Abstract
The inflammatory bowel diseases (IBD) Crohn’s disease and ulcerative colitis are common causes of morbidity in children and young adults in the western world. Here we report the results of a genome-wide association study in early-onset IBD involving 3,426 affected individuals and 11,963 genetically matched controls recruited through international collaborations in Europe and North America, thereby extending the results from a previous study of 1,011 individuals with early-onset IBD1. We have identified five new regions associated with early-onset IBD susceptibility, including 16p11 near the cytokine gene IL27 (rs8049439, P = 2.41 × 10−9), 22q12 (rs2412973, P = 1.55 × 10−9), 10q22 (rs1250550, P = 5.63 × 10−9), 2q37 (rs4676410, P = 3.64 × 10−8) and 19q13.11 (rs10500264, P = 4.26 × 10−10). Our scan also detected associations at 23 of 32 loci previously implicated in adult-onset Crohn’s disease and at 8 of 17 loci implicated in adult-onset ulcerative colitis, highlighting the close pathogenetic relationship between early- and adult-onset IBD.
Crohn’s disease and ulcerative colitis are chronic inflammatory disorders of the gastrointestinal tract that most commonly arise during the second and third decades of life. Incidence, family, twin and phenotype concordance studies suggest that IBD is highly heritable, albeit complex, spurring an ongoing search for genetic factors that confer susceptibility to this disease2,3. Genome-wide association studies (GWASs) applying high-density SNP array technology have greatly expanded the number of genetic factors implicated in IBD pathogenesis to include 32 loci associated with Crohn’s disease and 17 associated with ulcerative colitis, spanning pathways involved in adaptive (IL23R, IL10, IL12B, STAT3) and innate (CARD15, ATG16L1, IRGM) immunity4–7.
Most genetic analyses in IBD have been performed in adult-onset disease2,3. Early-onset IBD, however, has unique characteristics of phenotype, severity and familiality8,9, features that provide support for the search for loci that may be specific to early-onset disease. In addition, because early-onset IBD has a stronger familial component than the adult disease, studies targeting this subgroup potentially provide additional power to identify genes that contribute modest effects, as illustrated by the success of our previous scan in identifying 20q13 and 21q22 as IBD loci1.
We now report results from the largest GWAS conducted so far in early-onset IBD (Fig. 1). Our IBD discovery cohort (DC-IBD) comprised 2,413 individuals of European ancestry with IBD (cases), including 1,636 with Crohn’s disease (DC-CD), 724 with ulcerative colitis (DC-UC) and 53 with IBD of unclassified type (IBD-U), and 6,158 genetically matched controls, and was genotyped on the Illumina HumanHap550 platform. Affected individuals were recruited from multiple centers from four geographically discrete countries and diagnosed before their nineteenth birthday according to standard IBD diagnostic criteria (Supplementary Table 1). Our study extends a previous IBD GWAS that was based on a subset of these cases (1,011 IBD cases, including 647 with Crohn’s disease, 317 with ulcerative colitis and 47 with IBD-U; Supplementary Table 2)1. An independent replication cohort (RC1) of 482 early-onset IBD cases (289 with Crohn’s disease, 120 with ulcerative colitis and 73 with IBD-U) and 1,696 genetically matched controls was gathered from the Children’s Hospital of Philadelphia (CHOP) health system and collaborating centers. We refer to Crohn’s disease and ulcerative colitis subanalyses of data set RC1 as RC1-CD and RC1-UC, respectively. A second replication cohort (RC2-CD) of 531 Crohn’s disease cases diagnosed in childhood and 4,109 controls was assembled by the International IBD Genetics Consortium (IIBDGC). This cohort is based on subsets of data from genome-wide scans generated by the US National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)10, the Wellcome Trust Case Control Consortium (WTCCC)11 and a Belgian-French12 collaboration, which have been combined previously in a large-scale meta-analysis5. We computed P values in each cohort by comparing single-marker allele frequencies using χ2 statistics on SNPs that passed quality-control criteria. We conducted meta-analysis across multiple studies using a Z-score transformation. See Online Methods for more detailed descriptions of cohorts and methods used in this study.
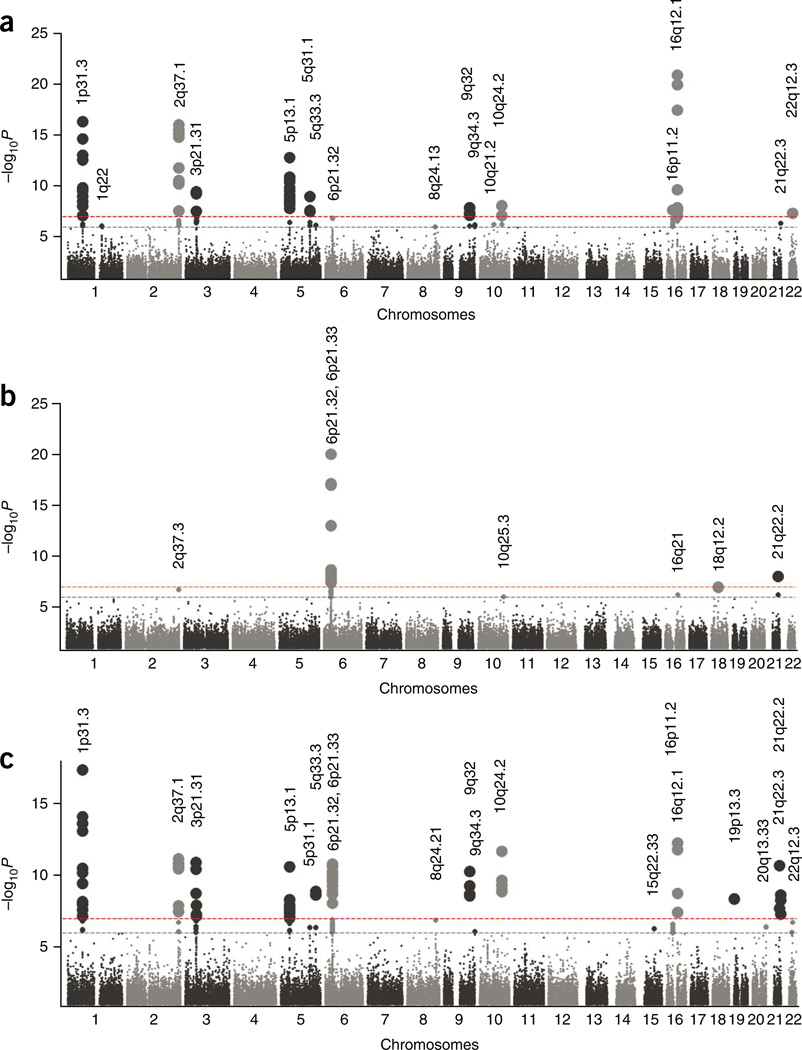
Figure 1.
Overview of genome-wide association results. (a–c) Chromosomal plots of –logP against genomic location for discovery scans in the early-onset DC-CD (a), DC-UC (b) and DC-IBD (c) cohorts. Red and blue dotted lines represent thresholds for genome-wide significant (P = 1 × 10−7) and suggestive (P = 1 × 10−6) signals, respectively. Loci on 16p11, 22q12 and 2q37 emerged from these analyses as suggestive signals and were validated in the follow-up cohorts RC1 and RC2-CD. The following signals did not replicate or show significance in the meta-analysis: 1q22 (CD), 10q25 (UC), 16q21 (UC), 18q12 (UC), 8q24 (IBD) and 15q22 (IBD). The remaining loci have been previously identified in majority adult-onset genome scans of Crohn’s disease and ulcerative colitis. Loci on 22q12 and 20q13 were identified in a previous early-onset IBD scan involving a subset of the cohort analyzed in this study1.
We first searched for previously unreported signals, including loci that met stringent genome-wide significant (P < 5 × 10−8) and suggestive (P < 1 × 10−6) thresholds in our three discovery scans (DC-CD, DC-UC and DC-IBD). For confirmation of these loci, we sought evidence for replication in two independent early-onset cohorts (RC1 and RC2-CD). Lastly, we combined our discovery and replication cohorts for Crohn’s disease (Table 1), ulcerative colitis (Table 2) and IBD (Table 3) in a meta-analysis.
Table 1.
Newly discovered loci significant in the GWAS of the DC-CD, RC1-CD and RC2-CD cohorts
| CD discovery (DC-CD) (n = 1,636 / 6,158) |
CHOP CD replication (RC1-CD) (n = 289 / 1,696) |
IIBDGC replication (RC2-CD) (n = 531 / 4,109) |
Combined (n = 2,456 / 11,963) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Band (Mb) | Genes | SNP | Allele | P | MAFA | MAFU | OR [95% CI] | P | MAFA | MAFU | OR [95% CI] | P | MAFA | MAFU | OR [95% CI] | P | Z |
| 10q22.3 (80.71–80.73) | ZMIZ1 | rs1250550 | A/T | 1.59E-05 | 0.28 | 0.32 | 0.83 [0.76–0.90] | 0.001 | 0.28 | 0.35 | 0.73 [0.60–0.88] | 0.005 | 0.20 | 0.24 | 0.82 [0.70–0.96] | 4.41E–10 | −6.24 |
| 16p11.2 (28.45–28.81) | IL27, SULT1A1, SULT1A2, EIF3C | rs8049439 | C/G | 2.72E-07 | 0.42 | 0.37 | 1.23 [1.13–1.32] | 0.28 | 0.40 | 0.38 | 1.10 [0.92–1.32] | 0.0014 | 0.42 | 0.39 | 1.14 [1.00–1.30] | 2.87E–09 | 5.94 |
| 22q12.2 (28.75–28.86) | HORMAD2, MTMR3, LIF | rs2412973 | A/T | 6.16E-06 | 0.50 | 0.46 | 1.19 [1.10–1.29] | 0.0095 | 0.52 | 0.46 | 1.26 [1.06–1.51] | 0.0160 | 0.50 | 0.46 | 1.15 [1.01–1.31] | 3.77E-08 | 5.50 |
MAF, minor allele frequency.
Table 2.
Newly discovered loci significant in the GWAS of the DC-UC and RC1-UC cohorts
| UC discovery (DC-UC) (n = 724 / 6,158) |
CHOP UC replication (RC1-UC) (n = 120 / 1,696) |
Combined (n = 844 / 7,854) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Band (Mb) | Genes | SNP | Allele | P | MAFA | MAFU | OR [95% CI] | P | MAFA | MAFU | OR [95% CI] | P | Z |
| 2q37.3 (241.21–241.42) | CAPN10, GPR35, KIF1A, RNPEPL1 | rs4676410 | A/T | 1.70 × 10−7 | 0.24 | 0.18 | 1.41 [1.24–1.61] | 0.0611 | 0.25 | 0.20 | 1.34 [0.99–1.82] | 3.64 × 10−8 | 5.51 |
Table 3.
Newly discovered loci significant in the GWAS of the DC-IBD, RC1 and RC2-CD cohorts
| IBD discovery (DC-IBD) (n = 2,413 / 6,158) |
CHOP IBD replication (RC1-IBD) (n = 482 / 1,696) |
IIBDGC replication (RC2-CD) (n = 531 / 4,109) |
Combined (n = 3,426 / 11,963) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Band (Mb) | Genes | SNP | Allele | P | MAFA | MAFU | OR [95% CI] | P | MAFA | MAFU | OR [95% CI] | P | MAFA | MAFU | OR [95% CI] | P | Z |
| 19q13.11 (38.44–38.45) | rs10500264 | A/T | 1.18E-05 | 0.21 | 0.18 | 1.21 [1.11–1.31] | 0.0034 | 0.22 | 0.18 | 1.30 [1.07–1.58] | 0.00064 | 0.12 | 0.12 | 1.08 [0.89–1.31] | 4.26E-10 | 6.24 | |
| 22q12.2 (28.75–28.86) | HORMAD2, MTMR3, LIF | rs2412973 | A/T | 9.14E-07 | 0.50 | 0.46 | 1.18 [1.11–1.26] | 0.0052 | 0.51 | 0.46 | 1.23 [1.05–1.43] | 0.016 | 0.50 | 0.46 | 1.15 [1.01–1.31] | 1.55E-09 | 6.04 |
| 16p11.2 (28.45–28.81) | IL27, SULT1A1, SULT1A2, EIF3C | rs8049439 | C/G | 2.38E-07 | 0.41 | 0.37 | 1.20 [1.12–1.28] | 0.34 | 0.40 | 0.38 | 1.08 [0.92–1.26] | 0.0014 | 0.42 | 0.39 | 1.14 [1.00–1.30] | 2.41E-09 | 5.97 |
| 10q22.3 (80.71–80.73) | ZMIZ1 | rs1250550 | A/T | 3.55E-05 | 0.29 | 0.32 | 0.86 [0.80–0.92] | 0.0016 | 0.29 | 0.35 | 0.78 [0.66–0.92] | 0.00461 | 0.20 | 0.24 | 0.82 [0.70–0.96] | 5.63E-09 | −5.83 |
Analysis of DC-CD identified a region on 16p11 as the single new genome-wide significant locus; the most significant SNP in the block of linkage disequilibrium (LD) containing this locus, rs1968752, yielded a value of P = 2.09 × 10−8, with the minor allele (A) conferring risk (odds ratio (OR) = 1.25 (1.16–1.36)). We observed nominal replication of rs1968752 in the RC2-CD data set (P = 0.036, OR = 1.09 (0.94–1.27)) and a trend for association in the combined analysis of the replication cohorts (RC1-CD and RC2-CD), using two-sided P values (P = 0.059; Supplementary Table 3).
Analysis of our combined IBD discovery scan yielded suggestive association at the 16p11 locus (rs8049439, P = 2.38 × 10−7, OR = 1.20 (1.12–1.28)). rs8049439 is located ~200 kb upstream of rs1968752, and the two loci are in strong LD (r2 = 0.796). rs8049439 showed a statistically significant association (P = 0.00144, OR = 1.14 (1.00–1.30)) in the RC2-CD data set, whereas meta-analysis of rs8049439 in the discovery data set and the two replication cohorts (RC1, RC2-CD) reached genome-wide significance in both IBD (P = 2.41 × 10−9) and Crohn’s disease (P = 2.87 × 10−9; Tables 1 and 3). In addition, analysis of the available data in a meta-analysis data set for adult-onset Crohn’s disease5 demonstrated that an LD proxy for these two SNPs, rs4788084 (r2 = 0.83 to rs1968752, and r2 = 0.86 to rs8049439), was also associated with Crohn’s disease (P = 0.0035; Supplementary Table 4). Of note, we found that the risk-conferring minor allele (G) at rs8049439 shares haplotypes with the risk-conferring ancestral allele (A) of the type 1 diabetes SNP rs4788084 (HapMap CEU, r2 = 0.864)13.
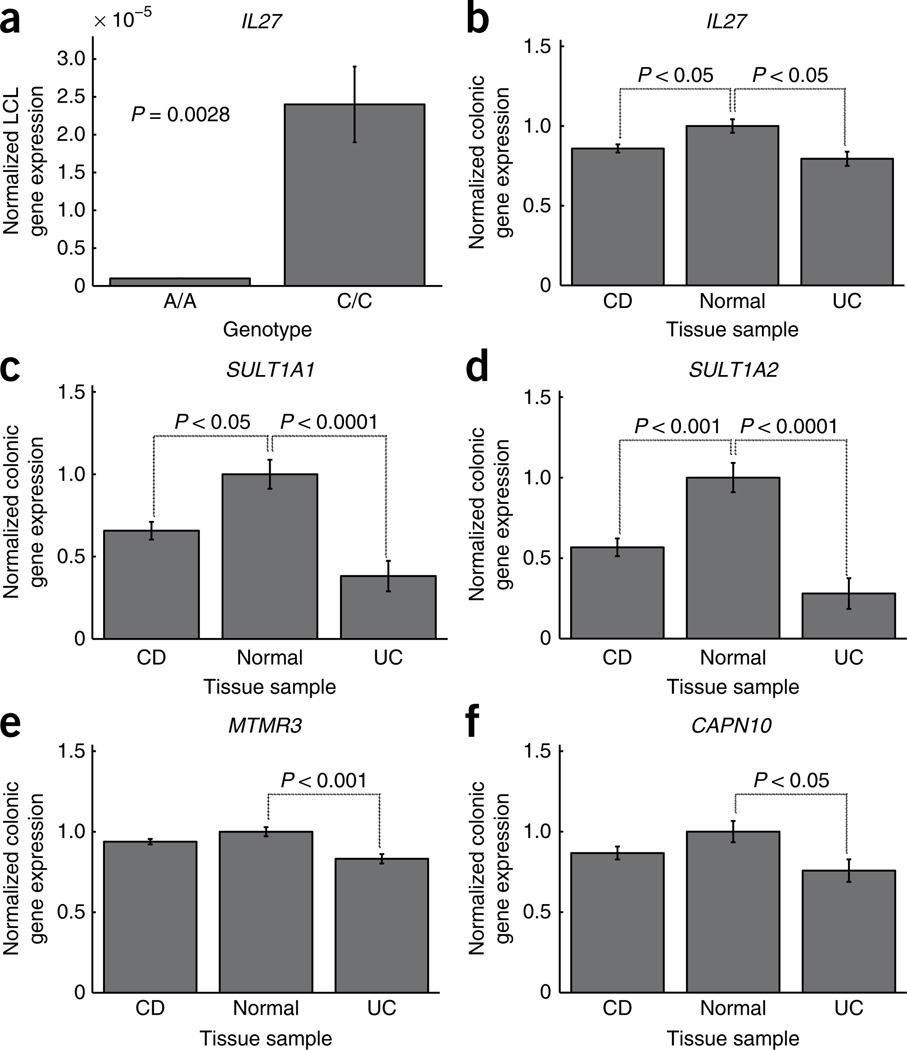
The LD block incorporating rs1968752, rs8049439 and rs4788084 contains multiple genes, including IL27, CCDC101, CLN3, EIF3C, NUPR1, SULT1A1 and SULT1A2. Of these, we considered IL27, which encodes an immunomodulatory cytokine that regulates adaptive immunity responses, to be the most plausible candidate gene for susceptibility to IBD. Analysis of IL27 gene expression in lymphoblastoid cell lines (LCLs) obtained from ten healthy individuals homozygous for rs1968752 (that is, either A/A or C/C) showed that those individuals with two copies of the risk allele (A/A) had a several-fold decrease in IL27 expression relative to those with two copies of the nonrisk allele (C/C; P = 0.0031; Fig. 2a), suggesting that this SNP may exert a regulatory effect on IL27 gene expression. In addition, colonic expression of IL27 was significantly lower in 30 individuals with early-onset Crohn’s disease than in 11 healthy controls (P < 0.05; Fig. 2b). These effects remained significant after correction for medication use and histological inflammation score.
Figure 2.
Colonic and LCL mRNA expression of genes in significantly associated loci. The A allele of rs1968752 confers risk in our early-onset Crohn’s disease and IBD cohorts (OR = 1.23 (1.12–1.40)). rs1968752 lies in a block of LD containing the IL27 gene. (a) Cell lines from individuals (n = 5) with the A/A genotype at rs1968752 have over 90% decrease in IL27 gene expression as compared with those with the C/C genotype. (b) IL27 colonic expression was significantly lower in samples obtained from individuals with early-onset Crohn’s disease and ulcerative colitis cases than in normal tissue. (c,d) Expression of both SULT1A1 (c) and SULT1A2 (d) genes (also in the IL27 locus) was also significantly lower in Crohn’s disease and ulcerative colitis tissue than in normal tissue. (e,f) Expression of putative IBD gene MTMR3 at 22q12 (e) and the putative ulcerative colitis gene CAPN10 at 2q37 (f) was significantly lower in ulcerative colitis tissue than in normal tissue. All significant expression effects were found to be independent of treatment and histological inflammation score (data not shown). Error bars represent s.e.m.
When examining the colonic expression of other genes at this locus, we also detected significantly lower expression of SULT1A1 and SULT1A2 in both early-onset Crohn’s disease (P < 0.05, P < 0.001) and ulcerative colitis (P < 0.0001, P < 0.0001) as compared with healthy controls (Fig. 2c,d). SULT1A1 and SULT1A2 encode sulfotransferases that catalyze sulfate conjugation of catecholamines, phenolic drugs and neurotransmitters. These biological functions make SULT1A1 and SULT1A2 less attractive as IBD candidate genes. We also observed a strongly expressed quantitative trait locus (eQTL) for EIF3C expression in LCLs at this locus (lod score = 5–8) on the basis of publicly available data14; however, we did not observe altered EIF3C expression in Crohn’s disease or ulcerative colitis cases relative to healthy controls, nor did we detect an eQTL for EIF3C in colonic tissue at SNPs in this region (Supplementary Figs. 1 and 2). EIF3C encodes a eukaryotic translation initiation factor that forms part of the basic translational machinery, which also makes EIF3C less likely to be an IBD candidate gene. Additional allele-specific expression effects were not observed for the other genes at this locus either in our LCL and colonic expression data sets (Supplementary Fig. 1) or in the public database14. Taken together, these results point most strongly to IL27 as a candidate gene associated with early-onset IBD; however, further functional and fine-mapping studies are warranted to confirm this and rule out the involvement of other genes at this locus.
The first of the two suggestive early-onset disease associations was identified in the DC-IBD cohort at 22q12 (rs2412973, P = 9.14 × 10−7, OR = 1.18 (1.10–1.26)). This SNP replicated in RC1 (P = 0.0052, OR = 1.23 (1.05–1.43)) and RC2-CD (P = 0.016, OR = 1.15 (1.01–1.31)), yielding P = 1.55 × 10−9 in a meta-analysis across all three early-onset IBD cohorts (Table 3). Meta-analysis of rs2412973 across the data sets DC-CD, RC1-CD and RC2-CD also reached genome-wide significance (P = 3.77 × 10−8). rs2412973 also showed association in the CD meta-analysis data set with an age of onset primarily in adulthood5 (P = 0.000953; Supplementary Table 4). rs2412973 is located within HORMAD2, an open reading frame whose putative functions include mitotic checkpoints, chromosome synapsis and DNA repair. The LD block incorporating rs2412973 also contains MTMR3, encoding myotubularin-related protein-3, and LIF, encoding leukemia inhibitory factor, a cytokine that stimulates differentiation in leukocytes. We observed a significant difference in colonic MTMR3 expression in biopsies from individuals with ulcerative colitis as compared with controls (P < 0.001), but not in those from individuals with Crohn’s disease (Fig. 2e), and we did not detect colonic eQTL for MTMR3 near rs2412973. Other genes in the LD block did not exhibit significant expression effects (Supplementary Fig. 3).
The second suggestive association was identified only in DC-UC at 2q37 (rs4676410, P = 1.70 × 10−7, OR 1.41 (1.24–1.61)). This SNP showed only a trend for replication in the small RC1-UC cohort (P = 0.0611, OR = 1.38 (1.08–1.77)), but yielded genome-wide significance in the combined analysis (P = 3.64 × 10−8; Table 2). rs4676410 lies within GPR35, which encodes an orphan receptor primarily expressed in the intestine of humans and rats. Other genes in the LD block of rs4676410 include CAPN10, KIF1A and RNPEPL1. CAPN10 colonic gene expression was significantly lower in ulcerative colitis cases than in controls (P < 0.05; Fig. 2f). CAPN10 encodes a Ca2+-regulated thiol-protease involved in cytoskeletal remodeling and signal transduction. We did not observe significant expression effects in the remaining genes (Supplementary Figs. 1 and 2).
We next combined our discovery DC-IBD and replication cohorts (RC1, RC2-CD) for a genome-wide meta-analyses of early-onset Crohn’s disease, ulcerative colitis and IBD. These analyses yielded two new loci achieving genome-wide significance. The first new signal is associated with IBD (rs10500264, P = 4.26 × 10−10) and is located at 19q13 in a small block of LD devoid of known genes lying within 50 kb of SLC7A10 and CEBPA. Notably, rs10500264 showed only nominal association in the adult-onset Crohn’s disease meta-analysis (P = 0.0217), suggesting that this locus may be weighted more toward early-onset disease. The second new signal, rs1250550, lies on 10q22 inside the ZMIZ1 gene and is associated with both Crohn’s disease (P = 4.41 × 10−10) and IBD combined (P = 5.63 × 10−9; Tables 1 and 3). In addition to showing significance in the early-onset meta-analysis, rs1250550 robustly associates in the majority adult-onset Crohn’s disease meta-analysis5 (P = 3.27 × 10−5). ZMIZ1 encodes a PIAS-like protein that interacts with Smad4 to regulate Smad3 transcription and modulate transforming growth factor-β signaling15. Despite achieving robust significance in our meta-analysis, these loci merit replication in an independent cohort.
We conducted a meta-analysis of our discovery and replication cohorts to determine association with the 49 previously reported IBD loci implicated in adult-onset disease, determining nominal (P < 0.05) and Bonferroni-corrected (P < 0.001, correcting for 49 hypotheses; Supplementary Table 5) significance. Of 32 previously confirmed loci associated with adult-onset Crohn’s disease, 29 were nominally significant and 21 were significant after Bonferroni correction in meta-analysis of DC-CD and RC1-UC data sets. Of eight additional Crohn’s disease loci that attained nominal significance (P < 0.05) in the previously reported majority adult-onset meta-analysis5, two showed significant association with early-onset Crohn’s disease, namely the IL18R1-IL18RAP locus on 2q12 (rs917997, P = 6.84 × 10−5, Z = 3.98) and the C-C motif chemokine (CCL) gene cluster on 17q12 (rs991804, P = 2.31 × 10−4, Z = –3.68; Table 4). We found that 13 of 17 previously identified adult-onset ulcerative colitis loci showed nominal significance, and 8 were significant after Bonferroni-corrected meta-analysis of DC-CD and RC1-UC data sets, including IL23R on 1p31 and IL26 on 12q15 (Supplementary Table 5). Our data also supported the association of loci on 20q13 and 21q22 with early-onset IBD, as previously reported1 on the basis of analyses of a subset of our discovery cohort (Supplementary Note).
Table 4.
Newly discovered effects of previously known or suggested adult IBD risk loci in early-onset IBDa
| CD (DC-CD, RC1-CD) (n = 1,925 / 7,854) |
UC (DC-UC, RC1-UC) (n = 844 / 7,854) |
IBD (DC-IBD, RC1) (n = 2,895 / 7,854) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CDkb | CDpb | UCkb | UCpb | Band | Mb | Genes | SNP | P | Z | P | Z | P | Z |
| • | 1q32.1 | 205.01 | IL10, IL19, IL20 | rs3024505 | 4.84 × 10−4 | 3.49 | 6.20 × 10−4 | 3.42 | 6.84 × 10−6 | 4.50 | |||
| • | 2q12.1 | 102.44 | IL18R1, IL18RAP | rs917997 | 6.84 × 10−5 | 3.98 | 0.17 | 1.38 | 5.98 × 10−5 | 4.01 | |||
| • | 17q12 | 29.61 | CCL11, CCL2, CCL7 | rs991804 | 2.31 × 10−4 | −3.68 | 2.78 × 10−2 | −2.20 | 6.64 × 10−5 | −3.99 | |||
| • | 17q12 | 35.29 | ORMDL3 | rs2872507 | 3.65 × 10−4 | 3.56 | 7.62 × 10−4 | 3.37 | 3.13 × 10−6 | 4.66 | |||
| • | 21q22.3 | 44.44 | ICOSLG1 | rs762421 | 3.32 × 10−7 | 5.10 | 2.54 × 10−5 | 4.21 | 3.19 × 10−10 | 6.29 | |||
Listed are 5 of 49 previously identified adult-onset IBD loci that were either (1) previously nominal adult IBD signals that are verified by our data or (2) validated adult signals previously shown to have an effect on early-onset UC (CD) and found in our study to have an effect on early-onset CD (UC).
Filled circles in the first four columns of the table specify whether the given row represents a known CD locus (CDk), putative/nominal CD locus (CDp), known UC locus (UCk) and/or putative/nominal UC locus (UCp). Loci replicating at a Bonferroni-corrected P < 0.05 (corrected for 49 hypotheses) are denoted in bold; newly discovered significant effects are denoted in bold italics.
We also evaluated previously reported loci associated with adult-onset Crohn’s disease for association with early-onset ulcerative colitis, and vice versa. Examining 32 known Crohn’s disease signals in our ulcerative colitis cohort implicated two loci that had not previously been associated with adult-onset ulcerative colitis susceptibility in early-onset ulcerative colitis: ICOSLG on 21q22 (rs762421, P = 2.54 × 10−5, Z = 4.21) and ORMDL3 on 17q12 (rs2872507, P = 7.62 × 10−4, Z = 3.37; Supplementary Table 5). When examining the association of early-onset Crohn’s disease with 17 previously reported adult-onset ulcerative colitis signals, we detected association only with the ulcerative colitis gene IL10 on 1q32.1 (rs3024505, P = 0.00048, Z = 3.49), suggesting that this locus may also play a role in early-onset Crohn’s disease susceptibility.
Our study adds insight into the pathogenic mechanisms mediating early-onset IBD and its close relationship with adult-onset disease. In particular, identification of IL27 as a candidate gene for Crohn’s disease susceptibility lends further support to the involvement of the T-helper 17 pathway16,17 in pathogenesis of Crohn’s disease, complementing gene discoveries in other genome scans (IL23R, STAT3, JAK2, IL12B)2,3,5. In addition, our discovery of five new IBD susceptibility loci through analysis of a genetically enriched early-onset disease cohort underscores the validity of this approach in the study of complex disease.
Methods
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturegenetics/.
Accession codes. Gene Expression Omnibus (GEO): colonic gene expression data set, GSE10616. Entrez Gene: IL27, 246779; SULT1A1, 171150; SULT1A2, 601292; EIF3C, 8663; HORMAD2, 150280; MTMR3, 8897; LIF, 159540; ZMIZ1, 57178; CAPN10, 11132; GPR35, 2859; KIF1A, 547; RNPEPL1, 57140.
Supplementary Material
ACKNOWLEDGMENTS
We thank all participating subjects and families. We thank the medical assistants, nursing staff and clinicians at CHOP who assisted with the recruitment of control subjects, which made this work possible; and members of the International HapMap and Wellcome Trust Case Control Consortiums for publicly providing data that were critically important for part of our analyses. The following physicians of the SIGENP (Italian Society of Pediatric Gastroenterology, Hepatology and Nutrition) contributed by providing DNA samples and clinical information from their patients: A. Andriulli, M.R. Valvano, O. Palmieri, F. Bossa, E. Colombo, M. Pastore, M. D’Altilia, O. Borrelli, C. Bascietto, A. Ferraris, B. Papadatou, A. Diamanti, P. Lionetti, E. Pozzi, A. Barabino, A. Calvi, G.L. de’ Angelis, G. Guariso, V. Lodde, G. Vieni, C. Sferlazzas, S. Accomando, G. Iacono, E. Berni Canani, A.M. Staiano, V. Rutigliano, D. De Venuto, C. Romano, G. Lombardi, S. Nobile, C. Catassi and A. Campanozzi. The following physicians of SPGHANG (Scottish Pediatric Gastroenterology, Hepatology and Nutrition Group) contributed by providing DNA samples and clinical information from their patients: W.M. Bisset, P.M. Gillett, G. Mahdi and P. McGrogan. This research was supported by the Children’s Hospital of Philadelphia, the Primary Children’s Medical Center Foundation and grants DK069513, M01-RR00064, M01 RR002172-26 and C06-RR11234 from the National Center for Research Resources. All genome-wide genotyping was funded by an Institute Development Award from the Children’s Hospital of Philadelphia. M.S. is funded by NIH/NIDDK grant DK062423 and the Gale and Graham Wright Research Chair in Digestive Diseases. T.W. received fellowship funding support jointly from the Canadian Association of Gastroenterology (CAG), Crohn’s Colitis Foundation of Canada (CCFC), Canadian Institutes of Health Research (CIHR) and Astra-Zeneca.
Footnotes
A full list of members appears in a Supplementary Note.
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
M.I., R.N.B., A.G., R.K.R., V.A., M. Dubinsky, S.K., S.F.A.G., M.S.S., J. Satsangi and H.H. participated in study conception and design. R.N.B., A.G., R.K.R., V.A., M. Dubinsky, S.K., T.D.W., S.S., R.G., M.C., A. Latiano, B.D., J. Stempack, D.J.A., K.T., B.K., J.L., J.E., R.G., M. Stephens, A. Levine, D.P., J.V.L., S.C., S.L.G., C.E.K., G.D.F., E.C.F., C.H., G.O., R.M.C., A.M., L.D., D.C.W., M.S.S., S.F.A.G., J. Satsangi, H.H., D.S.M., D.M. and M.B.H. recruited patients and directed sample collection. C.E.K., E.C.F., G.O., R.M.C. and J.T.G. performed genotyping and quality-control measures on all data sets and H.H. supervised all sample organization and genotyping. L.D., R.G. and A.M. supervised gene expression experiments. M.I. performed statistical analysis with supervision from H.H. M.I., J.P.B., P.S., K.W., H.Z, R.G., J.H.F. and M. Daly provided bioinformatics, database and statistical support. M.I. and H.H. wrote the manuscript. M.I., R.N.B., A.G., R.K.R., V.A., M. Dubinsky, S.K., S.F.A.G., M.S.S., J. Satsangi and H.H. participated in drafting and critical revision of the manuscript. All authors contributed to the final paper, with M.I., R.N.B., A.G., R.K.R., V.A., M. Dubinsky, S.K., M.S.S., J. Satsangi and H.H. playing key roles.
References
- 1.Kugathasan S, et al. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat. Genet. 2008;40:1211–1215. doi: 10.1038/ng.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Limbergen J, Wilson DC, Satsangi J. The genetics of Crohn’s disease. Annu. Rev. Genomics Hum. Genet. 2009;10:89–116. doi: 10.1146/annurev-genom-082908-150013. [DOI] [PubMed] [Google Scholar]
- 3.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat. Rev. Immunol. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 4.Fisher SA, et al. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn’s disease. Nat. Genet. 2008;40:710–712. doi: 10.1038/ng.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franke A, et al. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat. Genet. 2008;40:1319–1323. doi: 10.1038/ng.221. [DOI] [PubMed] [Google Scholar]
- 7.Silverberg MS, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat. Genet. 2009;41:216–220. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vernier-Massouille G, et al. Natural history of pediatric Crohn’s disease: a population-based cohort study. Gastroenterology. 2008;135:1106–1113. doi: 10.1053/j.gastro.2008.06.079. [DOI] [PubMed] [Google Scholar]
- 9.Van Limbergen J, et al. Definition of phenotypic characteristics of childhood-onset inflammatory bowel disease. Gastroenterology. 2008;135:1114–1122. doi: 10.1053/j.gastro.2008.06.081. [DOI] [PubMed] [Google Scholar]
- 10.Rioux JD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Libioulle C, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrett JC, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixon AL, et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Thyssen G, Beliakoff J, Sun Z. The novel PIAS-like protein hZimp10 enhances Smad transcriptional activity. J. Biol. Chem. 2006;281:23748–23756. doi: 10.1074/jbc.M508365200. [DOI] [PubMed] [Google Scholar]
- 16.Steinman L. A brief history of TH17, the first major revision in the TH1/TH2 hypothesis of T cell-mediated tissue damage. Nat. Med. 2007;13:139–145. doi: 10.1038/nm1551. [DOI] [PubMed] [Google Scholar]
- 17.Bettelli E, Oukka M, Kuchroo VKT. TH-17 cells in the circle of immunity and autoimmunity. Nat. Immunol. 2007;8:345–350. doi: 10.1038/ni0407-345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.