Abstract
Sensory adaptation in bacterial chemotaxis involves reversible methylation of specific glutamyl residues on chemoreceptors. The reactions are catalyzed by a dedicated methyltransferase and dedicated methylesterase. In Escherichia coli and related organisms, control of these enzymes includes an evolutionarily recent addition of interaction with a pentapeptide activator located at the carboxyl terminus of the receptor polypeptide chain. Effective enzyme activation requires not only the pentapeptide but also a segment of the receptor polypeptide chain between that sequence and the coiled-coil body of the chemoreceptor. This segment has features consistent with a role as a flexible and presumably unstructured linker and enzyme tether, but there has been no direct information about its structure. We used site-directed spin labeling and electron paramagnetic resonance spectroscopy to characterize structural features of the carboxyl-terminal 40 residues of E. coli chemoreceptor Tar. Beginning ∼ 35 residues from the carboxyl terminus and continuing to the end of the protein, spectra of spin-labeled Tar embedded in native membranes or in reconstituted proteoliposomes, exhibited mobilities characteristic of unstructured, disordered segments. Binding of methyltransferase substantially reduced mobility for positions in or near the pentapeptide but mobility for the linker sequence remained high, being only modestly reduced in a gradient of decreasing effects for 10–15 residues, a pattern consistent with the linker providing a flexible arm that would allow enzyme diffusion within defined limits. Thus, our data identify that the carboxyl-terminal linker between the receptor body and the pentapeptide is an unstructured, disordered segment that can serve as a flexible arm and enzyme tether.
Keywords: bacterial chemotaxis, disordered protein segments, sensory adaptation, EPR spectroscopy, receptor methyltransferase
Introduction
Sensory adaptation is central to the mechanism of bacterial chemotaxis.1,2 Adaptation is mediated by reversible covalent modification of chemoreceptors, methylation of specific glutamyl residues by methyltransferase CheR, and demethylation/deamidation of those methylesters by methylesterase/deamidase CheB. For Escherichia coli and its close relative, Salmonella enterica, efficient modification by these enzymes and thus effective chemotaxis requires an activity-enhancing pentapeptide, asparagine-tryptophan-glutamate-threonine or serine-phenylalanine (NWETF or NWESF in the single-letter code), at the receptor carboxyl terminus [Fig. 1(B)].3–14 This pentapeptide binds the two adaptation enzymes,5,10,15,16 and in doing so enhances rates of modification for the sequence-bearing receptor as well as for neighboring receptors in the same membrane via adaptational assistance.6,17,18 Related pentapeptide sequences are found in receptors of other proteobacteria19–21 and are likely to perform a similar role in enhancing adaptational modification.
Figure 1.
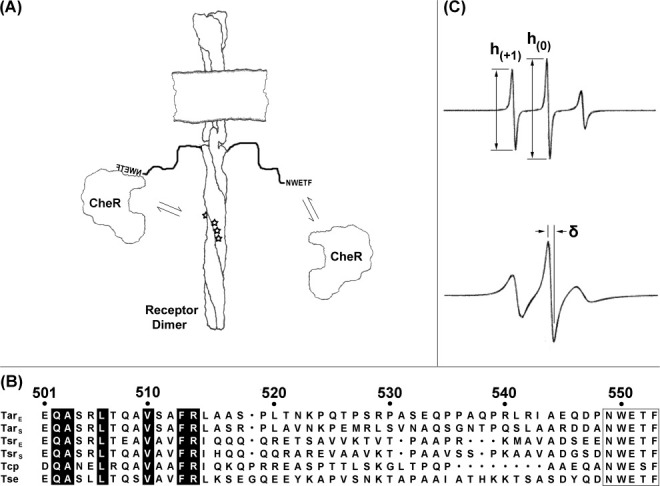
Chemoreceptors and EPR spectra. (A) Chemoreceptors and CheR. The cartoon shows a membrane-embedded chemoreceptor dimer with its carboxyl-terminal NWETF pentapeptide unoccupied (right) or bound to methyltransferase CheR (left). Equilibrium arrows indicate interactions of pentapeptide and CheR (right) or tethered CheR with methyl-accepting sites (stars; left). (B) Alignment of carboxyl-terminal sequences of pentapeptide-bearing receptors from E. coli and S. enterica. Exact matches for the six sequences are highlighted in black and the NWETF/NWESF pentapeptide is boxed. Position numbers are for E. coli Tar. (C) Example EPR spectra and illustrations of semiquantitative mobility parameters. The figure shows representative spectra of spin labels in an unstructured protein region (upper spectrum) and at a solvent exposed residue in an alpha helix (lower spectrum; spectra from Columbus and Hubbell, Biochemistry, 2004, 43, 7273–7287, © W.H. Freeman, reproduced by permission). The ratio of the low-field to central peak amplitudes (top spectrum) can be used to derive a semiquantitative mobility parameter h(+1)/h(0). The peak to peak width (δ) of the central line (bottom spectrum) is used to calculate the scaled mobility parameter MS (see Materials and Methods).
The pentapeptide is thought to enhance modification by acting as a high-affinity binding site for the methyltransferase5 and as an allosteric activator for the methylesterase.22 High-affinity binding to CheR would restrict diffusion of the enzyme, increasing its effective concentration and consequently the rate of modification.23 Allosteric activation of CheB by the pentapeptide would be expected to involve a ternary complex of pentapeptide, enzyme, and the side chain to be modified. Both mechanisms of enhancement imply that the modification enzyme binds the activating pentapeptide at the same time it binds its substrate residue. This simultaneous binding of distant sites on a chemoreceptor is thought to be possible because a linker of 30–35 residues between the helical coiled-coil body of the receptor24 and the carboxyl-terminal NWETF [Fig. 1(A)] could serve as a flexible arm. Flexibility would allow pentapeptide-tethered CheR to reach modification sites on the receptor to which it is bound as well as sites on neighboring receptors.23,25 It would also allow the pentapeptide to reach CheB docked at a modification site.22 Consistent with this notion, the ability of a carboxyl-terminal NWETF to enhance the action of either enzyme or to enable effective chemotaxis is dependent on the length of the linker.14 Many linker sequences contain several prolines and are enriched in polar residues, features commonly found in disordered protein segments.26 Furthermore, there is little conservation among linker sequences, even for the same receptor from closely related organisms, in contrast to substantial sequence conservation over much of the receptor cytoplasmic domain [Fig. 1(B)].
The importance of the carboxyl-terminal linker for chemoreceptor function and effective chemotaxis, coupled with the several lines of evidence for its role as a flexible arm, prompted us to investigate its structural features. As a flexible arm would in essence be an unstructured, disordered segment, we chose an experimental approach that would provide specific measurements for any residue of interest, whether in a structured or unstructured region and that could be applied to intact chemoreceptors in their native or reconstituted membrane environment. Thus, we chose site-directed spin labeling and electron paramagnetic resonance (EPR) spectroscopy, a combination that has already provided useful information about disordered protein regions,27–32 and applied this approach to characterize the carboxyl-terminal segment of the extensively studied aspartate receptor Tar from E. coli.
Results
Characterization of a putatively unstructured protein segment
Cysteines were introduced by site-specific mutagenesis into the 40-residue sequence at the carboxyl terminus of chemoreceptor Tar, a protein which otherwise lacks this amino acid, to create a collection of receptors with single cysteines at selected positions in that segment and thus provide a reactive side chain for introduction of a spin label by reaction with a methanethiosulfonate reagent carrying a nitroxide. Initially, cysteines were placed at five-residue intervals along the segment as well as in place of asparagine in the NWETF pentapeptide and as a one-residue extension following the native carboxyl terminus. Initial spectra indicated a transition from less mobile to more mobile ∼ 30 residues from the pentapeptide. To investigate the details of this transition, we introduced cysteines at every position from 514 to 522, although we were unable to analyze position 518 because Tar with a cysteine at that position was produced at too low a level.
Spectra were collected for spin-labeled Tar in two environments: isolated native cytoplasmic membrane vesicles and reconstituted proteoliposomes made with native E. coli lipids and pure receptor. Cytoplasmic membrane vesicles were prepared from cells producing high levels of each cysteine-containing Tar. Those membranes were labeled with a nitroxide using a methanethiosulfonate-based reagent. To obtain receptor for reconstitution into proteoliposomes, cysteine-containing forms of Tar were purified from cytoplasmic membrane vesicle preparations utilizing the subunit exchange that occurs between detergent-solubilized forms of the same chemoreceptor.33 We isolated Tar heterodimers with one cysteine-containing subunit and one subunit with a carboxyl-terminal six-histidine tag but no cysteine by mixing detergent-solubilized cytoplasmic membranes containing the two respective receptor forms and purifying these heterodimers (as well as homodimers with two tagged subunits but no cysteine) on a chelated nickel column. Purified receptors were treated with the methanethiosulfonate spin-labeling reagent and reconstituted into proteoliposomes. Formation of heterodimers by subunit exchange allowed purification of multiple cysteine-substituted forms of Tar using the histidine tag on one subunit in combination with characterization of spin-labeled sites on the subunit with a native carboxyl terminus, thus avoiding the presence of a six-histidine extension that could potentially perturb the linker region.34 The alternative, placing the affinity tag at the amino terminus, was not feasible because tags at this location resulted in low levels of protein production (A. Lilly, M. Li, and G.L. Hazelbauer, unpublished data). We assessed each preparation of spin-labeled, reconstituted Tar for structural perturbations by determining extents of deamidation and thus structural recognition by phosphorylated CheB. Approximately half the Tar for each variant (54 ± 3%) was modified, essentially the proportion expected to be accessible to added enzyme if the receptor was randomly oriented in reconstituted proteoliposomes and approximately the accessible proportion of vesicle-inserted Tar lacking a cysteine and a spin label. In any case, the data indicated that receptors with cysteines and thus spin labels at different position receptors were not differentially perturbed in the course of reconstitution and thus mobilities could be compared directly.
Patterns of EPR spectra for the carboxyl-terminal segment of chemoreceptor Tar
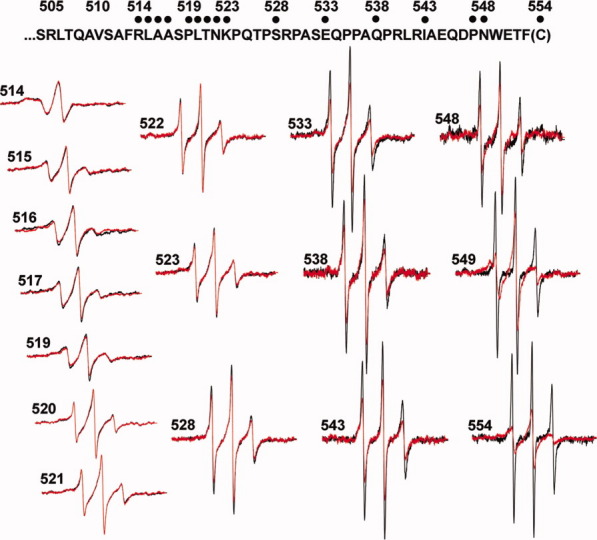
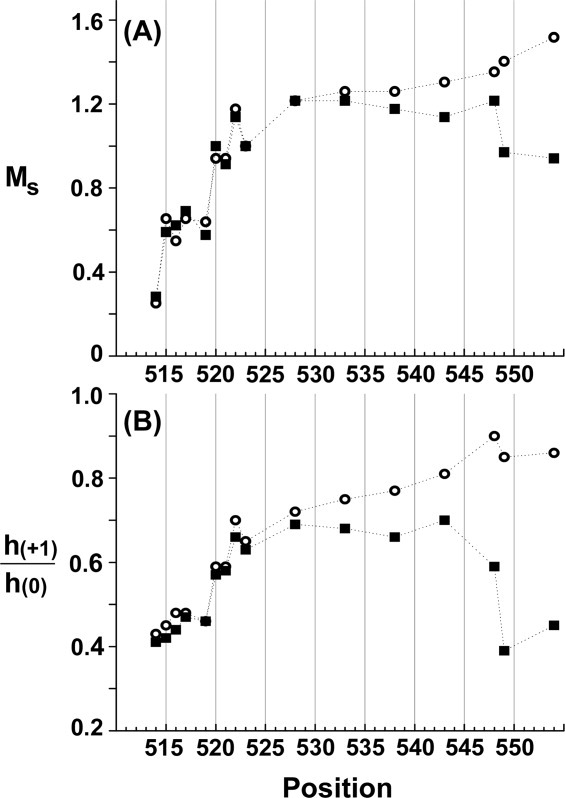
We collected EPR spectra for spin-labeled cysteines at 16 different positions in the final 40 residues of chemoreceptor Tar, including a one-residue carboxyl-terminal cysteine extension. Each spin-labeled receptor was characterized in the native, multiprotein environment of the cytoplasmic membrane (Fig. 2) and as the sole protein inserted in reconstituted proteoliposomes (Fig. 3). Spectra collected from Tar in native membrane included a low mobility component from spin labels attached to membrane proteins other than Tar (Fig. 2, spectra in dashed-line box). Fortunately this component was sufficiently different from the contributions of spin-labeled Tar that effects of varying the position of the spin label along the linker sequence were clearly evident in the respective spectra and these effects corresponded to those observed for purified Tar inserted in proteoliposomes (Figs. 2 and 3). In both native membrane and reconstituted proteoliposomes, spectra for positions 514 through 519 had features characteristic of the reduced mobility observed for side chains in structured regions of a protein. In contrast, positions beginning at 520 and extending over 30 residues to the carboxyl terminus had spectra characteristic of the high mobility of side chains in unstructured protein regions.27–30,35 The pattern of lower mobility for positions through 519 and high mobility thereafter is further illustrated in Figure 4 by two semiquantitative mobility parameters, Ms, the normalized central line width36 and h(+1)/h(0), the ratio of the heights of the low-field and central lines29 [Fig. 1(C)]. The slightly lower parameter values for Tar in native membrane versus proteoliposomes likely reflect the influence on the mobility parameters of the lower mobility background spectra in membrane. For positions 520 and beyond, the magnitudes of the Ms values [Fig. 4(A)] were comparable with those observed for highly mobile, unstructured segments in other proteins,37,38 as were the magnitudes of h(+1)/h(0) [Fig. 4(B)].29–32 Thus, both qualitative and quantitative assessment of the spectra indicated that the carboxyl-terminal sequence of Tar, beginning at approximately position 520 had the features of an unstructured and thus disordered segment.
Figure 2.
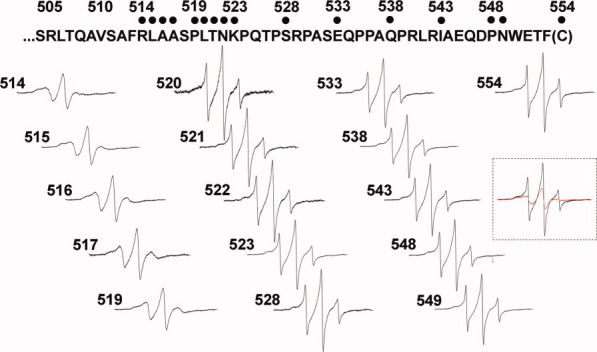
Spectra of spin-labeled Tar in native membrane vesicles. Normalized EPR spectra for 16 positions in the Tar carboxyl-terminal segment, identified with dots on the sequence above the spectra and position numbers, are shown for receptor embedded in isolated native membrane. The spectra in the dashed-line box compare un-normalized spectra from two native membrane preparations which were reacted with the sulfhydryl-reactive spin label reagent as described in Materials and Methods. One contained Tar with a cysteine at position 543 (black) and the other Tar devoid of cysteines (red), each with approximately equal amounts of total protein and Tar.
Figure 3.
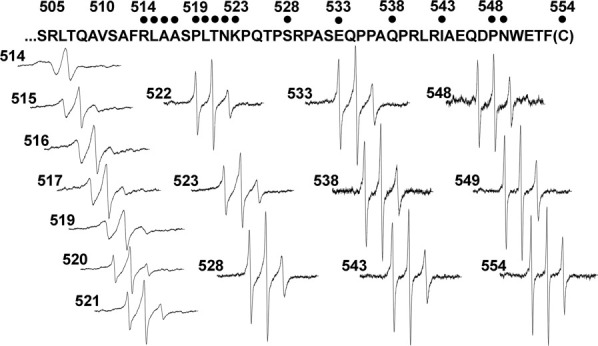
EPR spectra of purified, spin-labeled Tar reconstituted into proteoliposomes. Normalized EPR spectra, as in Figure 2, are shown for purified Tar reconstituted into proteoliposomes.
Figure 4.
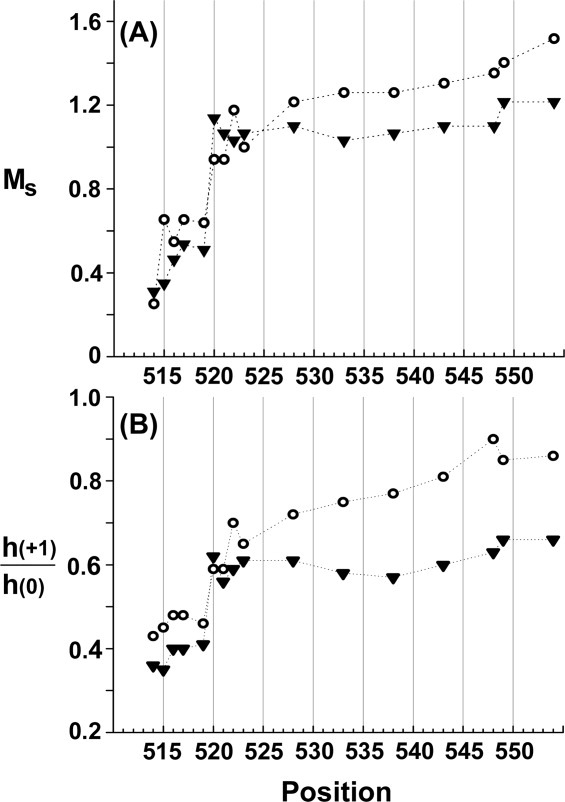
Mobility parameters as a function of spin label position for Tar in native vesicles or proteoliposomes. The parameters scaled mobility, MS (A) and h(+1)/h(0) (B), derived from the spectra in Figs. 2 and 3, are plotted as a function of spin label position for Tar in native vesicles (closed triangles) or proteoliposomes (open circles). See Materials and Methods for descriptions of the parameters. Dotted lines are provided to aid the eye.
Test for lack of structure: effects of denaturing urea
We tested the notion that the carboxyl-terminal sequence of Tar beginning at position 520 was essentially unstructured by exposing selected spin-labeled proteins embedded in their native membrane environment to 5M urea and comparing spectra before and after addition of denaturant. For positions 514, 515, and 517, locations at which spectra were characteristic of a structured region, denaturant generated major spectral changes, indicating significant increases in mobility, presumably reflecting loss of regular structure (Fig. 5, left-hand spectra). For positions 528, 549, and 554, locations at which the spectra were characteristic of lack of structure, urea generated little change (Fig. 5, right-hand spectra), consistent with those positions being in an unstructured, naturally disordered segment and thus not significantly affected.
Figure 5.
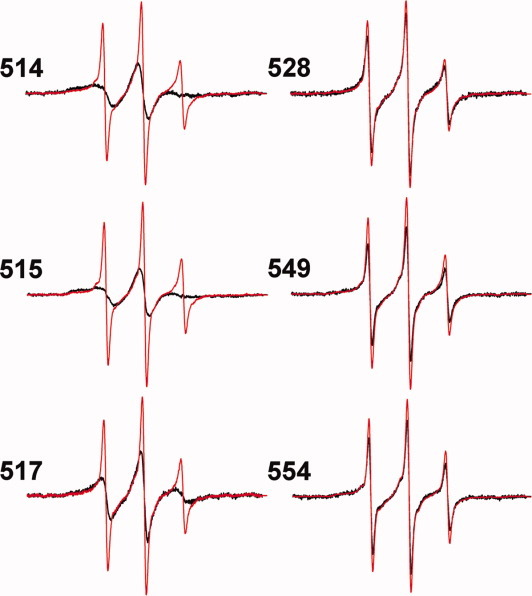
Effects of denaturant on EPR spectra of spin-labeled Tar. Normalized spectra for Tar in native membranes with spin labels at the indicated positions are shown in the absence (black) and presence (red) of 5M urea. Spectra were corrected for spin labels on other membrane proteins by subtracting the spectrum of native membrane vesicles containing Tar devoid of cysteine and treated with the spin-label reagent in the same way as vesicles containing the respective cysteine-containing form of Tar, scaled to total protein.
Effects of CheR on mobility of the carboxyl-terminal segment
The carboxyl-terminal linker is thought to enhance CheR action by serving as a flexible arm that restricts diffusion of pentapeptide-bound enzyme to a volume near the receptor but allows diffusion within that volume and thus increases the probability of enzyme interaction with substrate sites on the receptor body.5,23,25 We investigated effects of CheR binding on mobility of the carboxyl-terminal segment of Tar by collecting EPR spectra for our set of spin-labeled receptors in the presence of a high concentration of CheR. Pentapeptide interacts with CheR by becoming the fourth strand of a beta-sheet in an enzyme subdomain39 and thus should become significantly less mobile on binding. This was the case. CheR binding significantly broadened spectra of spin labels within (549) or at the carboxyl terminus (554) of the pentapeptide (Fig. 6) and reduced the mobility parameters Ms and h(+1)/h(0) (Fig. 7). We expected ∼ 50% of the population of spin-labeled Tar to be occupied by CheR because this proportion was accessible in proteoliposomes (see above). Fortunately, the magnitudes of the changes in mobility on CheR binding were sufficiently large to be easily detected even with ∼ 50% occupancy, although the strong influence of the most mobile component on the central line width meant that the proportional change was less for Ms than for h(+1)/h(0). Enzyme binding also reduced mobility of the linker, in a gradient of decreasing effects over 10–15 residues from the site of enzyme binding (Fig. 7), as might be expected for attaching the end of a flexible arm to a relatively large mass.
Figure 6.
Effects of CheR on EPR spectra of spin-labeled Tar. Normalized spectra for purified Tar in reconstituted proteoliposomes with spin labels at the indicated positions are shown in the absence (black) and presence (red) of CheR at a concentration sufficient to occupy ∼ 97% of accessible Tar-borne pentapeptide.
Figure 7.
Effects of CheR on mobility parameters of spin-labeled Tar. Mobility parameters MS (A) and h(+1)/h(0) (B) derived from the spectra in Figure 6 are plotted as a function of spin label position for Tar in proteoliposomes in the absence (open circles) or presence (filled squares) of excess CheR.
To determine whether the effects we observed on addition of CheR reflected specific binding to the carboxyl-terminal pentapeptide, we performed competition experiments using excess free peptide. This synthetic peptide, EENWETF, had the sequence of the final seven residues of E. coli chemoreceptor Tsr13 and was soluble at the required high concentrations, presumably due to the two amino-terminal glutamyl residues. Addition of a ∼ 10-fold excess of this free heptapeptide relative to receptor-borne pentapeptide essentially eliminated the otherwise drastic effects of CheR on spectra for positions 549 and 554 (Fig. 8), providing strong evidence that CheR was indeed binding at its physiologically relevant site. Excess competing peptide also eliminated the more subtle effect of CheR on a linker position, 543, which is outside the enzyme-binding site, indicating that effects on linker mobility were also the result of physiologically relevant binding.
Figure 8.
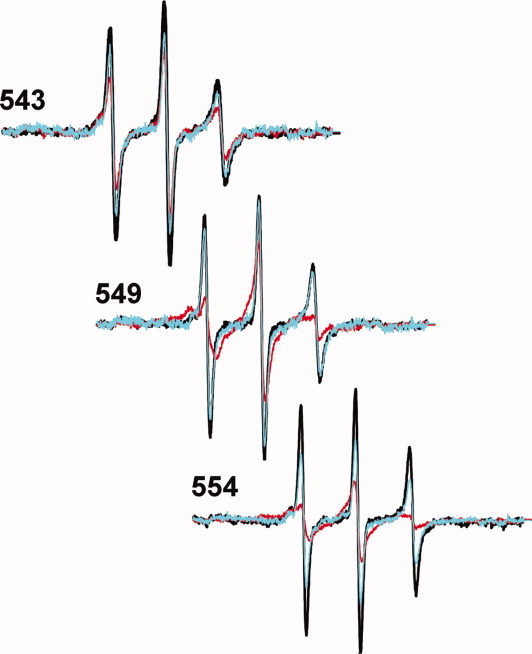
Effects of a competitor peptide on CheR-induced changes in spectra of spin-labeled Tar. Normalized spectra for purified Tar, with spin labels at the indicated positions, in reconstituted proteoliposomes are shown without additions (black), with CheR at a concentration calculated to occupy ∼ 97% of accessible Tar-borne pentapeptide NWETF (red) and with CheR at the same concentration plus peptide EENWETF at a 10-fold excess relative to Tar-borne pentapeptide (cyan).
Discussion
An unstructured, flexible linker
We used site-directed spin labeling and EPR measurements to investigate the carboxyl-terminal 40 residues of chemoreceptor Tar, a segment for which there was no direct structural information for any chemoreceptor. EPR spectra collected for 16 positions (Figs. 2 and 3) and semiquantitative parameters characterizing those spectra (Fig. 4) indicated that much of the segment was notably mobile, beginning at approximately position 520 and extending to the carboxyl terminus. We cannot exclude the possibility that introduction of spin-labeled cysteines at positions 520 and beyond disrupted weak structure or weak interactions, but the possibility seems unlikely to us, since a large body of data has revealed that site-directed spin-labeling seldom disrupts protein structure.35,40–42 Thus, we conclude that the final ∼ 34 carboxyl-terminal residues of Tar constitute an essentially unstructured, disordered region. This lack of stable structure means that the Tar carboxyl-terminal linker has the features of a flexible arm and enzyme tether between the coiled-coil receptor body and the NWETF recognition sequence at the carboxyl terminus (Fig. 9).
Figure 9.
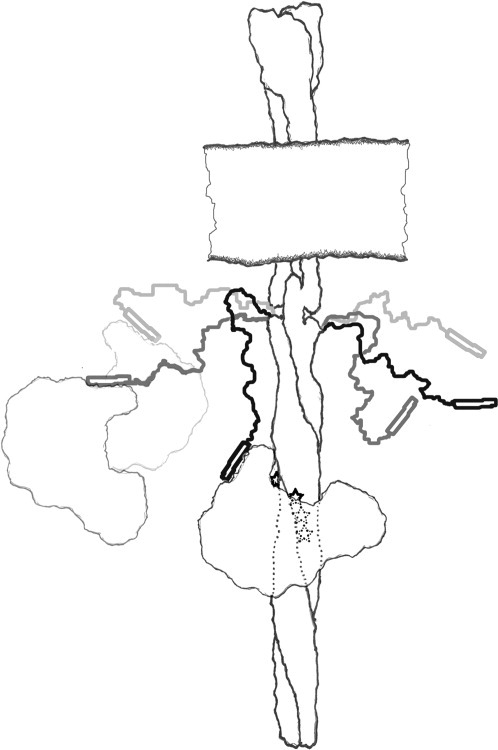
A flexible arm and enzyme tether. Cartoon of a membrane-imbedded chemoreceptor illustrating the principle conclusions of this study, that the carboxyl-terminal ∼ 35 residues of chemoreceptor Tar is a disordered, flexible arm (right-hand carboxyl-terminal segment, shown in three snapshots) that can act as a flexible tether for CheR bound to the carboxyl-terminal NWETF pentapeptide (open rectangle; left-hand carboxyl-terminal segment, shown in three snapshots bound to CheR).
The semiquantitative mobility parameters, Ms and h(+1)/h(0) highlighted an abrupt transition from structured to unstructured, occurring primarily between positions 519 and 520 (Fig. 4). For purified, spin-labeled Tar inserted in proteoliposomes and thus separated from the interfering background of spin labels on other membrane proteins, the parameters indicated that after the abrupt transition from lower to higher values, mobility continued to increase gradually, as would be expected for an unstructured polypeptide tethered at one end.37 In native Tar, the transition from structured to unstructured might well begin one residue prior to position 520 because position 519 in the natural sequence is a proline, often a helix breaker. In any case, a boundary between structured and unstructured segments at residues 519–520 is consistent with effects of a family of nested deletions within the Tar carboxyl-terminal linker, which all began at the amino terminus of the pentapeptide and extended toward the receptor body.14 Kinase activation was significantly reduced only for deletions extending past position 519, implying that only after this position did deletions disrupt the receptor and thus its ability to activate kinase.14 An additional correlation is provided by the crystal structure of a fragment of the cytoplasmic domain of E. coli chemoreceptor Tsr, in which the transition from resolved to unresolved residues occurred in the carboxyl-terminal segment at the position corresponding to Tar residue 518.24
Tethered CheR
The carboxyl-terminal, enzyme-binding pentapeptide in combination with the linker sequence between the pentapeptide and the coiled-coil receptor body are thought to act in concert to enhance action of methyltransferase CheR by binding enzyme and providing a flexible tether that restricts diffusion of bound CheR to a volume near methyl-accepting sites.23,25 CheR binding to pentapeptide at the Tar carboxyl terminus had the expected effect of substantially reducing mobility of a residue within and following the NWETF recognition sequence but reduced only modestly mobility of the linker, in a gradient of decreasing effects for approximately half its length (Figs. 6 and 7). This pattern supports the notion that NWETF-bound CheR is connected to the receptor body by a flexible tether that allows relatively unimpeded diffusion within specific limits and thus increases effective enzyme concentration near substrate sites and thus enzyme action.5,23,25
Experimental strategies
We characterized spin-labeled chemoreceptor Tar in two membrane environments: (1) native cytoplasmic membrane and (2) reconstituted proteoliposomes. The complementary features of these preparations provided internal checks on validity of our data and deductions. Characterization of Tar embedded in isolated cytoplasmic membrane had the advantage that receptor was in its native membrane environment but the disadvantage that EPR spectra included contributions from other spin-labeled proteins. Characterization of purified Tar reconstituted into proteoliposomes greatly reduced contributions by spin labels on other proteins but introduced the possibility that receptor was perturbed by purification and reconstitution. However, spectra for respective Tar positions shared many essential features, independent of environment and patterns as a function of spin label position were very similar for the two environments. This provided confidence that common spectral features were providing information about the structure of native chemoreceptor.
Subunit exchange among detergent-solubilized forms of the same chemoreceptor, first documented over 20 years ago,33 was effective in creating heterodimers with a targeted feature segregated to one of two respective subunits. For our purposes, these were an affinity tag and a single cysteine for spin labeling on a subunit that had an otherwise unchanged carboxyl terminus. Many other features would be possible, depending on the particular experimental design. Thus this strategy could be useful for future studies.
Functional roles for a disordered segment
Bioinformatic analyses indicate that sequences related to enzyme-activating, carboxyl-terminal pentapeptides of E. coli and Salmonella are found at carboxyl-termini in many proteobacterial chemoreceptors.20,21,43 Our analysis suggests that a disordered carboxyl terminus should be a common feature of these pentapeptide-bearing receptors. Outside of proteobacteria, few receptor sequences have a candidate carboxyl-terminal sequence, implying that development of such an enzyme-activating site is a relatively recent evolutionary development, one that should have provided a selective advantage. Identification of that advantage could provide further insights into the sophisticated feedback control of the bacterial chemosensory response. In a wider context, our data indicate that chemoreceptor Tar and presumably related receptors share the feature of a protein-interaction site at a terminus proceeded by a disordered region, a pattern observed for many proteins involved in signaling or complex formation.44,45
Materials and Methods
Strains, plasmids, and proteins
E. coli K-12 strains RP380846 and RP309847 carry, respectively, a deletion from cheA to cheZ that eliminates all che genes or a deletion from flhA to flhD that eliminates the presence or expression of all chemoreceptor and che genes. Plasmid pNT201 carries tar under control of a modified lac promoter and lacIq.48 A derivative, pAL533, codes for Tar with six histidines added to its carboxyl terminus and glutamines (Q in the single letter code) at all four sites of modification (Tar4Q-6H). Other derivatives, that code for Tar with the wild-type pattern of QEQE at the modification sites and a single introduced cysteine at the indicated position are pAL671 (R514C), pAL672 (L515C), pAL673 (A516C), pAL674 (A517C), pAL675 (P519C), pAL689 (L520C), pAL690 (T521C), pAL691 (N522C), pAL655 (K523C), pAL657 (S528C), pAL656 (E533C), pAL658 (Q538C), pAL659 (I543C), pAL660 (P548C), and pAL661 (Tar 554C).
CheR was purified from RP3808 containing pME43.49 Luria broth was inoculated at ∼ 5 × 107 cells per mL and incubated for ∼ 5.5 h at 35°C with vigorous aeration. The culture was chilled in an ice-water bath and centrifuged 8,000 rpm, 12 min, 4°C in a SLC 6000 rotor. Sedimented cells were suspended in 50 mM Tris-HCl (pH 7.5), 0.5 mM ethylenediaminetetraacetic acid (EDTA), 2 mM dithiothreitol (DTT), and 10% glycerol (TEDG), centrifuged as before, suspended in TEDG with additions of EDTA, PMSF, and lysozyme to 5 mM, 1 mM, and 0.15 mg/mL, respectively, incubated 40 min on ice and centrifuged 60,000 rpm, 2 h, 4°C in a Ti 60 rotor. The supernatant was concentrated in a Centriprep Ultracel YM-10 (Millipore, Billerica, MA) at 3,000 rpm and 4°C, dialyzed against TEDG overnight at 10°C and flash frozen in liquid nitrogen. Concentrated lysate was thawed, diluted 10-fold in 20 mM sodium phosphate (pH 7.0), 1 mM EDTA, 1 mM DTT, 0.5 mM phenylmethanesulfonylfluoride (PMSF) (column buffer) at 4°C and applied at 2.5 mL/min and 7°C to two 5-mL Hi Trap SP HP columns (GE Healthcare, Little Chalfont, UK) in tandem, equilibrated with five bed volumes of column buffer at 5 mL/min and coupled to a P-1 peristaltic pump (Pharmacia) and a UV monitor. The column was washed with column buffer at 5 mL/min until A280 returned to a baseline value and eluted with a step gradient of 40 mM, 80 mM, 120 mM, 160 mM, 200 mM, and 2M NaCl in column buffer with each higher salt concentration applied once A280 returned to a baseline value. Fractions containing CheR, as assessed using sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis, were pooled, concentrated with an Ultra15, 10 kDa molecular weight cutoff concentrator (Millipore, Billerica, MA) by centrifugation in an SS-34 rotor at 5000g, 9°C setting, brought to 10% w/v glycerol, dialyzed overnight against 50 mM Tris-HCl (pH 7.5), 0.5 mM EDTA, 2 mM DTT, 10% w/v glycerol at 7°C and stored at −80°C. The resulting preparation was ∼ 98% CheR as estimated by relative intensities of enzyme and contaminants on SDS polyacrylamide gels.
Isolation and spin labeling of receptor-containing cytoplasmic membranes
Cytoplasmic membrane vesicles enriched for each form of Tar were isolated from RP3098 harboring the appropriate plasmid, using cell culture conditions, osmotic lysis, and a sucrose gradient as described.50 Vesicles were suspended in 50 mM Tris-HCl (pH 7.5), 0.5 mM EDTA, 2 mM DTT, and 10% glycerol, flash frozen in liquid nitrogen and stored at −70°C. Tar content and total protein contents were determined by quantitative immunoblotting18 and BCA assays, respectively. Depending on the particular form of Tar and preparation, receptor was 20–40% of total membrane protein. Membranes containing 600 μg of Tar were diluted to 1 mL in 50 mM Tris-HCl (pH 7.5), 0.5 mM EDTA, 100 mM NaCl, 10% glycerol and centrifuged 15 min, 100,000 rpm in a TL100.2 rotor at 4°C. The pellet was suspended in 1 mL of the same buffer and the process repeated twice more to yield a calculated DTT concentration <1 μM. Methanethiosulfonate spin label reagent (Toronto Research Chemicals, North York, Canada) was added to 100 μM, a 10-fold molar excess over Tar, and the mixture incubated 1 h on ice in the dark. Unreacted spin label was removed by centrifugation and suspension as for the removal of DTT, and the labeled vesicles suspended to ∼ 50 μL of the same buffer.
Spectra for each respective spin-labeled forms of Tar were similar in native membrane and reconstituted proteoliposomes, and the same pattern of spectral features was evident in both conditions as a function of spin label position. This indicated that the spectra of spin-labeled native membrane provided information about the receptor, even though Tar was only 20–40% of total protein. In fact, comparison of spectra for the respective forms of Tar revealed that those from native membrane included a lower mobility component, likely from spin labels on non-Tar membrane proteins. This was confirmed by spectra of membranes, containing Tar with no cysteine, prepared and treated with the spin-labeling reagent in the same way as membranes with cysteine-containing Tar (Fig. 2, spectra in dashed-line box). We could not be confident of the precise extent of background contribution for any particular membrane preparation and thus subtraction of spectral features contributed by proteins other than Tar would have been arbitrary and might have distorted the data. Fortunately, because spin labels on the Tar linker were notably mobile, background subtraction was not necessary to observe distinct effects of varying the spin label position. Thus, in Figure 2, we show spectra that have not been processed to remove contributions from spin labels on nonreceptor proteins.
Protomer exchange, purification, and spin labeling
Two preparations of cytoplasmic membranes, one containing Tar4Q-6H and the other a cysteine-substituted Tar were mixed to provide a 1:1 receptor ratio and 4 mg at 1.5 mg/mL of each receptor in 50 mM Tris-HCl (pH 7.5), 10% glycerol, 2 μM pepstatin, 2 μM leupeptin, 5 μM TLCK, and 100 μM PMSF. Beta-d-octyl-glucoside was added to 5.5%, the mixture incubated 1 h on ice and centrifuged 15 min, 100,000 rpm, 4°C in a TL100.2 rotor. The supernatant was transferred to a new tube, aspartate added to 1 mM to block subunit exchange33 and the solution applied to a 4-mL bed volume Ni-NTA column equilibrated with four bed volumes 50 mM Tris-HCl (pH 7.5), 10% glycerol, 100 mM NaCl, 25 mM cholate, 30 mM imidazole, 1 mM aspartate (Buffer A). Four bed volumes of Buffer A and eight volumes of Buffer B (Buffer A with 300 mM imidazole) were passed through the column and 4 mL fractions collected. The first three fractions following application of Buffer B, which contained the majority of Tar, were combined, spin-label reagent in acetonitrile added at a fivefold molar excess over the maximum possible amount of cysteine-substituted Tar, the mixture incubated 1 h on ice in the dark, concentrated to <1 mL and buffer exchanged using a Nap10 column (GE Healthcare, Little Chalfont, UK) to 50 mM Tris-HCl (pH 7.5), 0.5 mM EDTA, 100 mM NaCl, 25 mM cholate, 10% glycerol, the solution concentrated to ∼ 200 μL, and stored at −80°C. The resulting Tar dimers were approximately two-thirds heterodimers, consisting of one cysteine-containing, spin-labeled protomer with a natural carboxyl terminus and one histidine-tagged protomer lacking a cysteine, and approximately one-third homodimer, consisting of two histidine-tagged subunits devoid of cysteines and thus no spin label. The proportion of the two Tar forms was determined by SDS polyacrylamide gel electrophoresis, exploiting their differential mobility. Purified Tar was quantified by comparison of intensities of Coomassie Brilliant Blue staining of bands on an SDS polyacrylamide gel to a Tar standard quantified by amino acid analysis.
Reconstitution into proteoliposomes
Detergent-solubilized, spin-labeled Tar was reconstituted into lipid bilayers by combining 1 mg of receptor with E. coli lipids (Avanti Polar Lipids, Alabaster, AL), solubilized in 150 mM cholate, in 1 mL of 50 mM Tris-HCl (pH 7.5), 0.5 mM EDTA, 100 mM NaCl, 10% glycerol, 80 mM cholate, 1 μM pepstatin, 1 μM leupeptin, 17 mM E. coli lipids and 17 μM spin-labeled Tar. Detergent was removed51 by adding 1 mL of SM-2 Biobeads (Bio-Rad, Hercules, CA) and incubating with rotation for 1.5 h, 10°C. Biobeads were removed as described,50 1 mL new Biobeads added and the mixture incubated and processed as before. Resulting proteoliposomes were sedimented by centrifugation 15 min, 100,000 rpm, 10°C in a TL100.2 rotor, suspended in 50 mM Tris-HCl (pH 7.5), 0.5 mM EDTA, 100 mM NaCl, 10% glycerol and frozen at −80°C. The ability of this procedure to incorporate Tar into proteoliposomes was verified by flotation in a metrizamide gradient. Each preparation of proteoliposome-inserted Tar was assayed for recognition and thus modification by phosphorylated CheB in conditions designed to modify all sites on all accessible and native Tar as described,14 except that receptor was 30 nM, CheB in the absence of DTT was 1.5 μM and phosphoramidate 50 mM.
EPR spectroscopy
X-band spectra were collected at 20 mW incident microwave power using a Brüker EMX spectrometer (Billerica, MA) equipped with a high-sensitivity resonator. Data for Tar imbedded in membrane vesicles or proteoliposomes were collected with a 100-kHz field modulation of 1 G or 1.2, 1.8, or 2.4 G, respectively. For spectra collected from spin-labeled Tar in proteoliposomes, a low background of nonspecific labeling was subtracted52 using spectra of proteoliposomes made with Tar lacking cysteine acquired for the same Tar concentration, conditions and field modulation as the respective experimental samples. Data processing was performed with Labview software (Christian Altenbach, University of California, Los Angeles, CA). For final data processing and comparisons, spectra were normalized to the same total spins.
For spectra in the presence of CheR, DTT in the CheR storage buffer was reduced to a calculated concentration of <50 nM by repeated concentration and dilution with the buffer in which proteoliposomes were stored using a 10 kDa-cutoff Nanosep concentrator (Pall Life Sciences, Port Washington, NY). CheR was added to spin-labeled Tar in proteoliposomes at a concentration calculated to occupy ∼ 97% of accessible, receptor-borne pentapeptides and thus ∼ 50% of total receptor, since ∼ 50% of spin-labeled Tar incorporated into proteoliposomes was accessible (see Results).
Semiquantitative measures of mobility
Ms is a measure of mobility of a nitroxide spin label at a position of interest normalized to a relatively mobile and a relatively immobile protein-coupled nitroxide. Specifically, Ms = (δexp−1 – δi−1)/(δm−1 – δi−1), where δi and δm are the central line-widths of a relatively immobile and a relatively mobile nitroxide spectra, respectively, and δexp is the central linewidth of the position under investigation.35,36 For the analysis of our data we utilized values of the constants previously used in characterization of an unstructured protein segment37: δm = 2.1 and δi = 8.4, corresponding, respectively, to values for a spin label near the end of the disordered carboxyl-terminal sequence of rhodopsin27 and the average value of an immobilized, buried residue in a protein undergoing slow rotational diffusion. The mobility parameter h(+1)/h(0), the ratio of amplitudes of the low-field and central spectral lines (Fig. 1C), has been used in characterization of unstructured protein segments.29,30,32
Acknowledgments
The authors thank Angela Lilly for construction of the plasmid-borne receptor genes used in this study, Wing-Cheung Lai for purification of CheR and Linda L. Randall for instruction, guidance, and advice in EPR spectroscopy.
References
- 1.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hazelbauer GL, Lai W-C. Bacterial chemoreceptors: providing enhanced features to two-component signaling. Curr Opin Microbiol. 2010;13:124–132. doi: 10.1016/j.mib.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engström P, Hazelbauer GL. Multiple methylation of methyl-accepting chemotaxis proteins during adaptation of E. coli to chemical stimuli. Cell. 1980;20:165–171. doi: 10.1016/0092-8674(80)90244-5. [DOI] [PubMed] [Google Scholar]
- 4.Yamamoto K, Macnab RM, Imae Y. Repellent response functions of the Trg and Tap chemoreceptors of Escherichia coli. J Bacteriol. 1990;172:383–388. doi: 10.1128/jb.172.1.383-388.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu J, Li J, Li G, Long DG, Weis RM. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- 6.Le Moual H, Quang T, Koshland DE., Jr. Methylation of the Escherichia coli chemotaxis receptors: intra- and interdimer mechanisms. Biochemistry. 1997;36:13441–13448. doi: 10.1021/bi9713207. [DOI] [PubMed] [Google Scholar]
- 7.Feng X, Baumgartner JW, Hazelbauer GL. High- and low-abundance chemoreceptors in Escherichia coli: differential activities associated with closely related cytoplasmic domains. J Bacteriol. 1997;179:6714–6720. doi: 10.1128/jb.179.21.6714-6720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng X, Lilly AA, Hazelbauer GL. Enhanced function conferred on low-abundance chemoreceptor Trg by a methyltransferase-docking site. J Bacteriol. 1999;181:3164–3171. doi: 10.1128/jb.181.10.3164-3171.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnakov AN, Barnakova LA, Hazelbauer GL. Comparison in vitro of a high- and a low-abundance chemoreceptor of Escherichia coli: similar kinase activation but different methyl-accepting activities. J Bacteriol. 1998;180:6713–6718. doi: 10.1128/jb.180.24.6713-6718.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnakov AN, Barnakova LA, Hazelbauer GL. Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation. Proc Natl Acad Sci USA. 1999;96:10667–10672. doi: 10.1073/pnas.96.19.10667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okumura H, Nishiyama S, Sasaki A, Homma M, Kawagishi I. Chemotactic adaptation is altered by changes in the carboxy-terminal sequence conserved among the major methyl-accepting chemoreceptors. J Bacteriol. 1998;180:1862–1868. doi: 10.1128/jb.180.7.1862-1868.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weerasuriya S, Schneider BM, Manson MD. Chimeric chemoreceptors in Escherichia coli: signaling properties of Tar-Tap and Tap-Tar hybrids. J Bacteriol. 1998;180:914–920. doi: 10.1128/jb.180.4.914-920.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai W-C, Barnakova LA, Barnakov AN, Hazelbauer GL. Similarities and differences in interactions of the activity-enhancing chemoreceptor pentapeptide with the two enzymes of adaptational modification. J Bacteriol. 2006;188:5646–5649. doi: 10.1128/JB.00497-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M, Hazelbauer GL. The carboxyl-terminal linker is important for chemoreceptor function. Mol Microbiol. 2006;60:469–479. doi: 10.1111/j.1365-2958.2006.05108.x. [DOI] [PubMed] [Google Scholar]
- 15.Djordjevic S, Stock AM. Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine. Structure. 1997;5:545–558. doi: 10.1016/s0969-2126(97)00210-4. [DOI] [PubMed] [Google Scholar]
- 16.Barnakov AN, Barnakova LA, Hazelbauer GL. Location of the receptor-interaction site on CheB, the methylesterase response regulator of bacterial chemotaxis. J Biol Chem. 2001;276:32984–32989. doi: 10.1074/jbc.M105925200. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Li G, Weis RM. The serine chemoreceptor from Escherichia coli is methylated through an inter-dimer process. Biochemistry. 1997;36:11851–11857. doi: 10.1021/bi971510h. [DOI] [PubMed] [Google Scholar]
- 18.Li M, Hazelbauer GL. Adaptational assistance in clusters of bacterial chemoreceptors. Mol Microbiol. 2005;56:1617–1626. doi: 10.1111/j.1365-2958.2005.04641.x. [DOI] [PubMed] [Google Scholar]
- 19.Perez E, Stock AM. Characterization of the Thermotoga maritima chemotaxis methylation system that lacks pentapeptide-dependent methyltransferase CheR:MCP tethering. Mol Microbiol. 2007;63:363–378. doi: 10.1111/j.1365-2958.2006.05518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexander RP, Zhulin IB. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci USA. 2007;104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wuichet K, Zhulin IB. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal. 2010;3:1–13. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barnakov AN, Barnakova LA, Hazelbauer GL. Allosteric enhancement of adaptational demethylation by a carboxyl-terminal sequence on chemoreceptors. J Biol Chem. 2002;277:42151–42156. doi: 10.1074/jbc.M206245200. [DOI] [PubMed] [Google Scholar]
- 23.Windisch B, Bray D, Duke T. Balls and chains—a mesoscopic approach to tethered protein domains. Biophys J. 2006;91:2383–2392. doi: 10.1529/biophysj.105.078543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim KK, Yokota H, Kim S-H. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 25.Muppirala UK, Desensi S, Lybrand TP, Hazelbauer GL, Li Z. Molecular modeling of flexible arm-mediated interactions between bacterial chemoreceptors and their modification enzyme. Protein Sci. 2009;18:1702–1714. doi: 10.1002/pro.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, Dunker AK. Comparing and combining predictors of mostly disordered proteins. Biochemistry. 2005;44:1989–2000. doi: 10.1021/bi047993o. [DOI] [PubMed] [Google Scholar]
- 27.Langen R, Cai K, Altenbach C, Khorana HG, Hubbell WL. Structural features of the C-terminal domain of bovine rhodopsa site-directed spin-labeling study. Biochemistry. 1999;38:7918–7924. doi: 10.1021/bi990010g. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Z, DeSensi SC, Stein RA, Brandon S, Dixit M, McArdle EJ, Warren EM, Kroh HK, Song L, Cobb CE, Hustedt EJ, Beth AH. Solution structure of the cytoplasmic domain of erythrocyte membrane band 3 determined by site-directed spin labeling. Biochemistry. 2005;44:15115–15128. doi: 10.1021/bi050931t. [DOI] [PubMed] [Google Scholar]
- 29.Morin B, Bourhis J-M, Belle V, Woudstra M, Carrière F, Guigliarelli B, Fournel A, Longhi S. Assessing induced folding of an intrinsically disordered protein by site-directed spin-labeling electron paramagnetic resonance spectroscopy. J Phys Chem B. 2006;110:20596–20608. doi: 10.1021/jp063708u. [DOI] [PubMed] [Google Scholar]
- 30.Belle V, Rouger S, Costanzo S, Liquière E, Strancar J, Guigliarelli B, Fournel A, Longhi S. Mapping α-helical induced folding within the intrinsically disordered C-terminal domain of the measles virus nucleoprotein by site-directed spin-labeling EPR spectroscopy. Proteins. 2008;73:973–988. doi: 10.1002/prot.22125. [DOI] [PubMed] [Google Scholar]
- 31.Kavalenka A, Urbancic I, Belle V, Rouger S, Costanzo S, Kure S, Fournel A, Longhi S, Guigliarelli B, Strancar J. Conformational analysis of the partially disordered measles virus NTAIL-XD complex by SDSL EPR spectroscopy. Biophys J. 2010;98:1055–1064. doi: 10.1016/j.bpj.2009.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pirman NL, Milshteyn E, Galiano L, Hewlett JC, Fanucci GE. Characterization of the disordered-to-α-helical transition of IA3 by SDSL-EPR spectroscopy. Protein Sci. 2011;20:150–159. doi: 10.1002/pro.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milligan DL, Koshland DE., Jr. Site-directed cross-linking. Establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J Biol Chem. 1988;263:6268–6275. [PubMed] [Google Scholar]
- 34.Lai W-C, Hazelbauer GL. Carboxyl-terminal extensions beyond the conserved pentapeptide reduce rates of chemoreceptor adaptational modification. J Bacteriol. 2005;187:5115–5121. doi: 10.1128/JB.187.15.5115-5121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Columbus L, Hubbell WL. A new spin on protein dynamics. Trends Biochem Sci. 2002;27:288–295. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 36.Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat Struct Mol Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 37.Columbus L, Hubbell WL. Mapping backbone dynamics in solution with site-directed spin labeling: GCN4-58 bZip free and bound to DNA. Biochemistry. 2004;43:7273–7287. doi: 10.1021/bi0497906. [DOI] [PubMed] [Google Scholar]
- 38.Kim M, Fanucci GE, Cafiso DS. Substrate-dependent transmembrane signaling in TonB-dependent transporters is not conserved. Proc Natl Acad Sci USA. 2007;104:11975–11980. doi: 10.1073/pnas.0702172104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Djordjevic S, Stock AM. Chemotaxis receptor recognition by protein methyltransferase CheR. Nat Struct Biol. 1998;5:446–450. doi: 10.1038/nsb0698-446. [DOI] [PubMed] [Google Scholar]
- 40.Altenbach C, Greenhalgh DA, Khorana HG, Hubbell WL. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proc Natl Acad Sci USA. 1994;91:1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hubbell WL, Gross A, Langen R, Lietzow MA. Recent advances in site-directed spin labeling of proteins. Curr Opin Struct Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 42.Hubbell WL, McHaourab HS, Altenbach C, Lietzow MA. Watching proteins move using site-directed spin labeling. Structure. 1996;4:779–783. doi: 10.1016/s0969-2126(96)00085-8. [DOI] [PubMed] [Google Scholar]
- 43.Wuichet K, Alexander RP, Zhulin IB. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol. 2007;422:3–31. doi: 10.1016/S0076-6879(06)22001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 46.Slocum MK, Parkinson JS. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: organization of the tar region. J Bacteriol. 1983;155:565–577. doi: 10.1128/jb.155.2.565-577.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parkinson JS, Houts SE. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borkovich KA, Kaplan N, Hess JF, Simon MI. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc Natl Acad Sci USA. 1989;86:1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simms SA, Stock AM, Stock JB. Purification and characterization of the S-adenosylmethionine:glutamyl methyltransferase that modifies membrane chemoreceptor proteins in bacteria. J Biol Chem. 1987;262:8537–8543. [PubMed] [Google Scholar]
- 50.Boldog T, Li M, Hazelbauer GL. Using Nanodiscs to create water-soluble transmembrane chemoreceptors inserted in lipid bilayers. Methods Enzymol. 2007;423:317–335. doi: 10.1016/S0076-6879(07)23014-9. [DOI] [PubMed] [Google Scholar]
- 51.Rigaud JL, Levy D, Mosser G, Lambert O. Detergent removal by non-polar polystyrene beads. Eur Biophys J. 1998;27:305–319. [Google Scholar]
- 52.Do Cao M-A, Crouzy S, Kim M, Becchi M, Cafiso DS, Pietro AD, Jault J-M. Probing the conformation of the resting state of a bacterial multidrug ABC transporter, BmrA, by a site-directed spin labeling approach. Protein Sci. 2009;18:1507–1520. doi: 10.1002/pro.141. [DOI] [PMC free article] [PubMed] [Google Scholar]


