Abstract
In Eastern Spain, almond trees have been cultivated in terraced orchards for centuries, forming an integral part of the Mediterranean forest scene. In the last decades, orchards have been abandoned due to changes in society. This study investigates effects of changes in land use from forest to agricultural land and the posterior land abandonment on soil microbial community, and the influence of soil physico-chemical properties on the microbial community composition (assessed as abundances of phospholipids fatty acids, PLFA). For this purpose, three land uses (forest, agricultural and abandoned agricultural) at four locations in SE Spain were selected. Multivariate analysis showed a substantial level of differentiation in microbial community structure according to land use. The microbial communities of forest soils were highly associated with soil organic matter content. However, we have not found any physical or chemical soil property capable of explaining the differences between agricultural and abandoned agricultural soils. Thus, it was suggested that the cessation of the perturbation caused by agriculture and shifts in vegetation may have led to changes in the microbial community structure. PLFAs indicative of fungi and ratio of fungal to bacterial PLFAs were higher in abandoned agricultural soils, whereas the relative abundance of bacteria was higher in agricultural soils. Actinomycetes were generally lower in abandoned agricultural soils, while the proportions of vesicular-arbuscular mycorrhyzal fungi were, as a general trend, higher in agricultural and abandoned agricultural soils than in forests. Total microbial biomass and richness increased as agricultural < abandoned agricultural < forest soils.
Keywords: Mediterranean soils; land use changes; phospholipids fatty acids, microbial community structure; biodiversity
1. Introduction
Understanding the biology and ecology of soils has been considered as a critical factor for the restoration and sustainable management of terrestrial ecosystems in the last years. Soil microorganisms play important roles in organic matter decomposition and nutrients cycles, and consequently provide an integrated vision of soil quality and health. Furthermore, microbiological properties are the most sensitive and rapid indicators of perturbations and land use changes. In this sense, the quantitative description of microbial community structure and diversity has aroused great interest in soil quality evaluation (Zelles, 1999).
In the mountainous region of the Mediterranean Basin of Spain, almond trees have been cultivated in terraced orchards for centuries. These crops are immersed in the Mediterranean forest scenery, part of a landscape mosaic where orchards are integrated into the forest areas. Cultivation of soils represent a type of land use with important effects on soil characteristics and microbiology. Continuous soil tillage leads to a great decrease in soil organic carbon, total nitrogen, cation exchange capacity, aggregate stability, water holding capacity and microbial biomass and activity in topsoils (Caldwell et al., 1999; Gardi et al., 2002). In addition, the incorporation of organic residues or fertilization can provoke alterations in the soil microbial community structure (Lundsquist et al., 1999; Calderón et al., 2000). In the last decades, almond orchards in Eastern Spain are being abandoned due to the low productivity of the crops, industrialization, or the development of tourism (Bonet, 2004). This phenomenon has led to an increase in vegetation cover (González-Bernáldez, 1991; Bonet, 2004), since abandoned fields are naturally colonized by the surrounding natural vegetation. In addition, after abandonment, soil properties such as organic matter content, soil structure and infiltration rate tend to improve, resulting in more effective protection again erosion (Kosmas et al., 2000). So, an improvement in soil quality is expected in these areas, as long as no erosive processes appear. However, there is little information about the impact of the abandonment of former arable soils on soil microbial community structure in the Mediterranean Basin, despite it being a common process in the entire region. Bound to the succession in vegetation, a succession in soil microbial communities should also be initialised. Thus, it is worth focusing on soil microbial community composition after land abandonment, since the functions of soil microbes are crucial for supporting plant growth, vegetation success and the reactivation of biogeochemical cycles, and, therefore, restoration studies should include below-ground community characterization. Furthermore, the understanding of the processes implied in the secondary forest succession initialised after crop abandonment is crucial to avoid degrading processes.
We used phospholipid fatty acid (PLFA) analysis to measure microbial community composition. PLFA analysis uses the lipids of the microbial membranes as biomarkers for specific groups of microorganisms, besides creating a profile or finger print of the community structure. As a consequence, rapid changes in soil microbial community structure can be detected by changes in the PLFAs pattern (Zelles, 1999). In addition, total concentration of PLFAs can be used as a measure of viable microbial biomass, since phospholipids are rapidly degraded after cell death (Zelles, 1997). The use of PLFA analysis has been used to study changes in soil microbial community structure across ecological succession as a consequence of different perturbations or management practices (Hedlund, 2002; Allison et al., 2005; DeGrood et al., 2005; Van der Wal et al., 2006).
An alternative and fast method to measure and monitor soil characteristics is the use of near infrared reflectance (NIR) spectroscopy. In the near infrared region, the radiation is absorbed by the different chemical bonds, such as C-H, N-H, S-H, C=O and O-H of the compounds present in the sample. Moreover, the radiation is absorbed in accordance with the concentration of these compounds. Thus, NIR spectra contain information about the compositional qualities of a soil sample. As a consequence, many authors have observed that NIR spectra contain information about physical, chemical and biological properties of soils, including variables related to the microbial community composition of the soil (Cozzolino and Moron, 2003; Chang and Laird, 2002; Rinnan and Rinnan, 2007; Zornoza et al., 2008).
In this paper, we selected four locations, with similar climate, vegetation composition, and land use history. All sites exhibit similar vegetation mosaic pattern, constituting forest, almond orchards and abandoned lands. Our study aims at investigating: (i) the effects of changes in land use from forest to arable and posterior land abandonment on soil microbial community, and (ii) the influence of soil physico-chemical properties on the microbial community structure.
2. Materials and Methods
2.1. Study sites and soil sampling
Samples were collected from four locations in the province of Alicante (SE Spain): Sierra de Crevillent (C), Sierra del Maigmó (M), Puig Campana (Pu) and Camara (Cam). An undisturbed forest soil was selected in each location. We selected native soils under natural and autochthonous vegetation, as close as possible to potential vegetation, which had not been recently disturbed by human action (>40 years). We also selected an agricultural soil dedicated to almonds, and a former almond orchard abandoned 10-15 years previous to sampling. Hence, for this study, three land uses were chosen: undisturbed forest (f), agricultural (a) and abandoned agricultural (ab). In all locations agricultural and abandoned agricultural soils were situated in the middle of wide forest zones, representing a landscape mosaic, characteristic of the mountainous areas of Eastern Spain. All forest soils have in common the presence of a tree stratum occupied by Pinus halepensis Miller (with Quercus rotundifolia Lam. in Pu). The understory vegetation is dominated by some shrub species and Brachipodium retusum (Pers.) Beauv. as the main herbaceous species. Abandoned almond orchards are characterized by various shrub species (dominated by Rosmarinus officinalis L., Rhamnus lycioides L., Helichrisum stoechas (L.) Moench, or Cistus albidus L.), belonging to the first stages of the succession of the vegetation series. All soils are situated on calcaric bedrock. An overview of the sites and soils characteristics is given in Table 1.
Table 1.
Sites and soils characteristics.
| Sitea | Land useb | Tmc | Pmc | Vegetation cover |
Soil type | Textured | pH | OMe | CO32− |
|---|---|---|---|---|---|---|---|---|---|
| °C | mm | % | SSS, 2006 | (% sand, silt, clay) | % | % | |||
| C | f | 95 | Calcixeroll | Loam (44, 43, 13) | 7.9 | 14.4 | 30 | ||
| C | a | 15.8 | 355 | 5 | Xerorthent | Clay (37, 34, 29) | 8.1 | 3.1 | 51 |
| C | ab | 30 | Xerorthent | Clay (37, 34, 29) | 8.1 | 3.8 | 52 | ||
| M | f | 70 | Xerorthent | Clay loam (56, 23, 21) | 7.9 | 11.0 | 59 | ||
| M | a | 18.2 | 302 | 5 | Xerorthent | Clay (51, 30, 29) | 8.2 | 1.5 | 66 |
| M | ab | 50 | Xerorthent | Clay loam (51, 32, 17) | 8.2 | 2.1 | 75 | ||
| Pu | f | 80 | Calcixeroll | Silty loam (46, 45, 9) | 7.6 | 17.9 | 25 | ||
| Pu | a | 15.6 | 424 | 5 | Xerorthent | Silty clay loam (17, 66, 17) | 7.7 | 1.9 | 59 |
| Pu | ab | 70 | Xerorthent | Silty loam (21, 70, 9) | 7.8 | 4.1 | 51 | ||
| Cam | f | 70 | Xerorthent | Clay loam (42, 35, 23) | 7.9 | 10.0 | 32 | ||
| Cam | a | 14.0 | 308 | 5 | Xerorthent | Clay (37, 36, 27) | 8.4 | 1.1 | 59 |
| Cam | ab | 50 | Xerorthent | Silty clay loam (27, 48, 25) | 8.3 | 1.9 | 54 |
C: Sierra de Crevillent; M: Sierra del Maigmó; Cam: Camara; Pu: Puig Campana.
Land uses: f (forest); a (agricultural); ab (abandoned agricultural).
Mean annual temperature (Tm) and mean annual precipitation (Pm).
Sand: 2-0.02 mm; silt: 0.02-0.002 mm; clay < 0.002 mm.
OM: organic matter.
A single sampling was carried out in July 2005. A plot of 200 m2 was defined for each land use at each location, where 5 soil samples (0-10 cm depth) were taken randomly from the mineral A horizon. Prior to soil analysis the samples were air-dried for a week. Afterwards, they were passed through a 2 mm mesh sieve, except for soil aggregate stability determination (4-0.25 mm). For all assays, the average value of two replicates per sample was used, and data have been expressed on an oven dry weight basis.
2.2. Analytical Methods
The following physical and chemical soil properties were assayed: pH and electrical conductivity (EC) were measured in deionised water (1:2.5 and 1:5 w/v, respectively); soil organic carbon (Corg) was determined by potassium dichromate oxidation (Nelson and Sommers, 1982); total nitrogen (N) was determined by the Kjeldahl method (Bremmer and Mulvaney, 1982); the soluble carbon (Csol) was extracted with 0.5 M K2SO4 and measured at 590 nm (Sims and Haby, 1971); cation exchange capacity (CEC) was measured by the method described by Roig et al. (1980); available phosphorus (P) was determined by the Burriel-Hernando method (Díez, 1982); water holding capacity (WHC) was assayed by the method exposed by Forster (1995); aggregate stability percentage (AS) was quantified using the method described by Roldán et al. (1994); available Ca, Mg, K and Na were extracted with 1N ammonium acetate (Knudsen et al., 1982) and measured by atomic absorption and emission spectrophotometry.
The following biochemical properties were assayed: microbial biomass carbon (MBC) was determined using the fumigation-extraction procedure (Vance et al., 1987); basal soil respiration (BSR) was monitored for 4 days at 55% WHC and 25°C with a multiple sensor respirometer (Micro-Oxymax, Columbus, OH, USA); urease activity was measured according to the method of Nannipieri et al. (1980); acid phosphatase activity was assayed by the method of Tabatabai and Bremmer (1969); the activity of β-glucosidase was determined according to Tabatabai (1982).
Phospholipid fatty acid (PLFA) analysis was carried out as described in Bossio et al. (1998). Briefly, fatty acids were extracted from 8g soil samples using chloroform:methanol:phosphate buffer. PLFAs were separated from neutral and glycolipid fatty acids on a solid phase extraction column (0.58 Si; Supelco Inc., Bellafonte, PA). After mild alkaline methanolysis, samples were analysed using a Hewlett Packard 6890 Gas Chromatograph with 25 m Ultra 2 (5% phenyl)- methylpolysiloxane column (J & W Scientific, Folsom, CA). Fatty acids were quantified by comparison of the peak areas with those of an internal standard 19:0 peak. The peaks were named using bacterial standards and identification software from the Microbial Identification System (Microbial ID, Inc., Newark, DE). Fatty acid nomenclature used was that described by Frostegard et al. (1993). The fatty acids i15:0, 15:0, a15:0, i16:0, 16:1ω7, i17:0, a17:0, cy17:0, 17:0, 18:1ω7 and cy19:0 were chosen to represent bacteria (Frostegard et al., 1993). The unsaturated PLFA 18:2ω6 was used as indicator of fungal biomass (Federle, 1986). PLFAs cy17:0, 18:1ω7c, cy19:0, 17:1ω9c, 16:1ω9c, 18:1ω9c and 15:1ω4c were chosen to represent Gram-negative [G−] bacteria (Zelles et al., 1994). The branched, saturated i14:0, i15:0, a15:0, i16:0, i17:0 and a17:0 were chosen to represent Gram-positive [G+] bacteria (Zelles et al., 1994). The PLFAs 10Me16:0, 10Me17:0 and 10Me18:0 were selected as indicators of actinomycetes biomass (Zelles et al., 1994). The PLFA 16:1ω5 was used as representative of vesicular-arbuscular mycorrhizal [VAM] fungi (Olsson et al., 1995). Two ratios were also calculated, fungal to bacterial PLFAs (fungi:bacteria) and Gram-positive:Gram negative bacteria (G−:G+ bacteria). The total biomass was estimated as the sum of all the extracted PLFAs [total PLFAs].
For NIR measurements we used a Fourier-Transform near infrared (FT-NIR) spectrophotometer (MPA, Bruker Optik GmbH, Germany), equipped with quartz beamsplitter and PbS detector. It is also equipped with an integrating macrosample sphere and rotating sample cup, allowing the scanning of large areas of the samples. NIR spectra of soil samples were obtained with the following procedure: aliquots of around 50 g of soil were placed in glass Petri-dishes, and scanned on reflectance mode from 12000 to 3800 cm−1. In each of the reflectance measurement, 64 scans were averaged. Samples were measured in duplicate. After this, they were averaged again. The time employed for the spectral measurement was approximately 1 min per sample. The resolution used for spectral analysis was 8 cm−1. Background corrections were made before each sample scan. The x-scale of each NIR spectrum was transformed from wavenumber to wavelengh, obtaining a spectrum with 1000 data of absorbance comprising from 830 to 2630 nm.
2.3. PLFA diversity indices
The richness (R) of the fatty acids was expressed as the total number of PLFAs present in each sample. The diversity of the fatty acids was calculated with the Shannon-index H:
| (1) |
where pi is the relative abundance of each fatty acid in the total sum, and R is the number of detected fatty acids. The equitability of the fatty acids was calculated with the Shannon’s evenness E (Shannon, 1948):
| (2) |
2.4. Statistical Analyses
PLFA data were converted to mol% of the total fatty acids concentration. Canonical Correspondence Analysis (CCA) was used to examine the relationship between the microbial community composition and soil characteristics. Samples with similar PLFA profiles have similar scores and thus will group closer together when plotted. Soil physical and chemical properties were tested for significant contribution to the explanation of the variation in the PLFA data with the Monte Carlo permutation test (P<0.05). Only soil properties that were significantly correlated with factor in the CCA were included in the plots. Soil properties are represented by vectors. Vectors of greater magnitude and forming smaller angles with an axis are more strongly correlated with that axis. CCA can be influenced by rare fatty acids. Fatty acids that only appear in a few samples are usually unreliably represented because they have values near the detection limit. Hence, fatty acids that were present in less than 25% of the samples were omitted.
We carried out a one-way ANOVA for each location separately to investigate the differences between land uses regarding microbial groups and ratios based on PLFA biomarkers and diversity indices. The separation of means was made according to Tukey’s honestly significant difference at P<0.05. We also developed Pearson correlations between the physical and chemical properties and the different PLFA microbial groups.
A discriminant analysis (DA) was independently performed for each set of soil variables: (1) soil physical, chemical and biochemical properties, (2) NIR data, and (3) PLFAs. The main purpose was to confirm that changes in land management had significantly changed soil microbial community structure, as well as to compare the sensitivity of PLFAs against other soil properties and soil spectral characteristics, so that soil samples can be correctly classified by land uses. The discriminant functions produced probability of membership of a given sample in each respectively land use class. A given sample was assigned to the land use class with the highest probability. With the aim of reducing the number of variables with NIR data, a previous principal component analysis (PCA) was applied to the NIR spectral matrix (60 samples ×1000 wavelengths), extracting the first 35 principal components (PCs), that explained the 99.99% of the variability. The factor scores of the first 35 PCs were used, reducing the size of the matrix (from 60 × 1000 to 60 × 35). Thus, the DA was performed on the 60 samples which represented 35 variables (the factor scores of these 35 PCs) in 3 groups or categories.
CCA was performed using CANOCO for Windows, Version 4.5, while ANOVA, correlations and DA were performed with the software SPSS for Windows, Version 13.0.
3. Results
3.1. Multivariate analysis
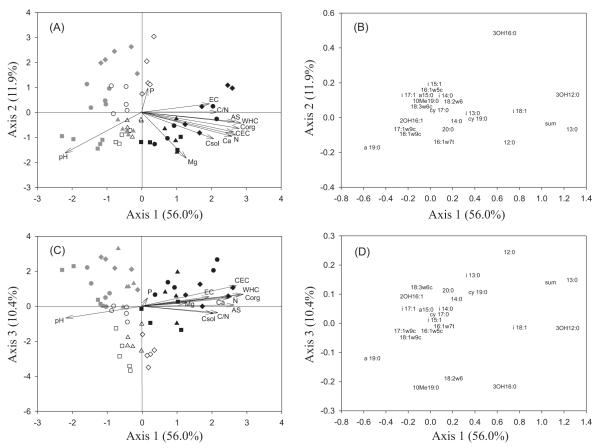
The CCA performed with the PLFA data showed that a 78.3% of total variation could be explained by the first three axes, and soil samples clearly clustered by land use (Fig. 1). Axis 1 separated forest soils from the other land uses, and explained 56.0% of the variation. Axis 2 explained 11.9% of the variation, and separated, only for agricultural and abandoned agricultural soils, C and Cam locations from M and Pu locations. Axis 3, which explained 10.4% of the variation, separated agricultural from abandoned agricultural soils. All soil characteristics, with the exception of Na and K, were significant (P<0.05) in explaining the variation in PLFA data. The variables Corg, N, C/N ratio, Csol, WHC, CEC, AS, EC, Mg and Ca showed a positive association with axis 1, and thus with forest soils, while pH was negatively associated with this land use. Thus, soil organic matter content largely accounts for variations in microbial community composition of forest soils, distributed in a positive direction along axis 1. Changes in microbial community composition along axis 2 were associated with both relatively higher values of P and lower values of Mg and pH. No soil property was related to the gradient along axis 3, differentiating between agricultural and abandoned agricultural soils.
Figure 1.
Samples and soil characteristics biplots (left) and loadings plots (right) from CCA performed on the relative concentration of PLFAs in all land uses: agricultural (grey symbols), abandoned agricultural (open symbols), and forest (black symbols). Locations: Sierra de Crevillent (triangles), Sierra del Maigmó (circles), Puig Campana (diamonds) and Camara (squares).
Corg: soil organic carbon; N: total nitrogen; Csol: soluble carbon; EC: electrical conductivity; WHC: water holding capacity; AS: aggregate stability; CEC: cation exchange capacity; sum: sum of 19:1ω11c, 19:1 and an unknown fatty acid.
Certain PLFAs were strongly associated with the different land uses. Forest soils were characterised by high concentrations of the saturated PLFAs 12:0, 13:0 and 3OH12:0, the monounsaturated i18:1, and the sum of the PLFAs 19:1ω11c, 19:1 and an unknown fatty acid. Agricultural soils (with positive values along axis 3) were associated with higher concentrations of the branched fatty acids a15:0 and i17:1, and the unsaturated fatty acids 2OH16:1 and 18:3ω6c. Abandoned agricultural soils (with negative values along axis 3) were characterised by high concentrations of the fungal PLFA 18:2ω6, and the branched saturated fatty acids 10Me19:0 and a19:0. The most positive loading score for axis 2 was found for the fatty acid 30H16:0, associated with samples from Pu and M locations.
3.2. PLFA biomarkers
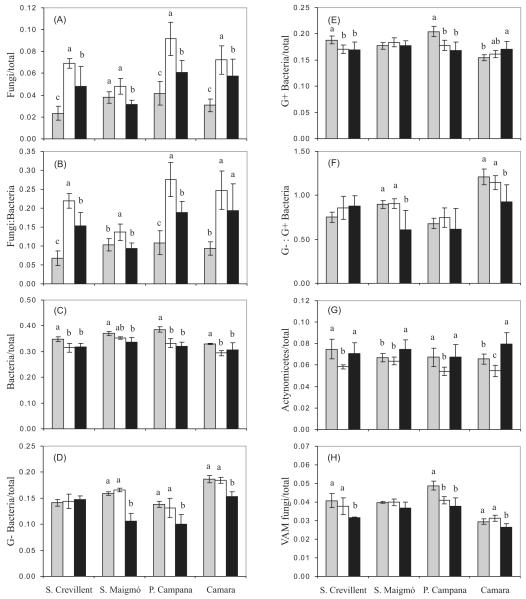
Microbial biomass, assessed as total PLFAs, showed the following trend in all locations: agricultural < abandoned agricultural < forest soil (Table 2). Specific PLFAs as biomarkers were quantified (Fig. 2). The proportion of the total PLFAs attributable to fungi was significantly higher (P<0.05, Fig. 2a) in abandoned agricultural soils, with a general decrease with abandoned agricultural soils > forest soils > agricultural soils. The ratio of fungal to bacterial PLFAs showed the same trend detected with the PLFA indicative of fungi (Fig. 2b). The relative abundance of bacteria was higher (P<0.05, Fig. 2c) in agricultural soils, whereas G− bacteria abundance was lower in forest soils, with no differences between agricultural and agricultural soils (Fig. 2d). We observed a high variability in the relative abundance of G+ bacteria in terms of location, although in all cases, there was no significant difference between forest and abandoned agricultural soils (P>0.05, Fig. 2e). The ratio G−:G+ bacteria (Fig. 2f) showed the same trend observed with G− bacteria. The specific PLFAs indicative of actinomycetes were generally lower in abandoned agricultural soils (Fig. 2g), while VAM fungi proportions were as a general trend higher in agricultural and abandoned agricultural soils (Fig. 2h).
Table 2.
Microbial biomass and PLFA diversity indices (means and standard deviation) for each land use and location
| Soil properties | Sierra de Crevillenta | Sierra del Maigmó | Puig Campana | Camara | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| f | a | ab | f | a | ab | f | a | ab | f | a | ab | |
| Total PLFA (nmol g−1) | 210 (57)a | 67 (24)b | 77 (16)b | 152 (53)a | 33 (8)c | 52 (9)b | 178 (82)a | 36 (16)c | 93 (15)b | 125 (38)a | 36 (13)c | 70 (14)b |
| Richness | 45 (3)a | 41 (3)b | 40 (1)b | 43 (3)a | 36 (2)c | 39 (2)b | 49 (3)a | 38 (2)b | 45 (2)a | 44 (3)a | 37 (3)c | 39 (1)b |
| Shannon index H | 3.20 (0.03) | 3.20 (0.05) | 3.12 (0.03) | 3.14 (0.05) | 3.18 (0.02) | 3.19 (0.03) | 3.16 (0.02)b | 3.27 (0.06)a | 3.12 (0.04)b | 3.16 (0.05) | 3.18 (0.03) | 3.12 (0.04) |
| Shannon Evenness E | 0.84 (0.02)b | 0.86 (0.02)a | 0.85 (0.01)a | 0.83 (0.03)c | 0.88 (0.01)a | 0.87 (0.02)b | 0.81 (0.02)b | 0.90 (0.03)a | 0.82 (0.02)b | 0.79 (0.07)c | 0.88 (0.02)a | 0.85 (0.01)b |
Land uses: f (forest), a (agricultural) and ab (abandoned agricultural).
Separately for each location, values with different letters indicate significant differences (P<0.05) among land uses after one-way ANOVA. Values with no letter are not significantly different (P>0.05).
Figure 2.
Values of various PLFA biomarkers (mean ± standard deviation) for all land uses in each location ( agricultural; □ abandoned agricultural; ■ forest). A one-way ANOVA and Tukey tests (P<0.05) were used to compare differences in land use for each location separately. Letters above the bars indicate significant differences between land uses in each location. Bars with no letter within the same location are not significantly different (P>0.05). G−: Gram-negative; G+: Gram-positive; VAM: vesicular-arbuscular mycorrhyzal.
agricultural; □ abandoned agricultural; ■ forest). A one-way ANOVA and Tukey tests (P<0.05) were used to compare differences in land use for each location separately. Letters above the bars indicate significant differences between land uses in each location. Bars with no letter within the same location are not significantly different (P>0.05). G−: Gram-negative; G+: Gram-positive; VAM: vesicular-arbuscular mycorrhyzal.
Total PLFAs were strongly correlated with most variables, the strongest correlation established with MBC (r=0.91; P<0.001) (Table 3). The relative abundance of bacteria was negatively correlated with Csol and Mg (P<0.001). The proportion of G− bacteria were negatively correlated with Corg, WHC, AS, CEC and MBC, but positively correlated with pH (P<0.001). The ratio G−:G+ bacteria was also positively correlated with pH.
Table 3.
Correlation coefficients (r values) for relationships between physical, chemical and biochemical properties, and the PLFA biomarkers.
| Total PLFA |
Fungi | Fungi:bacteria | Bacteria | G− | G+ | G− : G+ | Actinomycetes | VAM fungi | |
|---|---|---|---|---|---|---|---|---|---|
| Corg | 0.89* | 0.08 | 0.12 | −0.35 | −0.48* | −0.20 | −0.32 | 0.25 | −0.14 |
| N | 0.90* | 0.04 | 0.09 | −0.36 | −0.44 | −0.18 | −0.31 | 0.31 | −0.19 |
| C/N | 0.58* | 0.25 | 0.26 | −0.22 | −0.38 | −0.12 | −0.30 | 0.03 | 0.01 |
| Csol | 0.61* | 0.22 | 0.27 | −0.50* | −0.24 | −0.19 | −0.14 | 0.37 | −0.40 |
| pH | −0.53* | −0.24 | −0.18 | −0.11 | 0.54* | −0.30 | 0.56* | −0.14 | −0.37 |
| EC | 0.54* | 0.14 | 0.13 | 0.08 | −0.38 | 0.11 | −0.35 | 0.35 | −0.07 |
| WHC | 0.86* | 0.11 | 0.15 | −0.30 | −0.50* | −0.14 | −0.39 | 0.26 | −0.09 |
| AS | 0.70* | 0.10 | 0.11 | −0.21 | −0.49* | 0.06 | −0.37 | 0.31 | −0.01 |
| CEC | 0.87* | −0.02 | 0.02 | −0.28 | −0.49* | −0.13 | −0.36 | 0.31 | −0.11 |
| P | 0.06 | −0.03 | −0.10 | 0.29 | −0.06 | 0.22 | −0.12 | 0.08 | 0.17 |
| Ca | 0.84* | 0.05 | 0.10 | −0.39 | −0.34 | −0.19 | −0.31 | 0.28 | −0.16 |
| Mg | 0.59* | 0.02 | 0.11 | −0.50* | −0.03 | −0.27 | 0.07 | 0.51* | −0.61* |
| Na | 0.14 | 0.17 | 0.16 | −0.07 | −0.24 | 0.08 | −0.24 | 0.20 | −0.15 |
| K | 0.50* | 0.07 | 0.10 | −0.27 | −0.43 | −0.06 | −0.34 | 0.23 | −0.11 |
| MBC | 0.91* | 0.25 | 0.29 | −0.40 | −0.48* | −0.21 | −0.31 | 0.16 | −0.11 |
Corg: soil organic carbon; N: total nitrogen; Csol: soluble carbon; EC: electrical conductivity; WHC: water holding capacity; AS: aggregate stability; CEC: cation exchange capacity; MBC: microbial biomass carbon; G−: Gram-negative bacteria; G+: Gram-positive bacteria; VAM: vesicular-arbuscular mycorrhyzal.
Significant correlation at P<0.001.
3.3. Separation of land uses by means of Discriminant Analysis
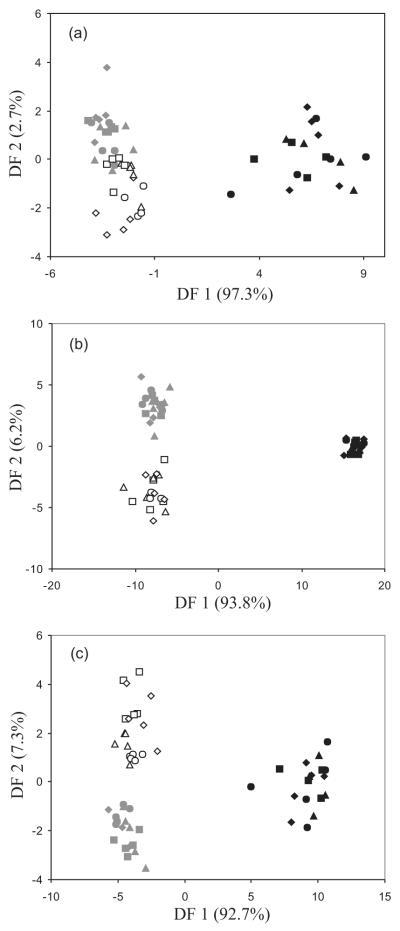
The DA performed on the set of physical, chemical and biochemical variables resulted in a good classification of samples in terms of the three land uses, and more than 80% of the samples were correctly classified. Using the DA with NIR data and PLFAs resulted in an excellent classification of land uses, and 100% of the samples were correctly classified (Fig. 3).
Figure 3.
Discriminant analysis with soil physical, chemical and biochemical properties (a), factor scores from NIR absorbance data (b), and PLFAs (c) of samples taken under three land uses: agricultural (grey symbols), abandoned agricultural (open symbols), and forest (black symbols). Locations: Sierra de Crevillent (triangles), Sierra del Maigmó (circles), Puig Campana (diamonds) and Camara (squares).
3.4. PLFAs diversity indices
As a general pattern, richness increased significantly as follows: agricultural soil < abandoned agricultural soil < forest soil, ranging from 36 in the agricultural soil from Sierra del Maigmó to 49 in the forest soil from Puig Campana (Table 2). The Shannon-index H showed no significant differences among land uses, except for the agricultural soil in Puig Campana, which was significantly the highest (3.27). The Shannon evenness E revealed an increasing trend with forest soil < abandoned agricultural soil < agricultural soil, with values ranging from 0.79 in the forest soil from Camara to 0.90 in the agricultural soil from Puig Campana.
4. Discussion
4.1. Soil microbial community structure regarding land uses
Soil organic carbon content increased with agricultural soil < abandoned agricultural soil < forest soil, owing to the highest inputs as a result of the highest vegetation cover. As a consequence, microbial biomass, which is strongly correlated with organic carbon, followed the same trend. The total concentration of PLFAs is strongly correlated with microbial biomass carbon determined by the fumigation-extraction method, which verifies the reliability of PLFAs as indicative of microbial biomass. Strong correlations between these two methods have been often reported. Nonetheless, the correlation coefficient achieved in this research (0.91) is high as compared with other studies, where r values ranged between 0.60 and 0.75 (Feng et al., 2003; Bünemann et al., 2004; Allison et al., 2005; Hackl et al., 2005).
Differences found among zones with regard to soil microbial communities are explained by the fact that the size, structure and activity of microbial communities depend on climatic, edaphic and topographic characteristics, as well as vegetation and management history, specific to each zone. However, our results suggest a substantial level of differentiation in microbial community structure according to land use, irrespective of location. In all locations, microbial biomass increased from agricultural soil to forest soil, accompanied by increases in richness and similar variations in microbial community structure. Multivariate analysis showed that the microbial community structure of forest soils was highly associated with soil organic matter content. Probably, microorganisms responded to an increment in organic matter levels in soil, involving in increases in carbon and nutrients availability and increments in the capacity for water retention. On the contrary, the differences in microbial community composition found between agricultural and abandoned agricultural soils were not explained by any physical or chemical soil property. Thus, the abandonment of the perturbation caused by agricultural activity per se, and the associated changes in vegetation, rather than direct changes in soil properties, may have led to shifts in soil microbial community structure. It has often been reported that plant species composition influences microbial communities by the release of radical exudates (Grayston et al., 1998; Yang and Crowley, 2000).
Abandoned agricultural soils had the highest relative fungi abundance. Fungi communities play a dominant role in fresh organic matter decomposition (Dilly et al., 2001). In abandoned agricultural soils, vegetation cover is higher than in agricultural soils. Thus, there is an increased incorporation of litter into the soil, which facilitates the development of fungi populations. As other authors have demonstrated previously, the abandonment of agricultural practices leads to increments in fungi dominance (Hedlund, 2002; Allison et al., 2005). Relative bacteria abundance was highest in agricultural soils. Bacteria tend to dominate decomposition and nutrient cycles in soils that are tilled and fertilized (Lovell et al., 1995; Allison et al., 2005).
The ratio of fungal to bacterial PLFAs increased after agriculture abandonment. When the mature ecosystem is reached, represented by forest, relative fungi abundance and the ratio fungi:bacteria descend. We think that increments in fungi abundances after land abandonment are related to the cessation of perturbations (tillage and fertilization), as often reported in other studies (Allison et al., 2005; van der Wall et al., 2006). Tillage provokes the break of hyphae. Hence, the interruption of this activity permits the development of fungi, stimulated by greater contributions of organic matter. However, succession towards a mature community may have caused a descent and stabilization in fungi abundance, probably as a consequence of the increment in species richness, and so, increases in competence. Allison et al. (2005) also observed that although in the first years after field abandonment fungi increased, in later successional stages (a tallgrass prairie), they started decreasing. The authors explained these findings by the fact that C inputs in the grassland mainly come from root exudates rapidly metabolised, which limit the development of fungi as this group use more recalcitrant sources of C. Nonetheless, this argument is not valid for explaining our results, since the incorporation of recalcitrant organic matter is high in the topsoil of conifer forests.
Although fungi are considered the major contributors to microbial biomass in forest soils as well as other soils with high organic matter content (Bailey et al., 2002), the fungal to bacteria ratio in this study is low, also reported in other studies that developed PLFA analysis (Bååth and Anderson, 2003; Certini et al., 2004; Allison et al., 2005; van der Wall et al., 2006), as compared with direct microscopic methods or selective respiratory inhibition (Alphei et al., 1995; Ananyeva et al., 2006; Busse et al., 2009). However, to be able to calculate actual biomass values from the ratio of fungal to bacterial PLFAs one needs reliable conversion factors. Such conversion factors will be different for fungi and bacteria, but also different for the PLFA and other techniques such as the selective inhibition technique. Since there are currently no reliable conversion factors, it is meaningless to compare the actual values of the fungal to bacterial ratios estimated with different methods (Bååth and Anderson, 2003). It is also probable that more basic research on the concentration of specific indicators PLFAs in soil bacteria and fungi is necessary to obtain the full quantitative information from this method. In fact, a major disadvantage of PLFA analysis is the fact that none of the indicator is fully specific for a certain microbial group (Joergensen and Wichern, 2008). However, changes in this ratio due to changes in land use should be reliable since the same analytical procedure was used and the same extraction efficiency was guaranteed.
The highest proportions of G− bacteria in agricultural and abandoned agricultural soils may be the result of higher pH values in these soils in comparison with forest soils (significant correlations have been found between G− bacteria and pH, Table 3). High values of pH favour the growth of bacteria populations (Nodar et al., 1992). Furthermore, Frostegard et al. (1993) observed that increments in soil pH lead to shifts in the bacterial community, with increments in G− bacteria.
The highest relative abundances in VAM fungi were found in agricultural soils, and in some cases in abandoned agricultural soils. In agricultural soils there is a higher proportion of herbaceous species, which generally form this type of mycorrhizal association (Rillig, 2004). Although no correlation was found between VAM fungi and aggregate stability with all soils, a significant positive correlation was observed in abandoned agricultural soils (r=0.72; P<0.001). This correlation seems to partly corroborate other researches which highlight the importance of VAM fungi hyphae in the stabilization of soil aggregates (Miller and Jastrow, 1990; Rillig, 2004; Allison et al., 2005), mainly in the first stages of succession after land abandonment. Mycorrhizal fungi directly favour the aggregation of particles by the extension of their mycelium or indirectly by the release of extracellular exudates (Miller and Jastrow, 1990). This relationship has only been observed in abandoned agricultural soils probably because in agricultural soils the continuous tillage provokes the breakage of aggregates. In forest soils, aggregates are mainly stabilised by the higher amount of organic matter.
The results of the DA carried out with the PLFAs supported the presence of different community composition among the three land uses, since all samples were correctly discriminated according to land use. Soil samples classification using PLFA data was better than that obtained with the set of physical, chemical and biochemical properties. Thus, these results confirm the sensitivity of parameters relating to soil microbial community composition in evaluating soil quality, perturbations and effects of management practices, as microbial communities respond very quickly to changes in land use, and give a result earlier than measurements of other variables of a different nature, including total microbial biomass or metabolic activity. Also excellent results were achieved with the NIR spectra. In fact, distances of each sample to the centroid of its group (forest, agricultural or abandoned agricultural) were lower when using NIR data than with PLFAs, mainly for forest soils (see Fig. 3), supporting the high sensitivity of NIR spectra to land use changes. In this sense, NIR spectra offer an integrated vision of soil conditions, since synthesize information regarding mineralogy, soil chemistry, soil biology, organic matter and physical attributes (Cohen et al., 2005). NIR spectrum could be considered as an integrated vision of soil quality, and as consequence offers an integrated vision of perturbations. As a consequence, NIR spectra have been satisfactorily used for land use classification (Velasquez et al., 2005; Elliot et al., 2007). In addition, Johnson et al. (2003) observed that the grouping of soil samples based on their soil reflectance properties was similar to the grouping based on DNA fingerprinting. Thus, since microbial community composition is related to reflectance properties, these techniques could be promising at providing insight into microbial distributions at the landscape scale.
4.2. PLFA diversity indices
Increases in microbial biomass were associated with increases in richness, with significant positive correlations among both parameters (r=0.76; P<0.001). DeGrood et al. (2005) also found increases in richness bound to increases in microbial biomass after the restoration of degraded serpentine soils. It has been reported that increments in biomass must be accompanied by increments in diversity in order to facilitate sustainable restoration on disturbed sites (Bradshaw, 1984). Furthermore, the increment of richness has been proposed as a basic objective in the management of degraded ecosystems (Bonet, 2004). Nevertheless, no correlation has been found between microbial biomass and the Shannon index. Thus, although forest soils showed the highest variety of PLFAs, and so a higher variety of groups of microorganisms, this did not entail highest biodiversity in terms of the relative abundance of each group. In agricultural and abandoned agricultural soils, even though having lower richness, microbial groups were better balanced, and there were no differences in the Shannon index between forest and agricultural soils. Furthermore, regarding the Shannon evenness, we observed that in the zones with lower richness and biomass (agricultural soils), the microbial groups were represented more equitably.
A possible explanation for these findings is that, in agricultural soils, microorganisms are mainly “r” strategists, fast colonisers with high growth rates, as they are characteristic of altered environments. Under these conditions, all microbial groups can colonise the environment with proportional increments in their abundance, according to the availability of substrates. On the contrary, with progressing succession, the detritus food webs would become more complex. Then, slower growing specialists, the “K” strategists, occupy specific niches. This may explain why the Shannon evenness was lower.
5. Conclusions
Different land uses support different microbial community structures. Soil organic matter content seems to be the main agent responsible for the different microbial community structure in forest soils. Agricultural soils have the highest relative abundance of bacteria, probably due to tillage and fertilisation. The abandonment of agriculture has led to increments in microbial biomass, richness and shifts in the microbial community structure, probably due to the cessation of tillage and the associated changes in vegetation, rather than direct changes in soil properties. The principal effect has been the increment in the ratio of fungal to bacterial PLFAs. The fact that increments in microbial biomass are accompanied by increments in richness contributes to reinforce the hypothesis of ecosystem recovery. Future studies should evaluate if the vegetation and microbial community composition of former agricultural soils with higher periods of abandonment move towards advanced successional stages, which would really guarantee the recovery of ecosystems after the cessation of agriculture in the mountainous areas of Eastern Spain.
Acknowledgements
This research was supported by the CICYT co-financed FEDER project CGL2006-011107-C02-01/BOS. Support for PLFA analysis was supported by the National Institute of Environmental Health Sciences (NIEHS), NIH. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of these agencies. R. Zornoza and V. Arcenegui acknowledge a grant from the Ministry of Education and Science of the Government of Spain and Caja de Ahorros del Mediterráneo, respectively.
Potential referees
C. Garcia. Department of Soil and Water Conservation and Organic Waste Management, Centro de Edafologia y Biologia Aplicada del Segura (CEBAS-CSIC), P.O. Box 4195, 30080 Murcia, Spain. Tel. +34 968 396 325. cgarizq@cebas.csic.es
M. Schloter. Institute of Soil Ecology, GSF-National Research Center for Environment and Health, Ingolstadter Landstr. 1, 85764 Neuherberg, Germany. Tel. +49 89 3187 2304. schloter@gsf.de
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allison VJ, Miller RM, Jastrow JD, Matamala R, Zak DR. Changes in soil microbial community structure in a tallgrass prairie chronosequence. Soil Sci. Soc. Am. J. 2005;69:1412–1421. [Google Scholar]
- Alphei J, Bonkowski M, Scheu S. Application of the selective inhibition method to determine bacterial:fungal ratios in three beechwood soils rich in carbon – optimization of inhibitor concentrations. Biol. Fertil. Soils. 1995;19:173–176. [Google Scholar]
- Ananyeva ND, Susyan EA, Chernova OV, Chernov IV, Makarova OL. The ratio of fungi and bacteria in the biomass of different types of soil determined by selective inhibition. Microbiology. 2006;75:702–707. [PubMed] [Google Scholar]
- Bååth E, Anderson T-H. Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol. Biochem. 2003;35:955–963. [Google Scholar]
- Bailey VL, Smith JL, Bolton H., Jr. Fugal-to-bacterial ratios in soils investigated for enhanced C sequestration. Soil Biol. Biochem. 2002;34:997–1007. [Google Scholar]
- Bonet A. Secondary succession of semi-arid Mediterranean old-fields in south-eastern Spain: insights for conservation and restoration of degraded lands. J. Arid Environ. 2004;56:213–233. [Google Scholar]
- Bossio DA, Scow KM, Gunapala N, Graham KJ. Determinants of soil microbial communities: effects of agricultural management, season, and soil type on phospholipid fatty acid profiles. Microb. Ecol. 1998;36:1–12. doi: 10.1007/s002489900087. [DOI] [PubMed] [Google Scholar]
- Bradshaw AD. Ecological principles and land reclamation practice. Landscape Plan. 1984;11:35–48. [Google Scholar]
- Bremmer JM, Mulvaney CS. Nitrogen total. In: Page AL, Miller RH, Keeney DR, editors. Methods of soil analysis, Part 2, Chemical and microbiological properties. American Society of Agronomy; Madison: 1982. pp. 595–624. [Google Scholar]
- Bünemann EK, Bossio DA, Smithson PC, Frossard E, Oberson A. Microbial community composition and substrate use in a highly weathered soil as affected by crop rotation and P fertilization. Soil Biol. Biochem. 2004;36:889–901. [Google Scholar]
- Busse MD, Sánchez FG, Ratcliff AW, Butnor JR, Carter EA, Powers RF. Soil carbon sequestration and changes in fungal and bacterial biomass following incorporation of forest residues. Soil Biol. Biochem. 2009;41:220–227. [Google Scholar]
- Calderón FJ, Jackson LE, Socw KM, Rolston DE. Microbial responses to simulated tillage in cultivated and uncultivated soils. Soil Biol. Biochem. 2000;32:1547–1559. [Google Scholar]
- Caldwell BA, Griffiths RP, Sollins P. Soil enzyme response to vegetation disturbance in two lowland Costa Rican soils. Soil Biol. Biochem. 1999;31:1603–1608. [Google Scholar]
- Certini G, Campbell CD, Edwards AC. Rock fragments in soil support a different microbial community form the fine earth. Soil Biol. Biochem. 2004;36:1119–1128. [Google Scholar]
- Chang C, Laird DA, Mausbach MJ, Hurburgh CR., Jr. Near-Infrared Reflectance Spectroscopy-Principal Components Regression Analyses of soil properties. Soil Sci. Soc. Am. J. 2001;65:480–490. [Google Scholar]
- Cohen MJ, Prenger JP, DeBusk WF. Visible-Near infrared reflectance spectroscopy for rapid, non-destructive assessment of wetland soil quality. J. Environ. Qual. 2005;34:1422–1434. doi: 10.2134/jeq2004.0353. [DOI] [PubMed] [Google Scholar]
- Cozzolino D, Morón A. The potencial of near-infrared reflectance spectroscopy to analyse soil chemical and physical characteristics. J. Agric. Sci. 2003;140:65–71. [Google Scholar]
- DeGrood SH, Claassen VP, Scow KM. Microbial community composition on native and drastically disturbed serpentine soils. Soil Biol. Biochem. 2005;37:1427–1435. [Google Scholar]
- Díez JA. Consideraciones sobre la utilización de la técnica extractiva de Burriel-Hernando para la evaluación de fósforo asimilable en suelos. An. Edafol. Agrobiol. 1982;41:1345–1353. [Google Scholar]
- Dilly O, Bartsch S, Rosenbrock P, Buscot F, Munch JC. Shifts in physiological capabilities in the microbiota during the decomposition of leaf litter in a black alder (Alnus glutinosa (Gaertn.)L.) forest. Soil Biol. Biochem. 2001;33:921–930. [Google Scholar]
- Elliott GN, Worgan H, Broadhurst D, Draper J, Scullion J. Soil differentiation using fingerprint Fourier transform infrared spectroscopy, chemometrics and genetic algorithm-based feature selection. Soil Biol. Biochem. 2007;39:2888–2896. [Google Scholar]
- Federle TW. Microbial distribution in soil. In: Megusar F, Gantar M, editors. Perspectives in microbial ecology. Slovene Society for Microbiology; Ljubljana: 1986. pp. 493–498. [Google Scholar]
- Feng Y, Motta AC, Reeves DW, Burmester CH, van Santen E, Osborne JA. Soil microbial communities under conventional-till and no till continuous cotton systems. Soil Biol. Biochem. 2003;35:1693–1703. [Google Scholar]
- Forster JC. Soil physical analysis. In: Alef K, Nannipieri P, editors. Methods in Applied Soil Microbiology and Biochemistry. Academic Press Inc.; San Diego, CA: 1995. pp. 106–111. [Google Scholar]
- Frostegard A, Bååth E, Tunlid A. Shifts in the composition of soil microbial communities in limed forests as revealed by phospholipid fatty acid analysis. Soil Biol. Biochem. 1993;25:723–730. [Google Scholar]
- Gardi C, Tomaselli M, Parisi V, Petraglia A, Santini C. Soil quality indicators and biodiversity in northern Italian permanent grasslands. Eur. J. Soil Biol. 2002;38:103–110. [Google Scholar]
- González-Bernáldez F. Ecological consequences of the abandonment of traditional land use systems in central Spain. Options Méditerranénnes. 1991;15:23–29. [Google Scholar]
- Grayston SJ, Wang S, Campbell RD, Edwards AC. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol. Biochem. 1998;30:369–378. [Google Scholar]
- Hackl E, Pfeffer M, Donat C, Bachman G, Zechmeister-Boltenstern S. Composition of the microbial communities in the mineral soil under different types of natural forest. Soil Biol. Biochem. 2005;37:661–671. [Google Scholar]
- Hedlund K. Soil microbial community structure in relation to vegetation management on former agricultural land. Soil Biol. Biochem. 2002;34:1299–1307. [Google Scholar]
- Joergensen RG, Wichern F. Quantitative assessment of the fungal contribution to microbial tissue in soil. Soil Biol. Biochem. 2008;40:2977–2991. [Google Scholar]
- Johnson MJ, Lee KY, Scow KM. DNA fingerprinting reveals links among agricultural crops, soil properties, and the composition of soil microbial communities. Geoderma. 2003;114:279–303. [Google Scholar]
- Knudsen D, Peterson GA, Pratt PF. Lithium, Sodium and Potassium. In: American Society of Agronomy and Soil Science Society of America Journal, editor. Methods of soil analysis. Vol. 2. Madison, WI: 1982. pp. 225–246. [Google Scholar]
- Kosmas C, Gerontidis St., Marathianou M. The effect of land use change on soils and vegetation over various lithological formations on Lesvos (Greece) Catena. 2000;40:51–68. [Google Scholar]
- Lovell RD, Jarvis SC, Bardgett RD. Soil microbial biomass and activity in long-term grassland: effects of management changes. Soil Biol. Biochem. 1995;27:969–975. [Google Scholar]
- Lundquist EJ, Scow KM, Jackson LE, Uesugi SL, Johnson CR. Rapid response of soil microbial communities from conventional, low input, and organic farming systems to a wet/dry cycle. Soil Biol. Biochem. 1999;31:11661–1675. [Google Scholar]
- Miller RM, Jastrow JD. Hierachy of roots and mycorrhizal fungi interactions with soil aggregation. Soil Biol. Biochem. 1990;22:579–584. [Google Scholar]
- Nannipieri P, Ceccanti B, Cervelli S, Matarese E. Extraction of phosphatase, urease, protease, organic carbon and nitrogen from soil. Soil Sci. Soc. Am. J. 1980;44:1011–1016. [Google Scholar]
- Nelson DV, Sommers LE. Total carbon, organic carbon, and organic matter. In: Page AL, Miller RH, Keeney DR, editors. Methods of soil analysis, Part 2. Chemical and biological methods. American Society of Agronomy and Soil Science of America; Madison, WI: 1982. pp. 539–579. [Google Scholar]
- Nodar R, Acea MJ, Carballas T. Microbiological response to Ca(OH)2 treatments in a forest soil. FEMS Microbiol. Ecol. 1992;86:213–219. [Google Scholar]
- Olsson PA, Bååth E, Jakobsen I, Söderström B. The use of phospholipid and neutral lipid fatty acids to estimate biomass of arbuscular mycorrhizal fungi in soil. Mycol. Res. 1995;99:623–629. [Google Scholar]
- Rillig MC. Arbuscular mycorrhyzae, glomalin, and soil aggregation. Can. J. Soil Sci. 2004;84:355–363. [Google Scholar]
- Rinnan R, Rinnan A. Application of near infrared reflectance (NIR) and fluorescence spectroscopy to analysis of microbiological and chemical properties of artic soil. Soil Biol. Biochem. 2007;39:1664–1673. [Google Scholar]
- Roig A, Romero M, Lax A, Fernández FG. Estudio comparativo de métodos de determinación de capacidad de cambio catiónica en suelos calizos. An. Edafol. Agrobiol. 1980;39:2021–2032. [Google Scholar]
- Roldán A, García-Orenes F, Lax A. An incubation experiment to determinate factors involving aggregation changes in an arid soil receiving urban refuse. Soil Biol. Biochem. 1994;26:1699–1707. [Google Scholar]
- Shannon C. A mathematical theory of communication. The Bell System Technical Journal. 1948;27:379–423. [Google Scholar]
- Sims JR, Haby VA. Simplified colorimetric determination of soil organic matter. Soil Sci. 1971;497:137–141. [Google Scholar]
- Soil Survey Staff . Keys to soil taxonomy. 10th ed NRCS; Washington DC: 2006. [Google Scholar]
- Tabatabai MA. Soil enzimes. In: Page AL, Miller RH, Keeney DR, editors. Methods of Soil Analysis, Part 2. 2nd ed American Society of Agronomy and Soil Science of America; Madison, WI: 1982. pp. 501–538. (Agronomical Monograph No. 9). [Google Scholar]
- Tabatabai MA, Bremner JM. Use of p-nitrophenyl phosphate in assay of soil phosphatase activity. Soil Biol. Biochem. 1969;1:301–307. [Google Scholar]
- Van der Wal A, van Veen JA, Smant W, Boschker HTS, Bloem J, Kardol P, van der Putten WH, de Boer W. Fungal biomass development in a chronosequence of land abandonment. Soil Biol. Biochem. 2006;38:51–60. [Google Scholar]
- Vance ED, Brookes PC, Jenkinson DS. An extraction method for measuring soil microbial biomass C. Soil Biol Biochem. 1987;19:703–707. [Google Scholar]
- Velasquez E, Lavelle P, Barrios E, Joffre R, Reversat F. Evaluating soil quality in tropical agroecosystems of Colombia using NIRS. Soil Biol. Biochem. 2005;37:889–898. [Google Scholar]
- Yang C, Crowley DE. Rhizosphere microbial community structure in relation to root location and plant iron nutritional status. Appl. Environ. Microb. 2000;66:345–351. doi: 10.1128/aem.66.1.345-351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelles L. Phospholipid fatty acid profiles in selected members of soil microbial communities. Chemosphere. 1997;35:275–294. doi: 10.1016/s0045-6535(97)00155-0. [DOI] [PubMed] [Google Scholar]
- Zelles L. Fatty acids patterns of phospholipids and lipopolysacharides in the characterisation of microbial communities in soil: a review. Biol. Fertil. Soils. 1999;29:111–129. [Google Scholar]
- Zelles L, Bai QY, Ma RX, Rackwitz R, Winter K, Beese F. Microbial biomass, metabolic quotient and nutritional status determined from fatty acid patterns and poly-hydroxybutyrate in agriculturally-managed soils. Soil Bio. Biochem. 1994;26:439–446. [Google Scholar]
- Zornoza R, Guerrero C, Mataix-Solera J, Scow KM, Arcenegui V, Mataix-Beneyto J. Near infrared spectroscopy for determination of various physical, chemical and biochemical properties in Mediterranean soils. Soil Biol. Biochem. 2008;40:1923–1930. doi: 10.1016/j.soilbio.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]