH-II RNA complex. No binding to the duplex was detected, and a single protomer associated with the ssRNA (Fig.1C). With ADP, a single protomer was bound to the RNA substrate and to the duplex, but no binding to the ssRNA was detected (Fig.1D).
Collectively, the density gradient centrifugation experiments provided three insights. First, NPH-II binds to the RNA substrate at all stages of the ATPase cycle tested here. This observation is consistent with the notion that NPH-II maintains uninterrupted substrate contact during unwinding initiation, despite the continued hydrolysis of hundreds of ATPs per substrate19. Second, NPH-II changes binding preferences between ssRNA and duplex RNA during the various stages of the ATPase cycle. This result explains the previously reported ability of NPH-II to maintain localization on the ss/duplex junction during initiation18. Third, the data revealed that NPH-II binds the RNA as a monomer, except in the ATP ground state mimicked by ADP-BeFx, where two protomers appear to bind.
NPH-II unwinds RNA duplexes as a monomer
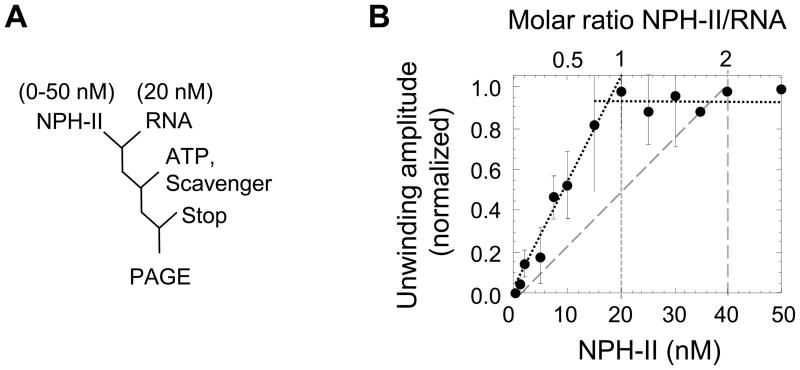
While instructive, the density gradient experiments left open the question whether binding of multiple protomers in the ATP ground state was required for unwinding, or merely reflected a propensity of NPH-II monomers to bind to duplex and single stranded RNA at this stage of the ATP hydrolysis cycle. To distinguish between these possibilities, we performed a functional stoichiometric titration (Fig.2). This experiment measures unwinding at substrate and enzyme concentrations that exceed the enzyme-substrate dissociation constant, and with substrate concentrations significantly higher than enzyme concentrations (Materials and Methods). The number of protomers needed for the unwinding reaction is reflected in the correlation between unwinding signal and protein/RNA ratio 6,26,27. To avoid complications in the data interpretation through multiple re-initiation events per substrate, we performed unwinding reactions under single cycle conditions, which preclude re-binding of the helicase to the substrate during the course of the reaction6,27. Under single cycle conditions, the final extent of the reaction for a given substrate (unwinding amplitude) corresponds to the fraction of NPH-II productively bound at the reaction start6.
Figure 2. Functional stoichiometric titration of NPH-II on RNA.
(A) Reaction scheme. (B) Plot of normalized unwinding amplitudes versus NPH-II concentration. Closed circles indicate the normalized reaction amplitude at each NPH-II concentration. Error bars indicate the standard deviation of multiple independent experiments. The lines with short dashes mark expected data for a single protomer of NPH-II. The line with larger dashes marks expected data for two protomers of NPH-II. The maximal reaction amplitude is reached at 16.9 ± 1.9 nM NPH-II, indicating a 1:1 ratio for [RNA]/[NPH-II].
Unwinding amplitudes increased linearly with the NPH-II concentration until an enzyme substrate ratio of 1:1. No further increase was seen at higher NPH-II concentrations (Fig.2B). This finding clearly shows that only one protomer of NPH-II per substrate is needed to reach the maximal unwinding amplitude. Oligomerization at any stage of the ATPase cycle would be reflected in a NPH-II substrate ratio greater than 1:1 26.
The data indicate that a single NPH-II is competent to catalyze the unwinding reaction. Binding of multiple NPH-II protomers in the ATP ground state is not required for unwinding. We therefore conclude that the binding of multiple protomers to the RNA substrate in the ATP ground state, seen by gradient centrifugation with ADP-BeFx, reflects a functionally independent association of protomers to both single strand and duplex regions.
Observation of RNA binding and duplex unwinding by NPH-II with single molecule FRET
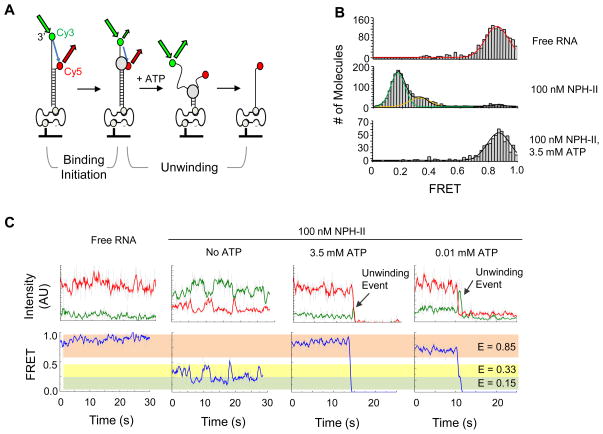
Having established that NPH-II unwinds as a monomer, we employed single molecule fluorescence resonance energy transfer (smFRET) to further characterize the initiation process. We used a TIR fluorescence detection setup (Materials and Methods) and fluorescently labeled RNA (19 bp duplex, 25 nt unpaired overhang 3' to the duplex), immobilized on a PEG-coated surface28,29 (Fig.3A). The labeling scheme allowed us to detect binding of unlabelled NPH-II to the single stranded region, conformational changes of the NPH-II -RNA complex, and unwinding events (Fig.3A).
Figure 3. NPH-II binding to RNA measured by smFRET.
(A) RNA construct design and principle of smFRET measurements. A 19 base pair duplex with a 24 nucleotide 3’ overhang was labeled with Cy3 (green circle) and a Cy5 (red circle). The RNA was biotinylated on the 3’ end of the Cy5-containing strand (grey circle) and attached via streptavadin to biotinylated PEG that was covalently linked to the glass slide. FRET is indicated by the grey arrow. NPH-II binding alters the smFRET values of free RNA. Unwinding leads to the separation of the strands, and thus results in a sharp decrease in FRET, followed by the disappearance of the Cy3 upon strand dissociation. (B) SmFRET histograms of NPH-II binding to RNA. Histograms were obtained by determining smFRET values for the number of molecules indicated on the right, averaged over five frames at the reaction start. In all histograms, the molecules with non-fluorescent Cy5 (FRET = 0) were subtracted. Lines indicate Gaussian fits of each FRET population, the black line marks the overall fit of the population with multiple peaks. Peak values were as follows; free RNA: E = 0.85 ± 0.17; with 100 nM NPH-II: E1 = 0.33 ± 0.15 and E2 = 0.15 ± 0.10; with 100 nM NPH-II and 3.5 mM ATP E = 0.87 ± 0.15. The errors mark one standard deviation of the Gaussian fit from the respective peak. (C) Representative smFRET timetraces. Green line: intensity of Cy3; red line: intensity of Cy5; blue line: FRET. Data were smoothed using adjacent averaging of the nearest 5 timepoints; grey lines show not averaged data. Shaded boxes mark the FRET states corresponding to the histograms in panel (B). Arrows indicate putative unwinding events.
We first recorded smFRET histograms for free RNA, and NPH-II bound to RNA with and without ATP (Fig.3B). The smFRET distribution of free RNA substrate showed a single peak at E = 0.85 (Fig.3B, upper panel). Addition of saturating amounts of NPH-II shifted the distribution to two peaks at E = 0.33 and E = 0.15 (Fig.3B, middle panel). This shift shows binding of NPH-II to the single stranded region and reveals two conformationally distinct NPH-II-RNA complexes.
NPH-II binding to ssRNA without nucleotide was not seen with density gradient centrifugation (Fig.1A), suggesting that NPH-II and ssRNA do not form complexes that are sufficiently persistent for detection by density gradient centrifugation. Alternatively, binding of NPH-II to ssRNA without ATP might require an adjacent duplex. Upon addition of NPH-II and ATP at concentrations where robust unwinding is seen6, the number of RNA complexes displaying FRET diminished over time, as expected, due to unwinding and corresponding removal of the CY3 label (data not shown). However, the smFRET distribution of the RNAs prior to unwinding was highly similar to that observed with free RNA (Fig.3B, lower panel). This finding suggests that ATP-dependent conformational changes of the NPH-II-RNA complex prior to unwinding occur at a timescale that exceeds the time resolution of the detection setup. Based on the high turnover number for ATP (kcat ~ 104 min−1 ref.19), the frequency of conformational transitions of the NPH-II-RNA complex driven by ATP hydrolysis events are in fact expected to be considerably greater than the time resolution of our smFRET setup (τ = 0.1s). Thus, only an average FRET value would be seen in each frame. Free RNA is also thought to sample a large number of different conformations over the time needed to acquire a single frame30, and it is therefore not unexpected that both free RNA and NPH-II bound to the RNA in the presence of ATP yield similar FRET values.
To gain further insight into the characteristics of NPH-II-RNA complexes with and without ATP, we examined smFRET timetraces (Fig.3C). While timetraces of individual free RNA substrates did not reveal interpretable patterns of dynamics (Fig.3C, left panel), addition of NPH-II without ATP showed interconversion between the two FRET states seen in the corresponding smFRET distribution (Fig.3C, second panel). Addition of ATP (3.5.mM) to the NPH-II RNA complex changed the FRET values again, consistent with the corresponding FRET histogram. No interpretable patterns of dynamics of the NPH-II-RNA complex could be elucidated, except for a spike in the FRET value after several seconds, which was followed by disappearance of the signal (Fig.3C, third panel). This signature is anticipated for a strand separation event (Fig.3A). Consistent with the notion that the FRET spike reflected a strand separation event, the duration of the spike was longer at lower ATP concentrations (0.01mM), where the strand separation process is expected to be slower (Fig.3C, right panel). Together, the timetraces with NPH-II and ATP thus directly reflect prolonged unwinding initiation (E ~ 0.85) and the much faster strand separation (smFRET spike). The absence of interpretable patterns of dynamics in the NPH-II-RNA complex before the strand separation further support the notion that ATP-dependent conformational changes of the NPH-II-RNA complex occur at a timescale that exceed the time resolution of the detection setup.
At lower ATP concentrations ([ATP] = 10 μM, Fig.3C, right panel), fluctuations in smFRET values before the unwinding event appeared more pronounced than at the higher ATP concentrations (Fig.3C, right panel). Nevertheless, we could not delineate statistically significant patterns of dynamics for the NPH-II-RNA complex preceding the strand separation. We were thus not able to determine whether NPH-II shuttles back and forth on the single stranded region, as seen for the Rep, BLM and HCV-NS3 helicases 31–33. Extensive NPH-II dissociation from the RNA at [ATP] < 10 μM precluded meaningful interpretations of timetraces at even lower ATP concentrations (data not shown).
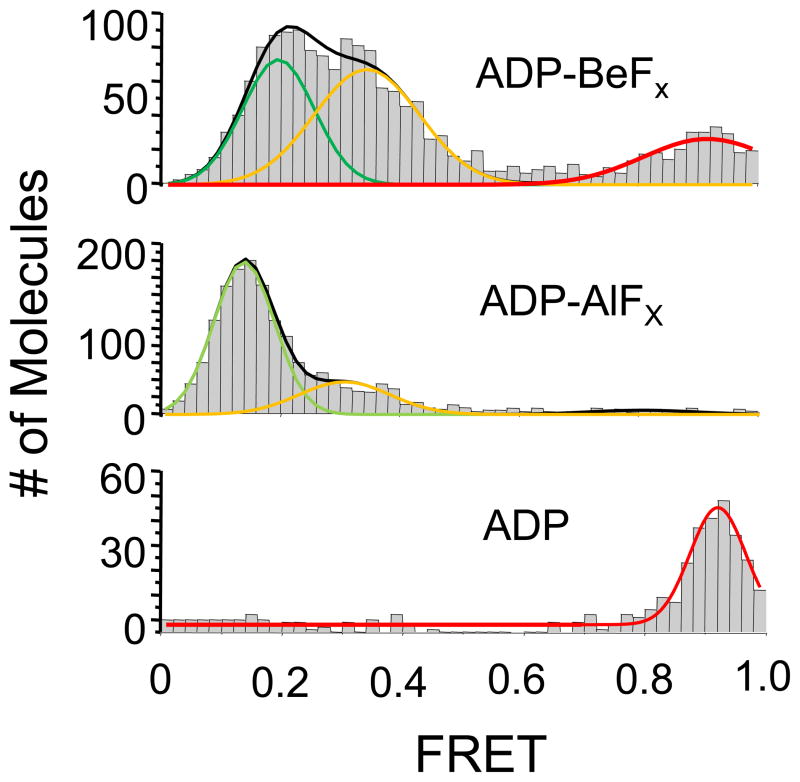
Formation of NPH-II RNA complexes with non-hydrolysable ATP analogs
Since we could not delineate clear dynamic patterns of the NPH-II-RNA complex in the presence of ATP during unwinding initiation, further characterization of this reaction step required the arrest of the ATP-hydrolysis cycle at defined stages. To accomplish this arrest, we used the non-hydrolyzable ATP ground state analog ADP-BeFx, the transition state analog ADP-AlFx, and ADP in binding reactions of NPH-II and RNA that we monitored by smFRET (Fig.4). SmFRET distributions for the NPH-II-RNA complex with ADP-BeFx showed three peaks at the highest experimentally accessible NPH-II concentrations (Fig.4, upper panel). Two peaks correspond to bound NPH-II, as seen without nucleotide (Fig.3B). The other peak corresponds to "free" RNA, and thus represents either RNA without NPH-II, or NPH-II bound to the duplex region, as suggested by the density gradient density centrifugation data (Fig.1B). With ADP-AlFx, we detected two peaks, corresponding to those representing NPH-II bound to the single stranded region (Fig.4, middle panel). With ADP, only one peak was seen, corresponding to either RNA without NPH-II, or, in line with the density gradient density centrifugation data, NPH-II bound to the duplex region (Fig.4, lower panel).
Figure 4. NPH-II binding to RNA with ATP analogs and ADP.
SmFRET histograms of NPH-II binding to RNA were obtained as described in Fig.3B (100 nM NPH-II: 100 nM, ADP-AlFx, ADP-BeFx, or ADP: 3.5 mM). Molecules with photobleached Cy5 (E = 0) were subtracted from the histograms. Lines indicate a Gaussian fit of each FRET population. SmFRET peak values were with ADP-AlFx: E1 = 0.29 ± 0.15, E2 = 0.14 ± 0.10; with ADP-BeFx: E1 = 0.34 ± 0.17 E2 = 0.19 ± 0.12, EFree = 0.91 ± 0.21, and with ADP: E = 0.92 ± 0.09. Errors mark one standard deviation of the Gaussian fit of the respective peak.
Collectively, the smFRET distributions without (Fig.3B) and with nucleotides (Fig.4) provide two insights. First, NPH-II binds to single stranded RNA regions of the substrate at all of the tested stages of the ATP hydrolysis cycle, except in the ADP-bound state. The smFRET data thus confirm and extend the findings obtained by density gradient centrifugation (Fig.1). Second, complexes formed between NPH-II and single stranded RNA at the interrogated stages of the ATP hydrolysis cycle appear to exist in at least two conformationally distinct complexes. The highly similar FRET peak values of the two NPH-II bound species suggest that these two conformations are similar at the tested stages of the ATP hydrolysis cycle .
Two distinct conformations of the NPH-II RNA complex reflect a single protomer bound to RNA
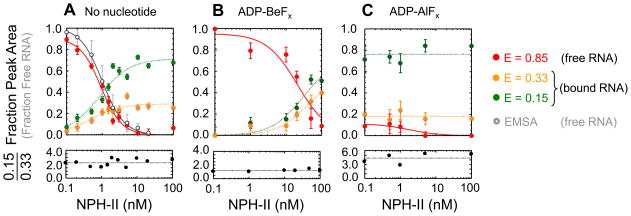
To further understand the two distinct conformations, we examined whether the two smFRET peaks were caused by successive binding of multiple NPH-II protomers. Although functional titration experiments had shown that a single NPH-II protomer was fully unwinding competent (Fig.2), multiple RNA-bound NPH-II protomers in the ATP ground state were seen by density gradient centrifugation (Fig.1B). Yet, even this method did not necessarily detect all NPH-II RNA complexes, as highlighted by NPH-II binding to ssRNA regions, which was not seen by density gradient centrifugation, but evident in the smFRET data (Fig.3B). To assess a possible oligomerization of NPH-II on the RNA by smFRET, we measured smFRET distributions at increasing NPH-II concentrations without nucleotide, with ADP-BeFx, and with ADP-AlFx (Fig.5). The fraction of the three smFRET peaks at E = 0.85 (free RNA), E = 0.33 and E = 0.15 (NPH-II bound RNA) was plotted as a function of the NPH-II concentrations (Fig.5).
Figure 5. Affinity of NPH-II to RNA without and with ATP analogs.
(A) NPH-II - RNA binding without nucleotide. Upper panel: fraction of each smFRET state vs. NPH-II concentration. Fractions were calculated from areas under the respective peak in the smFRET histograms (Figs.3B, 4). Colors mark the three smFRET states, as indicated on the right. Error bars indicate the standard deviation from the fit of the smFRET peaks to the Gaussian distribution (Figs.3B, 4). Grey open circles mark the fraction free RNA measured by EMSA (Suppl. Fig.S1). Lines show the fit to the Hill equation. Kd FRET = 0.91 ± 0.12 nM, n = 1.48 ± 0.22; Kd EMSA = 1.23 ± 0.59 nM, n = 1.63 ± 0.18. Lower panel: ratio of the fractions at E1 = 0.33 and E2 = 0.15. The dotted line marks the average ratio, R = 2.21 ± 0.51. (B) NPH-II - RNA binding with ADP-BeFx. Data were obtained as described in panel (A). Kd FRET = 20.8 ± 7.5 nM. R = 1.2 ± 0.2. (C) NPH-II - RNA binding with ADP-AlFx. Data were obtained as described in panel (A). Virtually complete binding occurred even at the lowest NPH-II concentrations (Kd FRET < 0.01 nM, R = 4.4 ± 1.1).
Without nucleotide, the fraction of the peak at E = 0.85 decreased with the NPH-II concentration, as expected for the binding reaction (Fig.5A). The binding constant calculated from these smFRET data was in excellent agreement with the constant obtained by non-denaturing gel shift electrophoresis (Fig.5A, Suppl. Fig.S1), indicating that the smFRET system faithfully reported quantitative aspects of NPH-II -RNA binding. Concomitant with the decrease of the peak at E = 0.85, the fraction of the peaks at E = 0.33 and E = 0.15 rose with the NPH-II concentration (Fig.5A). The ratio between the two peaks remained constant (Fig.5A, lower panel), indicating that neither peak arose from successive binding of multiple NPH-II protomers.
With ADP-BeFx, a similar trend was observed (Fig.5B). Free RNA (E = 0.85) decreased with increasing NPH-II concentrations, and bound RNA (E = 0.33 and E = 0.15) increased (Fig.5B). The ratio between the bound peaks remained constant. The binding constant for NPH-II was about one order of magnitude higher than without nucleotide.
With ADP-AlFx, almost no free RNA was seen, even at the lowest experimentally accessible NPH-II concentrations (Fig.5C). This observation shows that NPH-II was already bound at the lowest protein concentrations. Again, the ratio between the two peaks remained constant with increasing NPH-II concentrations (Fig.5C). Collectively, the data provided three insights. First, the two smFRET peaks reflecting bound NPH-II were not caused by successive binding of multiple protomers, and therefore correspond to distinct conformational states of a single NPH-II protomer bound to RNA. Second, the same conformational states exist at different stages of the ATP hydrolysis cycle. Third, the ratio between the states varies with the stage of the ATP hydrolysis cycle.
Interconversion between the conformations of the NPH-II RNA complex depends on the stage of the ATP hydrolysis cycle
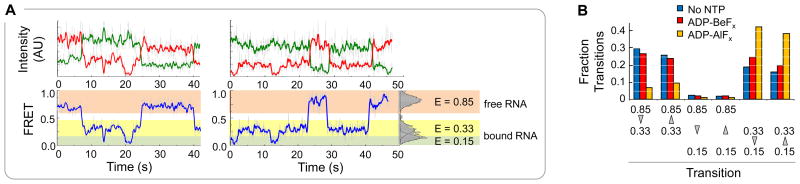
To further understand the different ratios between the two conformational states of the NPH-II RNA complex at different stages of the ATP hydrolysis cycle, we analyzed smFRET time trajectories (Fig.6). Without nucleotide, frequent transitions were seen between the NPH-II bound states E = 0.33 and E = 0.15 and between E = 0.33 and E = 0.85 and vice versa (Fig.6A, B). This pattern indicates frequent binding and dissociation events (E = 0.33 ↔ E = 0.85), and interconversion between the bound NPH-II states (E = 0.33 ↔ E = 0.15) at the experimentally accessible timescale. Time trajectories with ADP-BeFx showed a highly similar pattern and trajectories with ADP-AlFx showed transitions between the two lower FRET states, but only few transitions from and to the state at E = 0.85 (Fig.6B). This observation indicates that NPH-II binding and dissociation events occur frequently without nucleotide and with ADP-BeFx, but much less with ADP-AlFx, consistent with the stabilities of the respective complexes (Fig.5).
Figure 6. Interconversion between smFRET states.
(A) Representative smFRET timetraces (NPH-II: 1 nM, no nucleotide). Green line: Cy3; red line: Cy5; blue line: FRET. Data were smoothed by averaging of the nearest 5 timepoints; grey lines mark unaveraged data. Intensity is indicated as arbitrary units (AU). The histogram on the right shows the distribution of smFRET states, marked by the shaded boxes. (B) Fraction of transitions versus type of transition. Transitions between different smFRET states were counted manually (approximately 400 for each reaction condition) and the fraction of total transitions for each transition type was determined.
Transitions from E = 0.15 to E = 0.85 and vice versa were rare under all conditions (Fig.6B), suggesting that both NPH-II binding and dissociation proceeded virtually exclusively through the E = 0.33 state. Collectively, the data indicate a strongly preferred two step reaction path under all conditions, from E = 0.85 to E = 0.33 to E = 0.15, and reverse. The few transitions from E = 0.85 to E = 0.15 (and vice versa) may represent mostly events where the dwell time in the E = 0.33 state is shorter than the time resolution of the instrument, thus giving the appearance of a transition from the highest to the lowest FRET state.
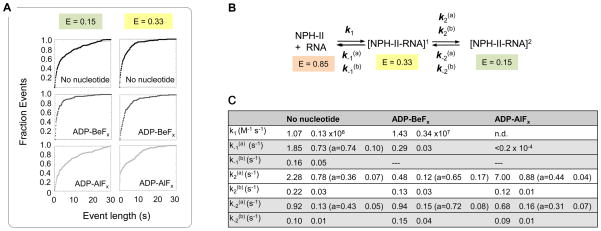
To illuminate dynamic aspects of the interconversion between the FRET states, we determined kinetic parameters from the dwell times in the respective states (Fig.7). The transition from E = 0.85 to E = 0.33, which represents the binding of NPH-II to the RNA, displayed single exponential kinetics (data not shown). Transitions from the two NPH-II bound states showed multi-exponential characteristics, indicating that each of these FRET state represents at least two kinetically discrete states. (Fig.7A). We did not detect molecular memory effects34 (Suppl. Fig. S2).
Figure 7. The kinetics of interconversion between smFRET states.
(A) Transitions from smFRET states corresponding to RNA-bound states. Cumulative plots of dwell times at E = 0.15 and E = 0.33 were fit to a sum of two exponentials using a maximum likelihood analysis (Materials and Methods). Error was assessed by bootstrapping. Cumulative plots of dwell times at E = 0.85 (free RNA) fit to single exponentials (data not shown). Dwell times at each FRET state were determined using custom Matlab software. 136 – 276 events for each reaction condition were used in the analysis. (B). Kinetic scheme for interconversion between smFRET states. Rate constants correspond to the transitions between each FRET state. (C). Rate constants for the smFRET transitions. Rate constants were calculated by globally fitting dwell time distributions to the kinetic model in panel (B) using maximum likelihood analysis. Errors were determined by bootstrapping. The amplitude of the first rate constant in bi-exponential fits is indicated as “a”. For ADP-AlFx, k1 and k-1 could not be determined by FRET, due to the high affinity of NPH-II for RNA. The value for k-1 is represented as a lower limit, as determined by EMSA (Suppl. Fig.S3) For ADP-BeFx, the transition from E2 = 0.33 to EFree = 0.85 (k-1) fit best to a single exponential.
Based on the two step reaction path (Fig.7B), we determined rate constants from the dwell times in the respective states (Fig.7C). Several rate constants were largely unaffected by the nucleotides, but notable nucleotide effects were seen for the dissociation of NPH-II from the RNA (k−1a). ADP-BeFx slowed NPH-II dissociation, compared to the reaction without nucleotide. ADP-AlFx slowed dissociation to an even greater extent, consistent with results obtained by EMSA (Suppl. Fig. S3). We discuss below how slowing of the dissociation rate constant by ADP-BeFx is reconciled with lower RNA affinity of NPH-II with this nucleotide. Significant nucleotide effects were also seen on the transition from E = 0.33 to E = 0.15 (k2a). The transition was slowed by ADP-BeFx , compared to the reaction without nucleotide. This slowing explains why the FRET state at E = 0.33 is more populated with ADP-BeFx (Fig.4). ADP-AlFx accelerated the transition from E = 0.33 to E = 0.15 (k2a), explaining the lower population of the FRET state at E = 0.33 (Fig.4). Collectively, the kinetic data show that the multiple FRET states seen for the NPH-II-RNA complex interconvert with all nucleotides tested as well as without nucleotide. The nucleotides alter the dynamics of the interconversion and modulate NPH-II dissociation, but they do not induce one particular state of the NPH-II-RNA complex.
DISCUSSION
In this analysis of the unwinding initiation process by NPH-II we have shown (i) that the helicase binds and unwinds RNA duplex substrates as monomer, (ii) that different stages of the ATP hydrolysis cycle modulate binding preferences for single stranded vs. duplex RNA, and (iii) that the NPH-II-RNA complex does not adopt a single, but multiple distinct, yet interconverting conformations. The NPH-II-RNA complex traverses these conformations at all tested stages of the ATP hydrolysis cycle. However, rate constants for certain transitions depend on the particular stage of the ATP hydrolysis cycle.
NPH-II functions as a monomer
Our data show that NPH-II unwinds RNA duplexes efficiently as a single protomer (Fig.2). Notwithstanding, multiple protomers can bind independently to the substrate (Fig.1), but these protomers do not functionally interact. The ability of NPH-II to function as a monomeric RNA helicase highlights a difference to the phylogenetically related HCV NS3, which requires oligomerization to achieve optimal unwinding activity35,36. Yet, HCV NS3 also functions as a monomer under certain conditions on some substrates37. The data presented here for NPH-II do not rule out the possibility that multiple NPH-II protomers bind to substrates with ssRNA tails longer than those used in this study. It also remains possible that binding of multiple protomers to longer single stranded regions synergistically enhance processivity over the monomeric protein, as has been observed for HCV NS3 and the SF1 DNA helicase Dda36,38. Effects of overhang lengths for unwinding by NPH-II remain to be examined.
Our data show that a NPH-II monomer is able to bind to both single and double stranded RNA (Fig.1). Although binding to double stranded RNA is not strictly required for a translocating helicase, other monomeric translocating DNA helicases, inlcuding PcrA, Rep, UvrD or Hel308, also contact duplex regions39–42. These helicases use auxillary domains, not the helicase core to bind the duplex RNA. While structural studies are required to determine how NPH-II binds to duplex and single stranded regions, one can speculate, based on analogies to other helicases, that the helicase core of NPH-II probably promotes binding to single stranded RNA. Duplex binding may be established by C- or N -terminal domains of NPH-II.
Different stages of the ATP hydrolysis cycle modulate RNA affinities of NPH-II
A central aim of our study was to illuminate connections between substrate binding and stages of the ATPase cycle during unwinding initiation. We found that affinities for both duplex and single stranded regions are modulated by the stages of the ATPase cycle. NPH-II binds to double stranded RNA without nucleotide, with the ATP ground state analog and with ADP, but not in the presence of the transition sate analog (Fig.1). Single stranded RNA is bound without nucleotide, and with ATP ground - and transition state analogs, but not with ADP (Fig.1). These findings show that NPH-II binds to RNA substrates with both single and duplex regions during the entire ATP hydrolysis cycle. This result explains how NPH-II can maintain contact to the substrate throughout many ATP turnovers during unwinding initiation. The ability of NPH-II to alternate binding between single strand and duplex region also rationalizes how the protein can remain localized at the junction throughout the initiation process18.
The analysis of NPH-II binding to a ssRNA region reveals interesting similarities to the closely related HCV NS3. Both, NPH-II and HCV NS3 bind single stranded nucleic acid tightly in the absence of nucleotide43 (Fig.5A). In the ATP ground state, the affinity decreases for both proteins25 (Fig.5B). Our data reveal an unanticipated facet of a seemingly straightforward result for NPH-II. While ADP-BeFx decreases the affinity of NPH-II for RNA (Fig.5), the dissociation rate constant of NPH-II from RNA is also markedly decreased, compared to the reaction without nucleotide (Fig. 7). This decrease is not offset by corresponding changes in the association rate constant. However, ADP-BeFx also alters the rate constant for interconversion between two conformations of the NPH-II RNA complex (E = 0.33, and E = 0.15), significantly favoring the population of the state at E = 0.33, from which NPH-II dissociates primarily, or even exclusively (Fig.7). Thus, dissociation events occur with a higher probability with ADP-BeFx than without nucleotide, even though the actual dissociation rate constant is decreased.
In contrast to the perhaps non-intuitive effects of the ATP ground state analog, the transition state analog ATP-AlFx impacts RNA binding largely as expected. It greatly stabilizes the interaction with RNA (Fig. 6C), similar to observations made with the DNA helicase Rep22. However, transitions between the two RNA-bound states of NPH-II still occur (Fig. 7B). This observation indicates that ADP-AlFx does not completely mimic the transition state, because no conversions are expected for a "true" transition state analog44. Notwithstanding, given that ADP-AlFx adopts a transition state - like geometry in numerous crystal structures of helicases20, the distinct characteristics of the NPH-II RNA complex with this analog render reasonable the interpretation that these characteristics correspond to features of the NPH-II RNA complex in the ATP transition state.
ADP promotes binding of NPH-II to duplex RNA, but greatly decreases the affinity of NPH-II for single stranded RNA (Figs.1,4). This decrease differs from the tight ssRNA binding induced by ADP for HCV-NS325. Thus, despite significant mechanistic similarities between NPH-II and HCV-NS3, the two proteins differ in the way how certain stages in the ATP hydrolyses cycle modulate RNA affinities.
The stages of the ATP hydrolysis cycle modulate interconversion between multiple conformations of the NPH-II RNA complex
Analysis of RNA binding by smFRET revealed unanticipated dynamics of the complex that NPH-II forms with single stranded RNA. Instead of adopting a single, defined conformational state at each stage of the ATP hydrolysis cycle, the NPH-II RNA complex is found in at least two distinct conformations (Figs.3, 5). The multiphasic kinetics observed for the transitions between these states suggest that states with identical FRET values might consist of even more kinetically distinct states. It is not clear to which extent the structure of NPH-II changes between the states, but the two distinct FRET values indicate at least minor differences in the bound RNA. However, the states have clearly different functional properties, reflected in the probability by which NPH-II dissociates from each state.
It is remarkable that the FRET values for the different states remained the same at the different stages of the ATPase cycle examined in our study. This observation suggests that the complex formed between NPH-II and ssRNA exists in a dynamic "equilibrium" of multiple bound conformations. The different stages of the ATP hydrolysis cycle direct rates by which the conformations interconvert, but they do not appear to induce a single specific conformation.
Similar, pre-existing equilibria that are altered by ligands have been observed with several other proteins. Examples include HIV Reverse Transcriptase, which displays two binding modes45,46. The "equilibrium" between these modes is altered by nucleotide or a non-nucleoside inhibitor. A further example is provided by the EF-G/ribosome interaction47. This complex also exists in two distinct, interconverting states whose ratio changes during different stages of translation47. Moreover, DNA polymerase I shifts rapidly between open and closed conformations in the absence of substrate, and this interconversion is changed in the presence of DNA or DNA and nucleotide48. In each of these cases, a pre-existing "equilibrium" of conformations is altered by ligands or other functional triggers, analogous to the change seen upon addition of ATP analogs to the NPH-II RNA complex.
The direct observation of distinct, interconverting states of the NPH-II RNA complex at defined stages of the ATP hydrolysis cycle highlights inherent dynamics of NPH-II bound to the RNA. As discussed above, functional consequences of these multiple states include an intricate control of substrate affinity. It is not clear to which extent the measured rate constants for the interconversion impact the NPH-II-RNA complex under ongoing ATP hydrolysis, given that ATP turnover is significantly faster than the interconversion kinetics measured with the non-hydrolyzable ATP analogs. Further research is thus needed to discern functional implications of the inherent dynamics of the NPH-II RNA complex. It is also important to investigate whether distinct conformational states of RNA-bound complexes exist for other helicases. If so, this feature would need to be considered in the functional interpretation of structural data.
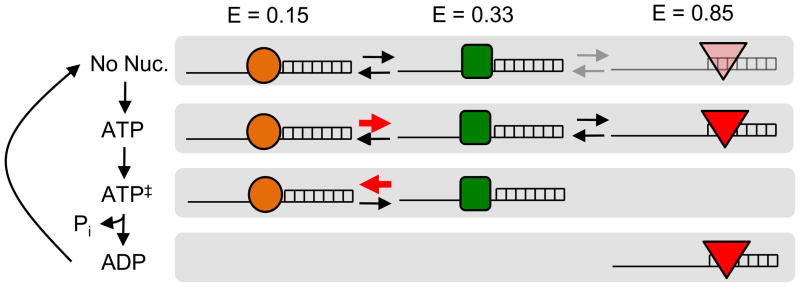
A basic, mechanistic model for unwinding initiation by NPH-II
Collectively, the data presented in this study suggest a basic model for the unwinding initiation process by NPH-II (Fig.8). Without nucleotide, NPH-II binds both, single strand and duplex regions. The complex between NPH-II and single strand RNA readily alternates between multiple conformations (E = 0.15, E = 0.33, Fig.7). ATP binding to NPH-II, as mimicked by ADP-BeFx, slows dissociation of NPH-II from the single stranded region and changes the kinetics of the transition between the multiple conformations of the NPH-II RNA complex. However, NPH-II is also able to bind the duplex region. In the ATP transition state, mimicked by ADP-AlFx, NPH-II no longer binds duplex RNA and binding to the single stranded region is essentially irreversible. Kinetics of the transition between the conformations of the NPH-II RNA complex change again, compared to the other stages of the ATP hydrolysis cycle. With bound ADP, i.e., after ATP hydrolysis and dissociation of the inorganic phosphate, NPH-II no longer binds the single stranded RNA, but associates with duplex RNA.
Figure 8. Basic model for unwinding initiation by NPH-II.
Different shapes mark the different states of NPH-II traversed during the initiation process.
This basic model explains the ability of NPH-II to maintain contact to the RNA during unwinding initiation, despite many ATP turnovers6,16,18. Binding to duplex RNA compensates for detachment from single stranded RNA and vice versa. The proposed initiation model does not explain why hundreds of ATP are often turned over prior to unwinding, and which specific events lead to the actual strand separation. We speculate that a certain conformation of the RNA duplex has to meet a specific conformation of the helicase, but that simultaneous occurrences of these arrangements are comparably rare.
Although we did not examine translocation by NPH-II, the proposed initiation model raises the attractive, albeit speculative possibility, that during directional duplex unwinding, NPH-II produces the forward step between the ATP transition state and the ADP-bound state, i.e., concurrent with the dissociation of inorganic phosphate. This scenario would be similar to the spring loaded translocation mechanism proposed for HCV-NS333.
MATERIALS & METHODS
Protein and RNA preparation
NPH-II was expressed in baculovirus infected insect cells and purified as described6. A bipartite RNA consisting of a 19 base pair duplex and a 24 nucleotide 3’ overhang was used in the sucrose gradient sedimentation and functional stoichiometric titration assays. The sequence of the overhang-containing strand, of which the first 19 bases were annealed to a complimentary RNA, was: 5'-AGCACCGUAAAGACGCAGCCAGCAUCAAUGACAUCAGCAUCAA. (duplex region underlined). For gradient sedimentation, the duplex RNA had the same sequence as the duplex region of this RNA, and the single-stranded substrate had the same sequence as the overhang. The substrate was radiolabeled with PNK and γP32-ATP. For smFRET experiments, the same bipartite substrate was used, except for two additional U’s at the 3’ end of the complimentary strand. The overhang-containing strand was labeled at the 3’ end with Cy3. The complimentary contained a biotin modification on the 3' end and a Cy5 on the 5' end. All RNAs were purchased from Dharmacon.
Sucrose Gradient Sedimentation
Reactions (40 mM Tris-HCl, pH 8.0, 0.5 mM MgCl2, 0.01% Nonidet P-40) containing 1 nM radiolabeled RNA, 100 nM NPH-II, and, where indicated, nucleotide - Mg2+ (3.5 mM) were loaded onto 4.6 mL 6-40% sucrose gradients. Where indicated, gradients also contained nucleotide-Mg2+ (3.5 mM). ADP-BeFx and ADP-AlFx were prepared as described 21. Gradients were centrifuged in a Beckman SW-55 Ti rotor at 42,000 rpm for 15.5 hours at 4ºC. 50 fractions (~ 90 μL each) were collected from the top of the tube. Size standards ovalbumin (3.6S, 45 kD) aldolase (7.35S, 149 kD), and catalase (11.3S, 240 kD) were added to a gradient with the same buffer conditions and centrifuged concurrently with the reactions.
Gradient fractions were analyzed by scintillation counter to determine the amount of RNA. Size standards were monitored by SDS-PAGE. To determine the relative amount of NPH-II in each fraction, gradient fractions were probed for ATPase activity. A volume of 7 μL of each fraction was incubated with reaction buffer (40 mM Tris-HCl pH 8.0, 0.5 mM MgCl2 and 0,01% Nonidet P-40) and 0.01 mM radiolabeled ATP for 10 minutes. Free phosphate was separated from ATP by TLC, and the fraction ATP hydrolyzed was determined with a PhosphorImager.
Functional Stoichiometric Titration
Unwinding reactions were performed in a buffer containing 40 mM Tris-HCl (pH 8.0), 0.5 mM MgCl2, and 0.01% Nonidet p-40. NPH-II (0.5 to 50 nM) was incubated with 20 nM RNA for 5 minutes, then ATP (3.5 mM final) and RNA scavenger (25 nt RNA, comprising the ss region of the substrate RNA, 1 μM final concentration) were added simultaneously to initiate single turnover unwinding. Aliquots were taken, the reaction was stopped in 2x Helicase Reaction Stop Buffer (50 μM EDTA, 1% SDS, 0.1 % BPB, 0.1% XC, 20% Glycerol) and samples were loaded onto a 15 % 29:1 Acryl:Bis gel containing TBE (4.5 mM Tris Base, 4.5 mM Boric Acid, 0.1 mM EDTA). The gel was run at 10 V/cm at room temperature, and subsequently dried, and exposed to a PhosphorImager screen. The fraction unwound substrate was quantified as described6. Unwinding amplitudes were determined by plotting the fraction unwound RNA over time and fitting the data to the integrated first order rate law, using the Kaleidagraph software.
Single Molecule FRET
Single molecule FRET (smFRET) measurements were performed with a custom-built TIR setup as described28. Cy3 and Cy5 labeled RNA samples were immobilized in a flow-cell coated with polyethylene glycol (PEG), which prevents non-specific protein adsorption. To generate the PEG-coating, a mixture of PEG-NHS (3,000–5,000 Da) and biotinylated PEG-NHS (3,000 Da) (SHEARWATER) was covalently attached to the flow-cells that had been amino-functionalized with Vectabond (VECTOR). Biotinylated RNA was immobilized via a streptavidin (MOLECULAR PROBES) link to the biotinylated PEG surface. Buffer conditions for the reactions were identical to those in binding and unwinding reactions, except that an oxygen scavenging system consisting of glucose oxidase (SIGMA) and catalase (SIGMA) was added with 5% glucose to prevent photobleaching49.
Fluorescent RNA molecules were excited using prism-based TIR49 and images were collected as described28 . Single-molecule time traces were collected at a rate of 10 frames per second using customized software. Fluorescence and corresponding FRET values were computed as described27. Dwell time analysis was performed with customized Matlab routines. Rate constants were determined from plots of cumulative dwell times (e.g., Fig.7A). Rate constants listed in Fig.7C were obtained by global fits of all plots against the kinetic scheme given in Fig.7B, using maximum likelihood analysis performed with Matlab. The errors represent the deviation of the obtained rate constants from the maximum likelihood analysis, assessed by a bootstrapping algorithm on the same data.
Supplementary Material
Acknowledgments
We thank the members of our laboratory for stimulating discussions. This work was supported by a grant from the NIH to E.J. (GM067700).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell. 2010;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 3.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gross CH, Shuman S. Vaccinia virions lacking the RNA helicase nucleoside triphosphate phosphohydrolase II are defective in early transcription. J Virol. 1996;70:8549–8557. doi: 10.1128/jvi.70.12.8549-8557.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raney KD, Sharma SD, Moustafa IM, Cameron CE. Hepatitis C virus non-structural protein 3 (HCV NS3): a multifunctional antiviral target. J Biol Chem. 2010;285:22725–22731. doi: 10.1074/jbc.R110.125294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 7.Kim DW, Gwack Y, Han JH, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem Biophys Res Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 8.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 9.Gwack Y, Kim DW, Han JH, Choe J. Characterization of RNA binding activity and RNA helicase activity of the hepatitis C virus NS3 protein. Biochem Biophys Res Commun. 1996;225:654–659. doi: 10.1006/bbrc.1996.1225. [DOI] [PubMed] [Google Scholar]
- 10.Serebrov V, Pyle AM. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature. 2004;430:476–480. doi: 10.1038/nature02704. [DOI] [PubMed] [Google Scholar]
- 11.Pang PS, Jankowsky E, Planet PJPAM. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 2002;21:1168–1175. doi: 10.1093/emboj/21.5.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor SD, Solem A, Kawaoka J, Pyle AM. The NPH-II helicase displays efficient DNA x RNA helicase activity and a pronounced purine sequence bias. J Biol Chem. 2010;285:11692–11703. doi: 10.1074/jbc.M109.088559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackintosh SG, Raney KD. DNA unwinding and protein displacement by superfamily 1 and superfamily 2 helicases. Nucleic Acids Res. 2006;34:4154–4159. doi: 10.1093/nar/gkl501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tai CL, Chi WK, Chen DS, Hwang LH. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3) J Virol. 1996;70:8477–8484. doi: 10.1128/jvi.70.12.8477-8484.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shuman S. Vaccinia virus RNA helicase. Directionality and substrate specificity. J Biol Chem. 1993;268:11798–11802. [PubMed] [Google Scholar]
- 16.Kawaoka J, Jankowsky E, Pyle AM. Backbone tracking by the SF2 helicase NPH-II. Nat Struct Mol Biol. 2004;11:526–530. doi: 10.1038/nsmb771. [DOI] [PubMed] [Google Scholar]
- 17.Beran RK, Bruno MM, Bowers HA, Jankowsky E, Pyle AM. Robust translocation along a molecular monorail: the NS3 helicase from hepatitis C virus traverses unusually large disruptions in its track. J Mol Biol. 2006;358:974–982. doi: 10.1016/j.jmb.2006.02.078. [DOI] [PubMed] [Google Scholar]
- 18.Kawaoka J, Pyle AM. Choosing between DNA and RNA: the polymer specificity of RNA helicase NPH-II. Nucleic Acids Res. 2005;33:644–649. doi: 10.1093/nar/gki208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shuman S. Vaccinia virus RNA helicase: an essential enzyme related to the DE-H family of RNA-dependent NTPases. Proc Natl Acad Sci U S A. 1992;89:10935–10939. doi: 10.1073/pnas.89.22.10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen B, et al. ATP ground- and transition states of bacterial enhancer binding AAA+ ATPases support complex formation with their target protein, sigma54. Structure. 2007;15:429–440. doi: 10.1016/j.str.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong I, Lohman TM. A two-site mechanism for ATP hydrolysis by the asymmetric Rep dimer P2S as revealed by site-specific inhibition with ADP-AlF4. Biochemistry. 1996;36:3115–3125. doi: 10.1021/bi9621977. [DOI] [PubMed] [Google Scholar]
- 23.Chakrabarti S, Jayachandran U, Bonneau F, Fiorini F, Basquin C, Domcke S, Le Hir H, Conti E. Molecular mechanisms for the RNA-dependent ATPase activity of Upf1 and its regulation by Upf2. Mol Cell. 2011;41:693–703. doi: 10.1016/j.molcel.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 24.Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci USA. 2010;107:521–528. doi: 10.1073/pnas.0913380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levin MK, Gurjar MM, Patel SS. ATP binding modulates the nucleic acid affinity of hepatitis C virus helicase. J Biol Chem. 2003;278:23311–23316. doi: 10.1074/jbc.M301283200. [DOI] [PubMed] [Google Scholar]
- 26.Harmon FG, Kowalczykowski SC. Coupling of DNA helicase function to DNA strand exchange activity. Methods Mol Biol. 2000;152:75–89. doi: 10.1385/1-59259-068-3:75. [DOI] [PubMed] [Google Scholar]
- 27.Matlock DL, et al. Investigation of translocation, DNA unwinding, and protein displacement by NS3h, the helicase domain from the hepatitis C virus helicase. Biochemistry. 2010;49:2097–2109. doi: 10.1021/bi901977k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Q, Fairman ME, Jankowsky E. DEAD-box-protein-assisted RNA structure conversion towards and against thermodynamic equilibrium values. J Mol Biol. 2010;368:1087–1100. doi: 10.1016/j.jmb.2007.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothenberg E, Ha T. Single-molecule FRET analysis of helicase functions. Methods Mol Biol. 2010;587:29–43. doi: 10.1007/978-1-60327-355-8_3. [DOI] [PubMed] [Google Scholar]
- 30.Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys J. 2004;86:2530–2537. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yodh JG, Stevens BC, Kanagaraj R, Janscak P, Ha T. BLM helicase measures DNA unwound before switching strands and hRPA promotes unwinding reinitiation. EMBO J. 2009;28:405–416. doi: 10.1038/emboj.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 33.Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu HP, Xun L, Xie XS. Single-molecule enzymatic dynamics. Science. 1998;282:1877–1882. doi: 10.1126/science.282.5395.1877. [DOI] [PubMed] [Google Scholar]
- 35.Levin MK, Patel SS. The helicase from hepatitis C virus is active as an oligomer. J Biol Chem. 1999;274:31839–31846. doi: 10.1074/jbc.274.45.31839. [DOI] [PubMed] [Google Scholar]
- 36.Tackett AJ, Chen Y, Cameron CE, Raney KD. Multiple full-length NS3 molecules are required for optimal unwinding of oligonucleotide DNA in vitro. J Biol Chem. 2005;280:10797–10806. doi: 10.1074/jbc.M407971200. [DOI] [PubMed] [Google Scholar]
- 37.Jennings TA, et al. NS3 helicase from the hepatitis C virus can function as a monomer or oligomer depending on enzyme and substrate concentrations. J Biol Chem. 2009;284:4806–4814. doi: 10.1074/jbc.M805540200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Byrd AK, Raney KD. Increasing the length of the single-stranded overhang enhances unwinding of duplex DNA by bacteriophage T4 Dda helicase. Biochemistry. 2005;44:12990–12997. doi: 10.1021/bi050703z. [DOI] [PubMed] [Google Scholar]
- 39.Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- 40.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 41.Richards JD, et al. Structure of the DNA repair helicase hel308 reveals DNA binding and autoinhibitory domains. J Biol Chem. 2008;283:5118–5126. doi: 10.1074/jbc.M707548200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levin MK, Patel SS. Helicase from hepatitis C virus, energetics of DNA binding. J Biol Chem. 2002;277:29377–29385. doi: 10.1074/jbc.M112315200. [DOI] [PubMed] [Google Scholar]
- 44.Schramm VL. Enzymatic transition states and transition state analog design. Annu Rev Biochem. 1998;67:693–720. doi: 10.1146/annurev.biochem.67.1.693. [DOI] [PubMed] [Google Scholar]
- 45.Liu S, Abbondanzieri EA, Rausch JW, Le Grice SF, Zhuang X. Slide into action: dynamic shuttling of HIV reverse transcriptase on nucleic acid substrates. Science. 2008;322:1092–1097. doi: 10.1126/science.1163108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abbondanzieri EA, et al. Dynamic binding orientations direct activity of HIV reverse transcriptase. Nature. 2008;453:184–189. doi: 10.1038/nature06941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, et al. Single-molecule structural dynamics of EF-G--ribosome interaction during translocation. Biochemistry. 2007;46:10767–10775. doi: 10.1021/bi700657d. [DOI] [PubMed] [Google Scholar]
- 48.Santoso Y, Joyce CM, Potapova O, Le Reste L, Hohlbein J, Torella JP, Grindley ND, Kapanidis AN. Conformational transitions in DNA polymerase I revealed by single-molecule FRET. Proc Natl Acad Sci U S A. 2010;107:715–720. doi: 10.1073/pnas.0910909107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.