Abstract
Human Dickkopf-1 (huDKK1), an inhibitor of the canonical Wnt-signaling pathway that has been implicated in bone metabolism and other diseases, was expressed in engineered Chinese hamster ovary cells and purified. HuDKK1 is biologically active in a TCF/lef-luciferase reporter gene assay and is able to bind LRP6 coreceptor. In SDS-PAGE, huDKK1 exhibits molecular weights of 27–28 K and 30 K at ∼ 1:9 ratio. By MALDI-MS analysis, the observed molecular weights of 27.4K and 29.5K indicate that the low molecular weight form may contain O-linked glycans while the high molecular weight form contains both N- and O-linked glycans. LC-MS/MS peptide mapping indicates that ∼ 92% of huDKK1 is glycosylated at Asn225 with three N-linked glycans composed of two biantennary forms with 1 and 2 sialic acid (23% and 60%, respectively), and one triantennary structure with 2 sialic acids (9%). HuDKK1 contains two O-linked glycans, GalNAc (sialic acid)-Gal-sialic acid (65%) and GalNAc-Gal[sialic acid] (30%), attached at Ser30 as confirmed by β-elimination and targeted LC-MS/MS. The 10 intramolecular disulfide bonds at the N- and C-terminal cysteine-rich domains were elucidated by analyses including multiple proteolytic digestions, isolation and characterization of disulfide-containing peptides, and secondary digestion and characterization of selected disulfide-containing peptides. The five disulfide bonds within the huDKK1 N-terminal domain are unique to the DKK family proteins; there are no exact matches in disulfide positioning when compared to other known disulfide clusters. The five disulfide bonds assigned in the C-terminal domain show the expected homology with those found in colipase and other reported disulfide clusters.
Keywords: human DKK1, N & O-linked glycans, cysteine-rich domains, disulfide mapping
Introduction
Dickkopf-1 (DKK1) is a member of the Dickkopf family of secreted proteins that play an important role as negative regulators of the canonical Wnt signaling pathway, which has a central role in bone formation and development.1–4 Canonical Wnt molecules bind to secreted frizzled-related receptor proteins (sFRP) and LRP5/6 coreceptors located in the plasma membrane and signal through a complex network of protein interactions.5–7 Activation of the canonical pathway results in cytosolic stabilization and the subsequent nuclear translocation of β-catenin, leading to the transcription of downstream target genes. LRP5/6 activity is directly inhibited by several molecules including members of the sclerostin and Dickkopf (DKK) families, which bind various domains on LRP5/6 and compete for Wnt binding.1,8,9 In addition, interaction between DKK1 and the LRP5/6 complex with Kremen proteins leads to internalization of the complex thus diminishing the number of Wnt coreceptors available for signaling.10,11 DKK1, through its modulation of Wnt-signaling, is involved in patterning and organogenesis that occurs during embryogenesis.1,4,12 The involvement of DKK1 in bone biology was first suggested by the evidence that gain-of-function mutations in LRP5 leads to high bone mass in humans with impaired DKK1 binding. Several reports confirmed that activation of Wnt/β-catenin signaling induced osteoblastogenesis and that these effects could be blocked by DKK1.2,3,8,13 In addition, it was shown that transgenic overexpression of DKK1 in bone caused osteopenia in mice14 and that DKK1 expression is increased in human diseases that manifest bone loss such as rheumatoid arthritis15 and multiple myeloma.16 Many reviews from recent studies concluded that antagonizing Wnt signaling via inhibition of DKK1 is a promising strategy for promoting bone formation.17–21
The DKK family consists of four molecules, DKK1, DKK2, DKK3, and DKK4, that share similar localization of their 20 cysteine residues clustered at the N- and C-terminal Cys-rich domains.4,22 Disulfide arrangement among these DKK molecules is therefore expected to be similar. Despite the similarity in the arrangement of the Cys-rich domains, the gene family has been demonstrated to exhibit functional and structural diversity.22 The DKK1 protein contains a signal sequence and an N-terminal linker sequence (L1), followed by N- and C-terminal Cys-rich domains with a linker sequence (L2) connecting the two domains. C-terminal domain of DKK1 has a similar cysteine localization and disulfide structure with colipase and other related proteins; and thus a predicted three dimensional model similar to colipase was proposed for the C-terminal domain.23 The disulfide structure of the N-terminal Cys-rich domain has not been reported and this region lacks similarity to the C-terminal Cys-rich domain of DKK1 or to other nonfamily members.
When expressed in 293 cells, human Dickkopf-1(huDKK1) was secreted as a molecule larger than 40K.22 A 35K human DKK1 homologue has also been isolated and characterized (10). These early reports indicated that secreted DKK1 may be glycosylated. There is a potential Asn-linked glycosylation site at the C-terminal end of the molecule, and the observed molecular mass, though varied in size, is much higher than the expected mass predicted from the DKK1 gene sequence. Murine DKK1 protein expressed in 293 cells has been confirmed to contain both N- and O-linked sugars. Deglycosylated murine DKK1 retained its biological function with reduced biological activity, suggesting that the carbohydrate moieties may contribute to DKK1 activity, but are not essential.24
To better elucidate the structure of DKK1, we report here the characterization of glycosylation, including the sites of glycosylation and proposed glycan structures, as well as the determination of overall disulfide structure of the two Cys-rich domains. These two domains were compared with each other regarding the disulfide motifs and with other peptides or proteins containing a Cys-Cys pair at adjacent sequence position. Moreover, the presence of Cys-Cys pairs has made routine peptide mapping analysis less informative. Strategies using Edman sequencing/chemical cleavage/Edman sequencing, as well as partial reduction/ NEM-labeling followed by HPLC and MALDI-MS, have been useful to providing confirmatory data for disulfide linkage assignment.
Results and Discussion
Biochemical Characterization of HuDKK1 and Glycosylation
Functional activities of HuDKK1
The functional activity of recombinant huDKK1 was evaluated utilizing a Wnt-responsive TCF/lef-luciferase reporter construct. Luciferase activity is induced when cells transfected with this construct are exposed to biologically active Wnt3a. Wnt3a-induced luciferase activity can be suppressed by adding recombinant huDKK1 protein to the cells containing this construct. Supporting Information Figure 1A is a dose-response titration curve for huDKK1-mediated inhibition of luciferase activity, indicating that the suppression of Wnt3a activity was huDKK1 concentration-dependent with an IC50 of 34.5 ng/mL. These data indicate that the purified recombinant huDKK1 is a biologically functional inhibitor of Wnt3a.
Figure 1.
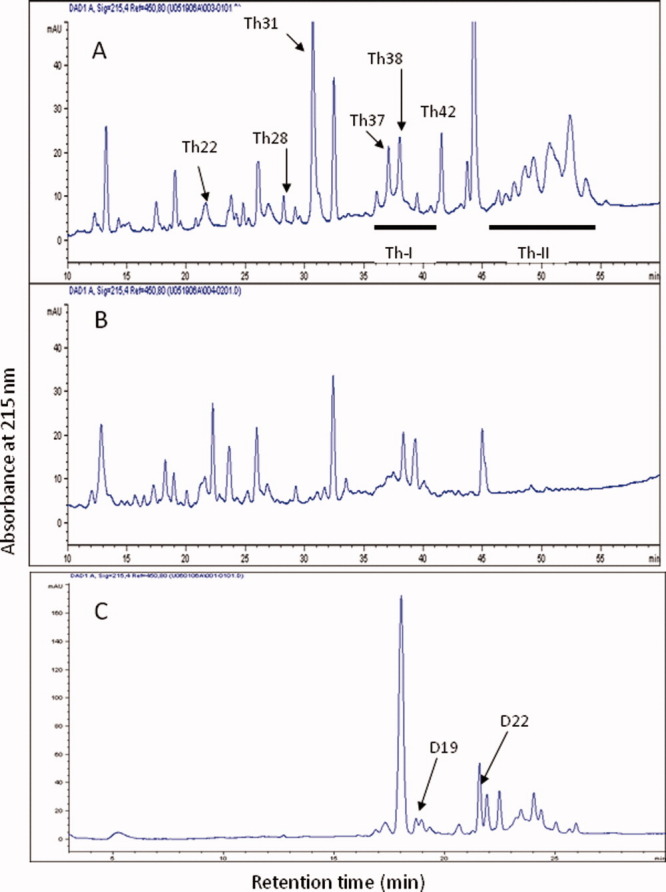
Panel A: RP-HPLC separation of huDKK1 peptides after thermolysin digestion. Underlined area (Th-I) was further treated with enzymes as described in Supporting Information Materials and Methods. Panel B: HPLC profile of the digest after reduction with dithiothreitol. Panel C: Endoproteinase Asp-N digest of fraction Th-I. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Purified huDKK1 was tested for its ability to bind the Wnt coreceptor, LRP6, using a Biacore binding assay. The results demonstrated that huDKK1 in solution can competitively bind LRP6 captured on a huDKK1-immobilized sensor chip surface (Supporting Information Fig. 1B). The competitive binding is huDKK1 concentration-dependent with an EC50 of 0.146 nM.
Sequence verification and molecular weight determination
Recombinant huDKK1 expressed in Chinese hamster ovary cells containing a C-terminal 8 amino acid flag tag sequence was purified by a three column process. N-terminal sequence analysis of purified huDKK1 confirmed that it contains a major N-terminal sequence, T-L-N-S-V-L-N-S-N----, matching the reported sequence following removal of the signal peptide.10 Therefore, the mature protein is properly processed to contain 243 amino acid residues. Including the C-terminal flag tag sequence of DYKDDDDK, huDKK1 without post-translational modifications has a theoretical molecular weight of 26,752.9 Da. SDS-PAGE shows that the final purified material has a major molecular weight of ∼ 30K (>90%) and a minor form migrated around 27–28 K (Supporting Information Fig. 2, inset). SDS-PAGE demonstrates that the molecular weight of huDKK1 is significantly higher than the calculated value. MALDI-MS analysis indicated that the majority of the huDKK1 molecule has a molecular weight of 29,537 Da, while a minor form of 27,402 Da is also present (Supporting Information Fig. 2). The data indicate that the high molecular weight form may contain an Asn-linked sugar and/or O-linked sugar (mass difference of 2,135 Da). The Asn-linked site is expected to reside at position 225, which is the only consensus Asn-linked sequence (N-S-S) in huDKK1. The lower molecular weight form of huDKK1 (27,402 Da) is 649 Da heavier than the predicted mature nonglycosylated form of huDKK1 (26,753 Da), suggesting the presence of a potential O-linked glycan. The molecular weight of DKK1 reported in earlier studies appeared to be higher than the data presented here.4,10,22
Assignment of Asn- and Ser-linked glycosylation sites and characterization of oligosaccharide structure
To characterize the glycan structures and potential N- and O-linked glycan attachment sites, huDKK1 was subjected to tryptic digestion followed by LC-MS/MS peptide mapping. The occurrence of carbohydrate oxonium ions (i.e., m/z= 204) at certain retention times in the source fragmentation scan were used to pinpoint multiply charged precursor ions corresponding to glycopeptides in the full MS scan of the LC-MS/MS run. CID fragmentation of different charge states of these glycopeptides was performed. Supporting Information Table 1 lists N-linked and O-linked glycopeptides found by LC-MS/MS peptide mapping and their respective proposed structures.
Table I.
Disulfide-Containing Thermolytic Peptides Obtained from HPLC Separation of huDKK1 Proteolytic Digest
| Sample | Sequence | Seq. position | Obs. mass (calc. mass) | Disulfidesa |
|---|---|---|---|---|
| Th28 | VLKEGQVCTKHRRKG | 182–196 | ||
| LSCR | 212–215 | 2219 (2217) | C189-C214 | |
| Th31 | LACRKRRKRCMRHAMCC PGNYCKNGICVSSDQNH | 81–114 | 3936 (3935) | (C83…C107)b |
| Th37 | IDNYQPYPCAEDEECGT DEYCASPTRGGDAG | 46–76 | ||
| IC | 79–80 | 3561 (3561) | (C54…C80) | |
| Th38 | IDNYQPYPCAEDEECGTD EYCASPTRGGDAGVQ | 46–78 | ||
| IC | 79–80 | 3789 (3785) | (C54…C80) | |
| Th42 | IDNYQPYPCAEDEECGTD EYCASPTRGGDAGVQIC | 46–80 | 3772 (3767) | (C54…C80) |
| Th44-45 | Domain 1 (undigested) | ND | (C54…C107) | |
| Th46-54 | Domain 2 (undigested) | ND | (C158…C232) | |
| Th22D20c,d | LACRK | 81–85 | ||
| MCCPGN | 95–100 | |||
| IC | 106–107 | 1317 (1318) | C83-C97; C96-C107 | |
| Th37-42D19c | DEECGT | 57–62 | ||
| DAGVQIC | 74–80 | 1358 (1355) | C60-C80 | |
| Th37-42D22 | IDNYQPYPCAE | 46–56 | ||
| DEYCASPTRGG | 63–73 | 2467 (2465) | C54-C66 |
Underlined disulfide linkages in bold face were assigned in this analysis. Disulfides in parenthesis were not determined here.
Disulfide bonds in Th31 were determined by Edman sequencing/CNBr cleavage/Edman sequencing as described in Figure 2.
Th22 and Th37-42 were subjected to Aps-N digestion and HPLC separation as described in “Materials and Methods.”
Sequence analysis of peptide Th22D20 showed a significant amount of di-PTH-cystine at cycles 2 and 3, suggesting disulfide linkages of C83-C97 and C96-C107.ND: not determined.
Three N-linked glycopeptides, namely peptides A, B, and C, elute together broadly at retention time around 19.7–20 min. Different charged states of the heterogenous glycopeptides are labeled in Supporting Information Figure 3A, while the deconvoluted masses are summarized in Supporting Information Table 1. When the glycopeptide fraction was subjected to N-terminal sequence analysis, a sequence of D219HHQASXSSR228 was assigned. The X at sequence position 225 should be PTH-Asn, however no PTH-amino acid sequence was recovered for this cycle. The missing PTH-Asn signal, together with the N-S-S consensus sequence, confirms the presence of Asn-linked carbohydrate in these peptides. All parent ions of different charged states as shown in Supporting Information Figure 3A were selected for CID fragmentation to generate MS/MS spectra for sequence confirmation. Supporting Information Figure 3B is an example of an MS/MS spectrum generated from mass ion of 1164.51 Da, a triply charged ion from glycopeptide B seen in Supporting Information Figure 3A. In the presence of N-linked glycan, CID fragmentation of the oligosaccharide is clearly dominant over fragmentation of the peptide bonds of the peptide. As a result, sequential fragmentation of monosaccharide sugar units can be observed and the truncated glycopeptides B with a mannose core structure (composed of two N-acetylglucosamine and three mannose sugar units) remains as the major, final product ion as indicated in Supporting Information Figure 3B (1190.37 Da; doubly charged). Likewise, MS/MS spectra generated from CID fragmentation of 2+ and 4+ charged ions (1746.08 Da and 873.32 Da, respectively) exhibit similar fragmentation profiles to that shown in Supporting Information S-Figure 3B (data not shown). These results conclude that peptide B is an N-linked glycopeptide containing a typical biantennary oligosaccharide with two terminal hexose (galactose) and two sialic acids.
Based upon the deconvoluted mass determined for glycopeptides A, B and C as well as MS/MS analysis of the precursor ions seen in Supporting Information Figure 3B, peptides A and B were confirmed to be linked with a biantennary oligosaccharide moiety containing one and two sialic acids respectively, while peptide C consists of a triantennary oligosaccharide moiety containing two sialic acids (see Supporting Information Table 1). Comparison of the base peak intensity of the same charge states of glycopeptides A- C suggests a relative distribution of the different glycan species of ∼ 23% peptide A, 60% peptide B, and 9% peptide C. The peptide, D219HHQASNSSR228, with no N-linked sugar present was also detected at a later retention time but the signal was estimated to be less than 8%.
Two O-linked glycopeptide forms (peptides D and E) coeluted at around 35–36 min by LC-MS/MS analysis and their parent masses were listed in Supporting Information Table 1. Partial sequence analysis of the collected peptide fraction confirms that they are from the same tryptic peptide [sequence position 13-42]. The theoretical average mass of this polypeptide chain alone is 2738.1 Da. The observed masses of peptides D and E exhibit a mass increase of 947 Da and 656 Da, respectively, suggesting the presence of typical mucin-type O-linked sugars. Their proposed sugar structures are N-acetyl galacosamine (sialic acid)-galactose-sialic acid and N-acetyl galacosamine-galactose-sialic acid, respectively. Based on base peak intensity estimated from both peptides at various charged states (2+, 3+ and 4+), the relative distribution of the 947 Da and 656 Da glycans is ∼ 60% and 35%, respectively. The remaining 5% appears to be the nonglycosylated peptide.
Ser27 and Ser30 of N13LPPPLGGAAGHPGSAVSAAPGILYPGGNK42 are both candidates to be the site of O-linked sugar attachment. Unsuccessful attempts were made using both CID and ETD fragmentation of peptides D and E to assign the glycosylation site (data not shown). When peptide D (4+; 922.1 Da) was selected for CID fragmentation, sequential fragmentation of the O-linked oligosaccharide was mainly observed, confirming the presence of the proposed glycan structure described above (Supporting Information Fig. 4A). A small number of low mass y-ions can be assigned to C-terminal portion of the sequence, but the fragment ions overlapping the serine residues do not exist for assignment of the O-glycosylation site. MS/MS data from other parent ions of both peptides D and E exhibited similar fragment ion profiles and contain no useful information for glycosylation site assignment.
β-elimination using sodium hydroxide treatment of the huDkk1 tryptic digest was performed to remove the O-linked glycan from either Ser27 or Ser30 of glycopeptides D and E. The β-eliminated peptide contains a dehydroalanine residue (-18 Da) at the site previously occupied by glycosylated Ser. The reaction is usually incomplete, however only a small percentage of the dehydroalanine/ -18 Da species needs to be formed before it can be targeted for MS2 or MS3 experiments. Supporting Information Figure 4B illustrates the result. The triply charged ion, 908.1 Da, of the β-eliminated peptide N13LPPPLGGAAGHPGSAVSAAPGILYPGGNK42 (MH1+= 2721.1) was selected for MS2 fragmentation. The most prominent MS2 fragment ion, 1247.7 Da, (2+ of 15-42; P15PPLGGAAGHPGSAVSAAPGILYPGGNK42) was then selected for MS3 fragmentation. The detection of the intense ions, [y13] and [y13]-NH3 (1227.49 and 1210.47 Da) indicate that Ser30 but not Ser27 has become dehydroalanine following β-elimination of the O-linked sugar. Thus, huDkk1 is glycosylated at Ser30 with the glycan structures described in Supporting Information Table 1.
Assignment of HuDKK1 disulfide structure
The two huDKK1 cysteine-rich domains are linked by the L2 domain that contains 50 amino acids and these two cys-rich domains are unique in structure and may play different roles in the function of huDkk1 molecule. The C-terminal cysteine-rich domain was proposed to share the identical disulfide structure with colipase22 based on the relative cysteine positions, and this domain binds LRP5/LRP6 molecules to modulate Wnt signaling.1,8,9 While the disulfide arrangement of the N-terminal cysteine-rich domain remains unknown and may be different from other known reported disulfide structures, the function of this disulfide-rich domain remains to be answered.
In the following experiments, we describe the assignment of disulfide structure for both domains. HPLC peptide mapping of huDKK1 was performed with samples derived from multiple sets of chemical and proteolytic digestions in conjunction with partial TCEP reduction/N-ethyl maleimide labeling. Edman N-terminal sequencing and MALDI-MS analysis as described below were performed to identify disulfide-linked peptides and to assign location of disulfide bonds in these peptides. Non disulfide-containing peptides isolated from HPLC peptide map analysis were excluded from further analysis. The numbering of the Cys positions is based on polypeptide sequence of signal peptide-processed huDKK1. In the HPLC UV profiles shown in Figures 1–3, non-disulfide-containing peptides will not be mentioned after their identification.
Figure 3.
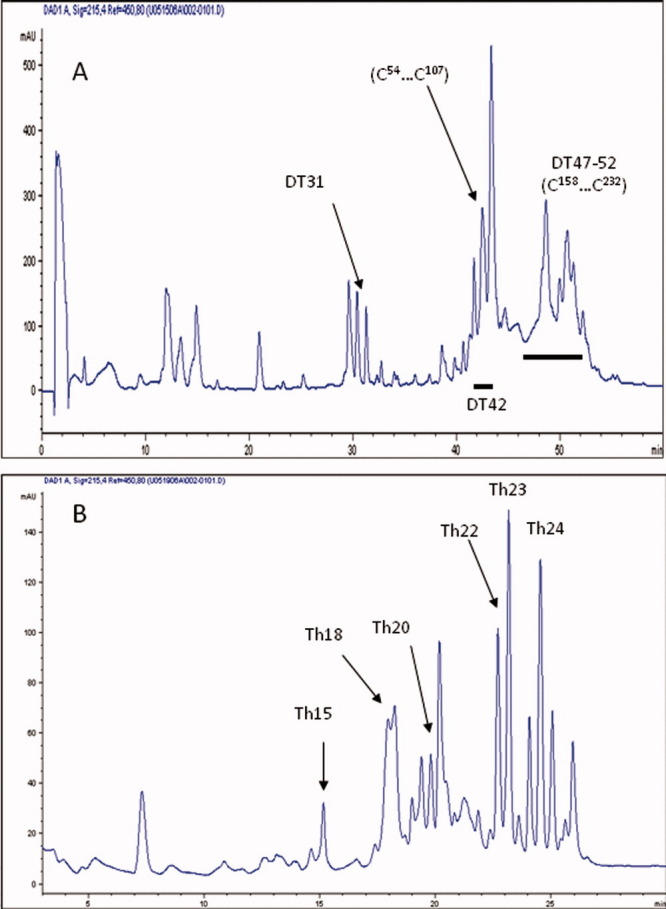
Panel A: RP-HPLC peptide map of huDKK1 peptides derived from sequential digestion of huDKK1 by endoproteinase Asp-N and trypsin. Underlined peaks (DT40-44) were combined and further treated with thermolysin. Panel B: HPLC separation peptides obtained from a second thermolysin digestion of fraction DT40-44 shown in panel A. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Thermolytic digestion
HuDKK1 was digested with thermolysin, a protease with low cleavage specificity, to generate smaller size disulfide-linked peptides before chromatography through a reversed-phase C18 HPLC column. A typical chromatogram is shown in Figure 1A and analysis of an identical digest but after DTT reduction is shown in Figure 1B. Six thermolytic peptides which disappeared upon reduction were readily identified. They were labeled as Th22, Th28, Th31, Th37, Th38 and Th42. Analysis of Th22 showed a complex mixture of disulfide linked peptides (Table I). Th22 was therefore further digested with endoproteinase AspN, which specifically cleaves peptide bonds before Asp, and rechromatographed. One peptide Th22D20 indicated that it consisted of 3 peptides linked by 2 disulfide bridges. They were identified to be L81ACRK85, M95CCPGN100, and I106C107 (measured mass = 1,317.0 Da; calculated = 1,318.0 Da). N-terminal analysis of this peptide showed good recovery of di-PTH cystine at both cycles 2 and 3. This can only be possible if C96 is linked to Cys107 and C83 is linked to Cys97. Automated Edman degradation of Th28 gave 2 sequences: V182LKEGQVCTKHRRKG196 and L212SCR215; MALDI-MS analysis of this peptide gave a measured mass of 2,219.0 Da and the calculated mass of these 2 peptides after disulfide bridge formation is 2,217.0 Da. Hence, C189 was concluded to be linked to C214.
Analyses of Th37, 38 and 42 showed that each peak consisted of 2 peptides linked by the same 2 pairs of disulfides (C54, C60, C66, and C80). They were generated because thermolysin is a relatively nonspecific protease. These peptides were therefore pooled together as Th-I and further digested with AspN. The chromatogram after this subdigestion is shown in Figure 1C. Peptide D19 gave D57EECGT62 and D74AGVQIC80 (measured mass = 1,358.0 Da; calculated mass of disulfide linked peptides = 1,355.0 Da). Hence C60 was concluded to be linked to C80. Peptide D22 also gave 2 sequences: I46DNYQPYPCAE56 and D63EYCASPTRGG73 (measured mass = 2,467.0 Da; calculated = 2,465.0 Da). Thus, C54 was linked to C66.
Peptide Th31 showed that it is a large peptide encompassing residues 81 to 114 and contains 6 cysteinyl residues forming 3 disulfide bridges: L81ACRKRRKRCMRHAMCCPGNYCKNGICVSSDQNH114. The peptide was first subjected to 1 cycle of automated Edman degradation on a glass fiber filter. The filter was removed and subjected to in situ CNBr cleavage, which was followed by subsequent sequence analysis. The data shown in Figure 2 (top panel) indicates that a high recovery of di-PTH cystine was detected at the second cycle, indicative that C83 is linked to C97. In a second experiment, peptide Th31 was subjected to N-terminal sequencing for 3 cycles, which was followed by in situ CNBr cleavage and resequencing, high recovery of di-PTH cystine was detected at both cycles 2 and 7 (Fig. 2, bottom panel). This therefore points to the linkages between C83 and C97, as well as between C90 and C102. Because peptide Th31 contains 3 disulfide bridges altogether, it therefore implies that C96 is linked to C107. The C96-C107 disulfide linkage has been verified by analyses of peptide Th22D20 as described below.
Figure 2.
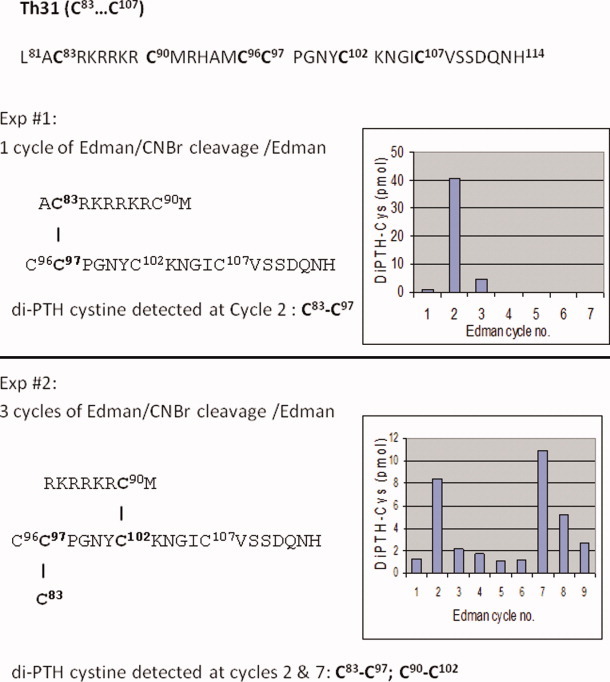
Assignment of Th31 disulfide bonds by detection of di-PTH cystine via Edman sequencing/in situ CNBr cleavage/Edman sequencing. Upper panel: Experiment 1 and lower panel: experiment 2. For experimental details, see text. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
As summarized in Table I, five disulfide bonds, four from the N-terminal domain (C54-C66; C60-C80; C83-C97; C96-C107) and one from the C-terminal domain (C189-C214), were assigned.
Endoproteinase Asp-N and trypsin digestion
The HPLC peptide map resulting from proteolytic digestion with endoproteinase Asp-N followed by trypsin, which specifically cleaves peptide bonds after Lys and Arg, is shown in Figure 3A. Disulfide-containing peptides were found in fractions DT31, DT42 and DT47-52. Characterization of these peptide fractions is summarized in Table II. Peptide DT31 was found to contain two peptides linked by a disulfide (D47NYQPYPCAE56 and D63EYCASPTR71) with a measure mass of 2,239 Da (theoretical mass: 2,238 Da), which provides an unambiguous assignment of C54 –C66 disulfide bond. The digestion using Asp-N and trypsin generated two large fragments DT42 and DT47-52. Fragment DT42 contains three peptides from the N-terminal Cys-rich domain (D47NYQPYPCAEDEECGTDEYCASPTR71, D74AGVQICLACRK85, and K88RCMRHAMCCPGNYCKNGICVSS110). A measured mass of 6,695 Da confirmed that the three peptide chains are linked by the five disulfide bonds (theoretical mass: 6,696 Da). DT47-52 is the undigested core of the C-terminal Cys-rich domain, and no mass was obtained perhaps due to the presence of an N-linked sugar.
Table II.
Disulfide-Containing Peptides Obtained from Endoproteinase AspN and Trypsin Double Digestion of huDKK1 and Thermolysin Subdigestion of Peptide DT42
| Peptide | Sequence | Seq. position | Obs. mass (calc.) | Disulfidesa |
|---|---|---|---|---|
| DT31 | DNYQPYPCAE | 47–56 | ||
| DEYCASPTR | 63–71 | 2239 (2238) | C54-C66 | |
| DT42 | DNYQPYPCAEDEECGTDEYCASPTR | 47–71 | ||
| DAGVQICLACRK | 74–85 | |||
| KRCMRHAMCCPGNYCKNGICVSS | 88–110 | 6695 (6696) | (C54…C107) | |
| DT47-52 | Domain II (undigested glycopeptide) | ND | (C158…C232) | |
| DT42Th15 | YCGEG | 207–211 | ||
| LHTCQRH | 229–235 | 1420 (1420) | C208-C232 | |
| DT42Th18 | LRSSDCASG | 159–167 | ||
| ICKP | 178–181 | 1354 (1355) | C164-C179 | |
| DT42Th20 | LACRK | 81–85 | ||
| RCMR | 89–92 | |||
| MCCPGNYCK | 95–103 | |||
| IC | 106–107 | 2405 (2406) | (C83…C107) | |
| DT42Th23 | DNYQPYPCAEDEECGTDEYCASPTR | 47–71 | ||
| IC | 79–80 | 3088 (3089) | (C54…C80) | |
| DT42Th24 | DNYQPYPCAEDEECGTDEYCASPTR | 47–71 | ||
| VQIC | 77–80 | 3314 (3315) | (C54…C80) |
See footnotes 1 in Table 1.
Fraction DT42 was further digested with thermolysin, and the digested sample was separated by reverse-phase HPLC as shown in Figure 3B. Two fractions, DT42Th15 and DT42Th18, were confirmed to contain a single disulfide bond, C208-C232 and C164-C179, respectively. DT42Th20 contains four peptides linked by three disulfides (from C83 to C107), while DT42Th23 and DT42Th24 are the fragments composed of two peptides with identical sequence of different length connected by 2 disulfide bonds including C54, C60, C66, and C80. These fractions were not further analyzed.
As summarized in Table II, endoproteinase Asp-N and trypsin digestion of huDKK1 and thermolysin digestion of peptide DT42 allows assignment of three disulfide bonds, C54–C66, C164-C179, and C208-C232. This digest further confirms the C54–C66 disulfide bond that had been identified previously in peptide Th37-42D22 (Table I).
CNBr cleavage, thermolysin digestion, and partial TCEP reduction/NEM alkylation of disulfide-containing peptides
An approach using CNBr cleavage and thermolytic digestion in tandem was useful to obtain several important peptides for disulfide assignment. Cleavage of huDKK1 after methionyl bonds using chemicals such as CNBr was effective to generate suitably large fragments. Acid-induced denaturation of the protein in 70% formic acid allowed the CNBr-treated sample to become more accessible to proteases in subsequent digestions with several proteases such as endoproteinase Lys-C, which specifically cleaves peptide bonds after Lys, and chymotrypsin, which cleaves peptide bonds after Tyr, Phe, Trp, Leu and His with lower specificity. Figure 4A shows the resulting peptide map from a huDKK1 CNBr and thermolysin digestion. Characterization of isolated disulfide peptides is summarized in Table III. Isolation from HPLC, Edman sequencing and MALDI-MS analysis of CBTh22 (V188CTKHRRKG196 and L212SCR215), CHTh23 (L207HTCQRH211 and Y229CGEG235) and CBTh26.4 (L159RSSDCASG167 and I177CKP181) verified that each contains two peptides, and the measured peptide mass confirms the existence of a single disulfide bond. The data thus provided valuable information on immediate assignment of three disulfide bonds, C189-C214, C208-C232, and C164-C179. C208-C232 and C164-C179 disulfides have also been confirmed by analysis of peptides DT42Th15 and DT42Th18 as summarized in Table III.
Figure 4.
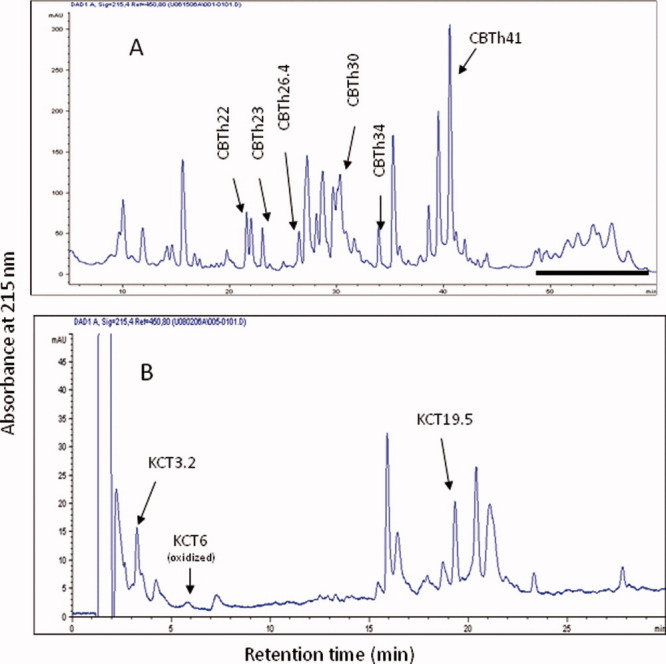
Panel A: RP-HPLC peptide map of huDKK1 peptides generated by CNBr cleavage followed by thermolytic digestion. Disulfide-containing peptides are indicated. Underlined fractions were combined and further treated with endoproteinase Lys-C and chymotrypsin. Panel B: RP-HPLC separation of peptides generated from combined endoproteinase Lys-C and chymotrypsin digestion of CBTh30 fraction as seen in panel A. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Table III.
Disulfide Containing-Peptides from CNBr Cleavage and Thermolysin of huDKK1 and from Lys-C and Chymotrypsin Digestion of CBTh30
| Sample | Sequence | Seq. position | Obs. mass (calc.) | Disulfidea |
|---|---|---|---|---|
| 1. CBTh22 | VCTKHRRKG | 188–196 | ||
| LSCR | 212–215 | 1561(1560) | C189-C214 | |
| 2. CBTh23 | LHTCQRH | 207–211 | ||
| YCGEG | 229–235 | 1419(1420) | C208-C232 | |
| 3. CBTh26.4 | LRSSDCASG | 159–167 | ||
| ICKP | 177–181 | 1355 (1355) | C164-C179 | |
| 4. CBTh30 | LACRKRRKRCM | 81–91 | ||
| CCPGNYCKNGICVSSDQNH | 96–114 | 3431(3433) | (C83…C107) | |
| 5. CBTh34b | VC | 157–158 | ||
| LCC | 168–170 | |||
| IFQRC | 202–206 | 1221 (1220) | C158-C170; C169-C206 | |
| 6. CBTh41 | IDNYQPYPCAEDEEC GTDEYCASPTRGGDAG | 46–76 | ||
| IC | 79–80 | ND (3405) | (C54…C80) | |
| 7. CBTh30KCT3.2 | RCMc | 89–91 | ||
| CK | 102–103 | 656 (657) | C90-C102 | |
| 8. CBTh30KCT6 (oxidized) | RCMRH | 89–93 | ||
| CK | 102–103 | 965 (949+16) | C90-C102 | |
| 9. CBTh30KCT19.5b | LACRK | 81–85 | ||
| CCPGNY | 96–102 | |||
| IC | 106–107 | 1477 (1476) | C83-C97; C96-C107 |
See footnote 1 in Table 1.
Disulfide linkages determined by TCEP/NEM method (see “Materials and Methods”).
Methionine converted to homoserine or homoserine lactone.
CB, cyanogen bromide; Th, thermolysin; K, endoproteinase LysC; CT, chymotrypsin.
Peptide CBTh30 contains two peptide chains with six cysteines, in which two adjacent Cys residues are present in one of the peptide chains. CBTh30 was digested with endoproteinase Lys-C for 20 hours and then with chymotrypsin for another 20 hours. The digested sample was separated by reversed phase HPLC as shown in Figure 4B. Peptides CBTh30KCT3.2 and CBTh30KCT6 elute at earlier retention time and contain a single disulfide C158-C170. Peptide KCT6 contains an unclipped oxidized Met residue. This generates a CNBr-resistant Met sulfoxide form that was detected by MALDI-MS. Peptide CBTh30KCT19.5 contains three peptides linked by two disulfide bonds consisting of C83, C96, C97, and C107, which were also found in peptide Th22D20 derived from the Asp-N digestion of Th22 seen in Table I. Due to its high recovery yield, this particular peptide fraction was very useful for the partial reduction experiment described below.
Both CBTh30KCT19.5 and CBTh34 contain three peptides linked by two disulfides, with two adjacent cysteine residues being present in one of the polypeptide chain. It has been very difficult to make definitive assignment for disulfide bonds including these cysteine residues. To address this, we performed partial reduction of these two samples with TCEP followed by alkylation with NEM as described in Supporting Information Materials and Methods. TCEP reduction at pH 4.5 generated a useful, partially reduced intermediate. The intermediate form is composed of two peptide chains with a single intact disulfide that links two peptide chains together: one peptide contains a single cysteine, while the other contains an unlabeled cysteine with an adjacent NEM-labeled cysteine. The third peptide chain containing an NEM-labeled cysteine was separated from this intermediate form after reduction. NEM-modified samples were then subjected to HPLC separation. Supporting Information Figure 5A shows the HPLC chromatogram for peptide CBTh30-KCT19.5 after TCEP reduction and NEM modification. The nonreduced peptide and the partially reduced and NEM-alkylated form were sequenced and the molecular weight determined by MALDI-MS. The unmodified disulfide-linked peptide (L81ACRK85, C96C97PGNY101, and I106C107) exhibited a mass of 1,477 Da, while the NEM-labeled intermediate showed a mass of 1,369 Da, corresponding to only two of the peptides (L81ACRK85 and C96C97PGNY101) which are connected by one disulfide bond (Supporting Information Fig. 5B, spectra 1 & 2, respectively). The data indicates that either C96 or C97 has been labeled with NEM. The NEM-labeled derivative was then subjected to one cycle of Edman degradation, the sequenced sample was extracted from PVDF membrane (see Supporting Information Materials and Methods), and immediately analyzed by MALDI-MS. The peptide derivative with an observed mass of 1,162 Da (Supporting Information Fig. 5, spectrum 3) corresponds to two peptides, A82CRK85 (-PTC) and C97PGNY101, linked by a disulfide bond, where the Lys85 ɛ-amino group reacts with the Edman reagent to form the phenylthicarbamyl (PTC) derivative. Therefore, C83 and C97 form a disulfide bond, and by default, C96 forms the disulfide bond with C107 (both were labeled with NEM following partial reduction).
Figure 5.
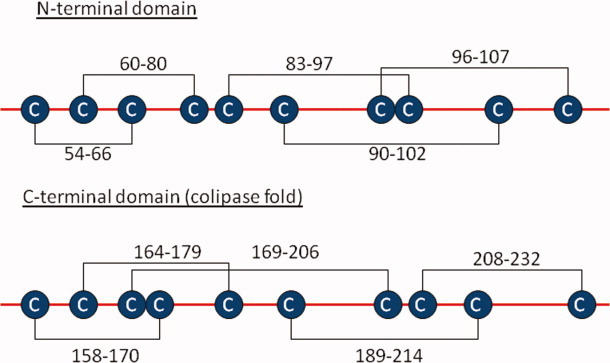
Assignment of disulfide bonds in huDKK1 indicated in the N- and C-terminal domains. The connection positions are shown for the assigned disulfide pairs. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Peptide CBTh34 also contained three peptides (V157C158, L168CC170, and I202FQRC206) connected by two disulfide bonds. Following TCEP partial reduction/NEM labeling, the sample was subjected to RP-HPLC for isolation of labeled products (Supporting Information Fig. 6A). The unlabeled peptide containing 0 NEM moiety has a mass of 1,220.7 Da (theoretical mass: 1,220.3 Da), while the derivative labeled with 1 NEM had a mass of 1128 Da [Supporting Information Fig. 6(B,C)]. This indicates that the disulfide bond linking to the V157C158 peptide had been preferentially reduced by TCEP and C158 labeled with NEM, leaving behind disulfide-linked peptides L168CC170 and I202FQRC206 with 1 NEM on one of the cysteine residues. The NEM derivative was subjected to three cycles of Edman degradation. NEM-labeled PTH-Cys was detected in the 3rd sequencing cycle but not at the 2nd cycle, indicating that C170 contains the NEM-label. This data establishes the C158-C170 and C169-C206 disulfide bonds in peptide CBTh34.
HuDKK1 disulfide structure and comparison with other Cys-rich clusters
Figure 5 shows the 10 disulfide bonds in the huDKK1 molecule: 5 in the N-terminal Cys-rich domain and 5 in the C-terminal Cys-rich domain. Several disulfide bonds have been confirmed more than once using different sets of isolated peptides and different characterization methods including Edman sequencing, MALDI-MS, and partial reduction/alkylation.
The three disulfide bonds that are sequentially contained within the Cys-Cys sequence pairs were compared between the N- and C-terminal domains. Despite the different lengths of the disulfide loops between Cys residues, the disulfide topographic arrangement of the C83-C97, C96-C107, and C90-C102 bonds in the N-terminal domain and for the C158-C170, C169-C206 and C164-C179 bonds in the C-terminal domain appear to be identical (Fig. 5). However, the overall disulfide pattern when considering all five disulfide bonds per domain is clearly different, suggesting that they must play a different role in the biological function of DKK1. As demonstrated by Brot and Sokol,25 the C-terminal Cys-rich domains of DKK1 and DKK2 are both necessary for Wnt inhibition; both associate with LRP6 and stimulate LRP6-dependent embryonic axis induction. In contrast, the N-terminal domain appears to play a regulatory role in these interactions. Further studies are required to unravel the role of the N-terminal domain structure on the interaction between DKK1 and its binding partners.
Comparison of disulfides C83-C97, C96-C107, and C90-C102 in the N-terminal domain with several known peptides and proteins such as omega conotoxin GV1A peptides26 and AGRP27,28 demonstrates a similar disulfide pairing pattern. These peptides, and other antimicrobial peptides, all contain common features in disulfide structure and the characteristic Cys–Cys sequence pair, similar to those three disulfides in the N-terminal domain of DKK1, even though the toxin peptides and DKK1 are not biologically related to each other. For example, omega conotoxin is a neurotoxin that strongly inhibits calcium channels.29 Moreover, the disulfide motif of DKK1 N-terminal domain as described above is similar to that of omega conotoxin GV1A, and their disulfide pairing pattern is also identical, suggesting that their refolding mechanism may be quite similar.
The disulfide structure of DKK1 C-terminal domain has been confirmed to belong to a “colipase fold”, typical of a wide range of small proteins including scorpion phospholipase, snake toxins, and proteases inhibitors.24,30 Several hydrophobic residues are conserved between the carboxy-terminal domain of DKK and colipases. These exposed hydrophobic residues are required for the interaction of lipases with hydrophobic lipid micelles during the process of lipid digestion, which suggest that the carboxy terminal, colipase-like domain of Dkk may play a role on the Wnt function via membrane interaction.24,31
Acknowledgments
The authors appreciate the assistance from Dr. Li Yang for providing human DKK1 purification protocol. They also thank the management support from Dr. Margaret Karow and from Amgen metabolic disorders therapeutic area team.
Abbreviations
- 4-HCCA
4-cyanohydroxycinnamic acid
- AGRP
agouti-related protein
- CID
collision-induced dissociation
- DKK1
Dickkopf-1
- ETD
electron transfer dissociation
- LRP
lipoprotein receptor-related protein
- MALDI
matrix-assisted laser desorption ionization
- NEM
N-ethylmaleimide
- PTH-
phenylthiohydantoinyl derivative of amino acid
- PVDF
polyvinylidene difluoride
- TCEP
tris (2-carboxyethyl)-phosphine
- TFA
trifluoroacetic acid
Supplementary material
References
- 1.Semenov MV, Tamai K, Brott BK, Kuhl M, Soko S, He X. Head inducer Dokkopf-1 is a ligand for Wnt coreceptor LRP6. Curr Biol. 2001;11:951–961. doi: 10.1016/s0960-9822(01)00290-1. [DOI] [PubMed] [Google Scholar]
- 2.Li X, Liu P, Liu W, Maye P, Zhang J, Zhang Y, Hurley M, Guo C, Boskey A, Sun L, Harris SE, Rowe DW, Ke HZ, Wu D. Dkk2 has a role in terminal osteoblast differentiation and mineralized matrix formation. Nat Genet. 2005;37:945–952. doi: 10.1038/ng1614. [DOI] [PubMed] [Google Scholar]
- 3.Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- 4.Semenov MV, He X. LRP5 mutations linked to high bone mass disease cause reduced LRP5 binding and inhibition by SOST. J Biol Chem. 2006;281:38276–38284. doi: 10.1074/jbc.M609509200. [DOI] [PubMed] [Google Scholar]
- 5.Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 6.Westendorf JJ, Kahler RA, Schroeder TM. Wnt signaling in osteoblasts and bone diseases. Gene. 2004;341:19–39. doi: 10.1016/j.gene.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 7.Krishnan V, Bryant HU, MacDougald OA. Regulation of bone mass by Wnt signaling. J Clin Invest. 2006;116:1202–1209. doi: 10.1172/JCI28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Zhang Y, Kang H, Liu W, Liu P, Zhang J, Harris SE, Wu D. Scleorostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J Biol Chem. 2005;280:19883–19887. doi: 10.1074/jbc.M413274200. [DOI] [PubMed] [Google Scholar]
- 9.Bourhis E, Tam C, Franke Y, Bazan JF, Ernst J, Hwang J, Costa M, Cochran AG, Hannoush RN. Reconstitution of Frizzled8-Wnt3a-LRP6 signaling complex reveals multiple Wnt and DKK1 binding sites on LRP6. J Biol Chem. 2010;285:9172–9179. doi: 10.1074/jbc.M109.092130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fedi P, Bafico A, Soria AN, Burgess WH, Miki T, Bottaro DP, Kraus MH, Aaronson SA. Isolation and biochemical characterization of the human DKK1 homologue, a novel inhibitor of mammalian Wnt signaling. J Biol Chem. 1999;274:19465–19472. doi: 10.1074/jbc.274.27.19465. [DOI] [PubMed] [Google Scholar]
- 11.Mao B, Wu W, Li Y, Hoppe D, Stannek P, Glinka A, Niehrs C. LDL receptor-related protein 6 is a receptor for Dkk proteins. Nature. 2001;411:321–325. doi: 10.1038/35077108. [DOI] [PubMed] [Google Scholar]
- 12.Mukhopadhyay M, Shtrom S, Rodriguez-Esteban C, Chen L, Tsukui T, Gomer L, Dorward DW, Glinka A, Grinberg A, Huang SP, Niehrs C, Belmonte JCI, Westphal H. Dickkopf1 is required for embryonic head induction and limb morphogenesis in the mouse. Dev Cell. 2001;1:423–434. doi: 10.1016/s1534-5807(01)00041-7. [DOI] [PubMed] [Google Scholar]
- 13.Morvan F, Boulukos K, Clement-Lacroix P, Roman SR, Suc-Royer I, Vayssiere B, Ammann P, Martin P, Pinho S, Pognonec P, Mollat P, Niehrs C, Baron R, Rawadi G. Deletion of a single allele of the DKK1 gene leads to an increase in bone formation and bone mass. J Bone Mineral Res. 2006;21:934–945. doi: 10.1359/jbmr.060311. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Sarosi I, Cattley RC, Pretorius J, Asuncion F, Grisanti M, Morony S, Adamu S, Geng ZP, Qiu WR, Kosetnuik P, Lacy DL, Simonet S, Bolon B, Qian X, Shalhoub V, Ominsky MS, Ke HZ, Li X, Richards WG. DKK1-mediated inhibition of Wnt signaling in bone results in osteopenia. Bone. 2006;39:754–766. doi: 10.1016/j.bone.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 15.Diarra D, Stolina M, Polzer K, Zwerina J, Ominsky MS, Dwyer D, Korb A, Smolen J, Hoffmann M, Scheinecker C, der Heide D, Landewe R, Lacey D, Richards WG, Schett G. Dickkopf-1 is a master regulator of joint remodeling. Nat Med. 2007;13:156–163. doi: 10.1038/nm1538. [DOI] [PubMed] [Google Scholar]
- 16.Tian E, Zhan FH, Walker R, Rasmussen E, Ma YP, Barlogie B, Shaughnessy JD. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. New England J Med. 2003;349:2483–2493. doi: 10.1056/NEJMoa030847. [DOI] [PubMed] [Google Scholar]
- 17.Hoeppner LH, Secreto FJ, Westendorf J. Wnt signaling as a therapeutic target for bone diseases. Exp Opin Ther Targets. 2009;13:485–496. doi: 10.1517/14728220902841961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MaCarthy HS, Marshall MJ. Dickkopf-1 as a potential therapeutic target in Paget's disease of bone. Exp Opin Ther Targets. 2010;14:221–230. doi: 10.1517/14728220903525720. [DOI] [PubMed] [Google Scholar]
- 19.Kubota T, Michigami T. Wnt signaling in bone metabolism. J Bone Miner Metab. 2009;27:265–271. doi: 10.1007/s00774-009-0064-8. [DOI] [PubMed] [Google Scholar]
- 20.Choi Y, Arron JR, Townsend MJ. Promising bone-related therapeutic targets for rheumatoid arthritis. Nature Rev Rheumatol. 2009;5:543–548. doi: 10.1038/nrrheum.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinzone JJ, Hall BM, Thudi NK, Vonau M, Qiang Y-W, Rosol TJ, Shaughnessy JD. The role of Dickkopf-1 in bone development, homeostasis, and disease. Blood. 2009;15:517–525. doi: 10.1182/blood-2008-03-145169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krupnik VE, Sharp JD, Jiang C, Robison K, Chickering TW, Amaravadi L, Brown DE, Guyot D, Mays G, Leiby K, Chang B, Duong T, Goodearl ADJ, Gearing DP, Sokol SY, McCarthy SA. Functional and structural diversity of the human Dickkopf gene family. Gene. 1999;238:301–313. doi: 10.1016/s0378-1119(99)00365-0. [DOI] [PubMed] [Google Scholar]
- 23.Aravind L, Koonin EV. A colipase fold in the carboxy-terminal domain of the Wnt antagonists –the Dickkopfs. Nature. 2000;8:R477–R478. doi: 10.1016/s0960-9822(98)70309-4. [DOI] [PubMed] [Google Scholar]
- 24.Fleury D, Gillard C, Lebhar H, Vayssiere B, Touitou R, Rawadi G, Mollat P. Expression, purification and characterization of murine DKK1 protein. Protein Exp Purif. 2008;60:74–81. doi: 10.1016/j.pep.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 25.Brott BK, Sokol SY. Regulation of Wnt/LRP signaling by distinct domains of Dickkopf proteins. Mol Cell Biol. 2002;22:6100–6110. doi: 10.1128/MCB.22.17.6100-6110.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olivera BM, Rivier J, Clark C, Ramilo CA, Corpuz GP, Abogadie FC, Mena EE, Woodward SR, Hillyard DR, Cruz LJ. Diversity of Conus neuropeptides. Science. 1990;249:257–263. doi: 10.1126/science.2165278. [DOI] [PubMed] [Google Scholar]
- 27.Bures EJ, Hui JO, Young Y, Rohde Chow DT, Katta V, Rohde MF, Zeni L, Rosenfeld RD, Stark KL, Haniu M. Determination of disulfide structure in Agouti related protein (AGRP) by stepwise reduction and alkylation. Biochemistry. 1998;37:12172–12177. doi: 10.1021/bi981082v. [DOI] [PubMed] [Google Scholar]
- 28.Young Y, Zeni L, Rosenfeld RD, Stark K, Rohde MR, Haniu M. Disulfide assignment of the C-terminal cysteine knot of Agouti-related protein (AGRP) by direct sequence analysis. J Peptide Res. 1999;54:514–521. doi: 10.1034/j.1399-3011.1999.00126.x. [DOI] [PubMed] [Google Scholar]
- 29.Pruneau D, Angus JA. [Omega]-Conotoxin GVIA, the N-type calcium channel inhibitor, is sympatholytic but not vagolytic: Consequences for hemodynamics and autonomic reflexes in conscious rabbits. J Cardio Pharmacol. 1990;16:675–680. doi: 10.1097/00005344-199010000-00022. [DOI] [PubMed] [Google Scholar]
- 30.Hubbard TJP, Murzin AG, Brenner SE, Chothia C. SCOP: a structural classification of proteins database. Nucleic Acids Res. 1997;25:236–239. doi: 10.1093/nar/25.1.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frieda Reichsman F, Moore HM, Cumberledge S. Sequence homology between Wingless/Wnt-1 and a lipid-binding domain in secreted phospholipase A2. Curr Biol. 1999;9:R353–R355. doi: 10.1016/s0960-9822(99)80225-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


