Abstract
Interest in generating lignocellulosic biofuels through enzymatic hydrolysis continues to rise as nonrenewable fossil fuels are depleted. The high cost of producing cellulases, hydrolytic enzymes that cleave cellulose into fermentable sugars, currently hinders economically viable biofuel production. Here, we report the crystal structure of a prevalent endoglucanase in the biofuels industry, Cel5A from the filamentous fungus Hypocrea jecorina. The structure reveals a general fold resembling that of the closest homolog with a high-resolution structure, Cel5A from Thermoascus aurantiacus. Consistent with previously described endoglucanase structures, the H. jecorina Cel5A active site contains a primarily hydrophobic substrate binding groove and a series of hydrogen bond networks surrounding two catalytic glutamates. The reported structure, however, demonstrates stark differences between side-chain identity, loop regions, and the number of disulfides. Such structural information may aid efforts to improve the stability of this protein for industrial use while maintaining enzymatic activity through revealing nonessential and immutable regions.
Keywords: cellulase, endoglucanase, cellulose, biofuel, Hypocrea jecorina, Cel5A, crystal structure
Introduction
Lignocellulosic biofuels have enjoyed recent popularity as sustainable energy alternatives to fossil fuels. In current enzymatic conversion schemes, a pretreatment step with high temperatures or extreme pH conditions removes indigestible lignin from feedstock materials. Cellulase cocktails then break cellulose polymers into component sugars suitable for fermentative fuel production. To achieve efficient digestion, three types of cellulases must exist in the preparation: (1) exoglucanases to cleave cellobiose molecules from cellulose strand termini, (2) endoglucanases to cleave strands internally, and (3) β-glucosidases to cleave cellobiose into glucose monomers.1 Few known organisms adequately produce cellulases from all three classes. Consequently, the filamentous fungus Hypocrea jecorina (Trichoderma reesei), a prodigious source of each cellulase class, enjoys wide-spread use in the biofuels industry.2 Enzyme production costs, however, still constitute a limiting factor to wide-scale bioethanol synthesis. Although advances in all areas of enzyme production have decreased costs up to 20–30 cents per gallon of ethanol, less-sustainable, corn-derived fuel remains the cheaper alternative at 3–4 cents per gallon.3 One strategy for further reducing enzymatic costs involves extending cellulase lifetimes through enhanced stability. As some protein engineering strategies utilize atomic-resolution models to guide the design process, obtaining crystal structures of each cellulase may significantly aid such endeavors. Thus far, efforts to crystallize H. jecorina cellulases have resulted in catalytic domain structures of exoglucanases Cel6A (CBHII)4 and Cel7A (CBHI)5 and endoglucanases Cel7B (EGI)6 and Cel12A (EGIII).7 Cel5A (EGII), however, accounts for as much as 55% of H. jecorina endoglucanase activity,8 yet has resisted previous crystallographic solution. Here we provide the crystal structure of H. jecorina Cel5A (Hj_Cel5A) resolved to 2.05 Å.
Results
With the exception of Cel12A, most H. jecorina cellulases consist of a heavily O-glycosylated linker tethering a small cellulose binding domain (CBD) to a larger catalytic domain. CBDs of this organism share ∼70% sequence identity9 and a solution structure of the Cel7A CBD has been solved.10 To minimize sample inhomogeneity resulting from glycosylation, the isolated H. jecorina Cel5A catalytic core was expressed in Escherichia coli BL21 (DE3) cells. The protein was crystallized, data were collected to 2.05 Å, and the structure solved and refined with an Rwork/Rfree of 16.3/20.5% (Table I and Supporting Information Fig. S1).
Table I.
Data Collection and Refinement Statistics
| Hj_Cel5A | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 83.0, 84.6, 90.1 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 39–2.05(2.16–2.05) |
| Rsym | 0.081(0.268) |
| Mn(I)/sd | 19.2(2.8) |
| Completeness (%) | 98.8(92.4) |
| Redundancy | 12.5(9.8) |
| Refinement | |
| Resolution (Å) | 40–2.05 |
| No. reflections | 39858 |
| Rwork/Rfree (%) | 16/21 |
| No. atoms | |
| Protein | 4966 |
| Ligand/ion | 74 |
| Water | 503 |
| B-factors | 23 |
| Protein | 22 |
| Ligand/ion | 40 |
| Water | 30 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.2 |
| Ramachandran map analysis | |
| Most favored regions | 87.2 |
| Additional allowed regions | 12.8 |
| Generously allowed regions | 0 |
| Disallowed regions | 0 |
Data were collected from one crystal.
Values in parentheses are for highest-resolution shell.
Hj_Cel5A adopts a (α/β)8 TIM-barrel fold common to other family 5 glycoside hydrolases [Fig. 1(A)]. The general topology bears a striking resemblance to Cel5A from Thermoascus aurantiacus (Ta_Cel5A, RMSD of 1.4 Å11) [Fig. 1(B)] with 29% sequence identity and 65% sequence similarity (Supporting Information Fig. S2). While both proteins demonstrate similar placement of most secondary structure elements, the H. jecorina homolog exhibits extensions in the β1-α1, β3-α3, and α5-β6 loops (see Supporting Information Fig. S3 for secondary structure numbering). The β1-α1 loop projects towards the active site, forming a relatively shallow substrate binding groove. In addition to eight canonical β-strands, the structure also contains a protruding β-hairpin consisting of residues 308 to 315. Sidechain densities along the tip of the loop could not be resolved, suggesting flexibility of the region. Tryptophan 314, however, appears to anchor the C-terminal region of the hairpin to the face of the protein as it rejoins the globular region to form a truncated α8 helix. Although similar β-hairpins appear in the structures of Thermotoga maritima Cel5A12 (Tm_Cel5A) (3MMW, residues 295–302) and Clostridium cellulovorans endoglucanase D (3NDY, residues 324–331), it remains unclear whether this hairpin assumes a functional role. A series of hydrophobic residues (F4, Y98, W142, F177, I214, L287) shields the active site from solvent rather than a short 2–3 β-strand13 and/or the small N-terminal α-helix plug observed in homologous structures.12
Figure 1.
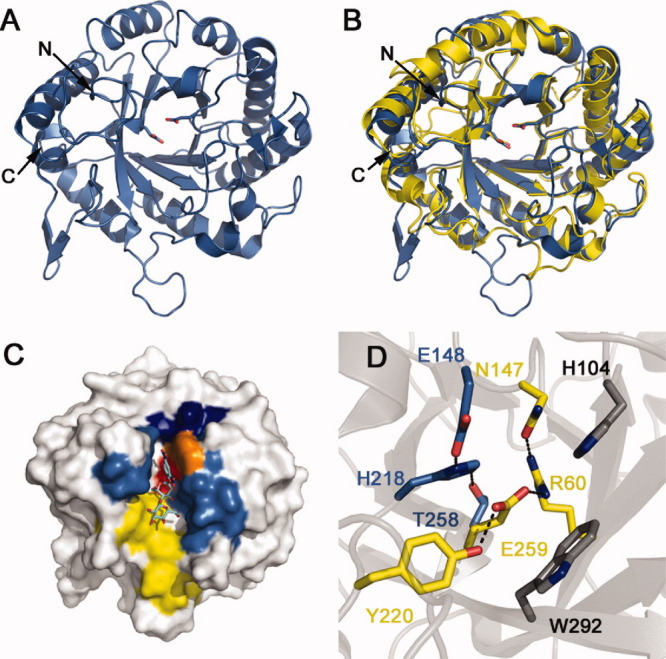
Structure of Hj_Cel5A. (A) Hj_Cel5A shown in cartoon representation with catalytic glutamates shown as sticks. (B) Superposition of Hj_Cel5A (blue) and Ta_Cel5A (yellow) generated in PyMOL using the align function. (C) Hj_Cel5A in surface representation highlighting the hydrophobic substrate docking patch (yellow), sugar-stacking base W185 at site −1 (orange), active site (red), substrate binding groove walls (light blue), and helical ridge composed of residues 183 to 187 (dark blue). The protein is modeled in complex with substrate mimic 2,4-dinitrophenyl-2-deoxy-2-fluoro-β-D-cellobioside from the structure of the Bacillus agaradhaerens Cel5A (PDB 4A3H). Sugar superpositioning was achieved through aligning Ba_Cel5A to Hj_Cel5A in PyMOL. (D) The active site of Hj_Cel5A depicting hydrogen bonding networks between the catalytic base (E148) and nucleophile (E259), as well as other conserved residues (gray).
Glycosylation
Mass spectrometry studies demonstrate that Hj_Cel5A contains a single GlcNAc N33-linked glycosylation when expressed in the organism of origin.14 The structure contains no discernable density compatible with such a modification, as expected for a bacterially-expressed protein. N33 is, however, solvent exposed and does not preclude previous findings.
Active site architecture
Consistent with structural studies of other GH5 endoglucanases, the substrate binding pocket consists of a deep catalytic cleft within a shallow binding groove. The deeper cleft contains a hydrophobic patch (F14, V27, Y28, Y40, F34, W292, A294, F297, Y301) surrounded by the β1-α1 loop (residues 15–22), the sidechain of W185, residues 104–107, residues 146–150, and the β6-α6 loop (residues 225–229) [Fig. 1(C)]. A short α-helical ledge (residues 183–187) abruptly terminates this hydrophobic groove in a manner that superficially appears incompatible with endoglucanase function—internal cellulose cleavage might require that the substrate thread through the deep cleft to access the active site. The ledge itself, however, forms a shallower hydrophilic groove. This architecture suggests that an extended cellulose chain initially binds to the shallow groove in a noncatalytic manner. Crystallographic studies of the Bacillus agaradhaerens Cel5A suggest that the Michaelis complex subsequently forms as the −1 site sugar adopts a 1S3 skew-boat conformation.15 W185 facilitates formation of this catalytic conformation through stacking with the −1 site sugar ring [Fig. 1(C)]. The resulting ∼110°–115° kink allows the substrate to pass over the helical ledge into solvent allowing for the internal cleavage of long cellulose strands. Previous studies characterize Hj_Cel5A as a promiscuous enzyme that generates a wide range of products including glucose, cellobiose, and cellotriose.16 The noncatalytic binding groove appears more hydrophilic and shallower than that of Ta_Cel5A. Further testing may reveal whether product inhomogeneity results from scant interaction between Hj_Cel5A and the reducing end of the chain beyond the active site.
The obtained Hj_Cel5A structure depicts an active enzyme as determined by comparison to homologous structures. Like other retaining cellulases, Hj_Cel5A hydrolyzes internal β-1,4-glycosidic cellulosic bonds through a double-displacement mechanism involving two carboxylates.15 First, a general acid/base catalyst protonates the glycosidic bond to promote cleavage. A second carboxylate then forms a covalent glucosyl-enzyme intermediate through an oxocarbonium ion transition state, displacing a newly-generated nonreducing cellulose terminus. The apo enzyme finally forms through a second oxocarbonium ion transition state. In Hj_Cel5A, the terminal oxygen atoms of the general base (E148) and nucleophile (E259) are separated by ∼5 Å, typical of retaining β-glycosidases.17 These residues were identified through homology with Ta_Cel5A and confirmed as necessary to catalysis through site-directed mutagenesis (Supporting Information Fig. S4). Residues T258, H218, and E148 form a type A catalytic triad involved in raising the pKa of the donor carboxylate to promote more efficient substrate protonation18 [Fig. 1(D)]. A hydrogen-bonding network around E259 also exists. R60 and Y220 position the nucleophilic glutamate for catalysis through contacting OE2 and OE1, respectively. N147 in turn tethers R60 in place. Although H104 and W292 are conserved across GH5 cellulases and reside near the active site, these residues appear to assist with substrate binding rather than influence the catalytic machinery.11
Disulfide bonding
Hj_Cel5A contains eight cysteines, all of which are involved in the formation of disulfide bridges [Fig. 2(A,B)]. The covalent link between C16 and C22 tethers the C-terminal and N-terminal regions of the β1-α1 loop that forms one wall of the substrate binding pocket. Near the C-terminal region, residues 273 and 323 anchor the final α-helical segment to the adjacent α7 helix. Hj_Cel5A exhibits a relatively high apparent Tm of 69.5°C (Supporting Information Fig. S5) that may be due in part to stability conferred by disulfide bonding. The hyperthermostable Ta_Cel5A exhibits two higher melting transitions at 77°C, and 81°C,19 yet contains a single disulfide bond at a location homologous to the linkage between C232 and C268. Observations from homologous structures, however, suggest that the thermostability of Ta_Cel5A may largely arise due to the truncation of loops, a highly pronounced feature in the Ta_Cel5A homolog.12 Our attempts to mutate several disulfide-bonded cysteines to serines resulted in insoluble protein expression (data not shown).
Figure 2.
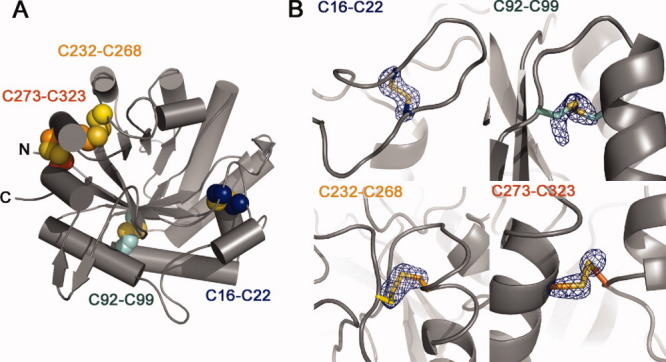
Disulfide bonding patterns in Hj_Cel5A. (A) Cartoon representation of the protein highlighting positions of the four intramolecular disulfide bonds detected in the electron density. (B) Fo-Fc cysteine sidechain omit maps contoured to 5 σ. Sidechain atoms from the Cβ to the end of the sidechain were deleted from the model before map generation.
Discussion
Hj_Cel5A constitutes only 1–10% of the total cellulase protein in H. jecorina, yet accounts for 55% of the total endoglucanase activity.8,20 The structural data presented here shows that the protein differs in sidechain identity and loop placement from its most similar crystallographically-probed homolog, Ta_Cel5A. Additionally, the structure reveals four disulfide bonds, in direct contrast with a previous report suggesting the absence of such elements.21 While an attempt to engineer Hj_Cel5A for optimum catalytic efficiency at a particular pH has met with some success, this effort relied on a highly inaccurate homology model built from Ta_Cel5A coordinates.22 The information presented here may better inform future efforts to rationally engineer Hj_Cel5A for various needs, as well as understand the wild-type activity of the protein.
Materials and Methods
Protein expression and purification
The catalytic domain of Hj_Cel5A (Genbank JN172972) was expressed in BL21(DE3) cells and purified as described in the Supporting Information. Cultures were grown at 37°C to an optical density of ∼0.5 in LB, induced, then allowed to express protein at 16°C for 24 hours. Purification was achieved through His-tag affinity chromatography and proteins were buffer exchanged into storage buffer (10 mM acetate pH 4.8, 100 mM NaCl) at a final concentration of 5.3 mg/mL.
Crystallization, data collection, and structure determination
Hexagonal plate crystals grew in 21 days by the sitting-drop vapor diffusion method in 0.1 M sodium citrate, 1 M magnesium sulfate, and 1 mM cellobiose. Crystals were flash frozen in cryoprotectant and shipped to beamline 12-2 at the Stanford Synchrotron Radiation Lightsource where a 2.1 Å data set was obtained. Phases were obtained through molecular replacement using a 1H1N mixed model generated with SCWRL.23 Following molecular replacement, model building and refinement were accomplished with the AutoBuild Wizard in PHENIX24/COOT25 and PHENIX,26 respectively. NCS restraints were applied to all refinement steps. Final coordinates were deposited in the Protein Data Bank with the code 3QR3. Data collection and refinement statistics are listed in Table I.
Acknowledgments
The authors acknowledge the use of beamline 12-2 at the Stanford Synchrotron Radiation Lightsource (SSRL) in Menlo Park, CA operated by Stanford University and supported by the Department of Energy and National Institutes of Health. They additionally acknowledge Jens Kaiser and Pavle Niklovski at the California Institute of Technology for their advice. They thank the Gordon and Betty Moore Foundation for support of the Molecular Observatory at Caltech.
Supplementary material
References
- 1.Kumar R, Singh S, Singh O. Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives. J Ind Microbiol Biotechnol. 2008;35:377–391. doi: 10.1007/s10295-008-0327-8. [DOI] [PubMed] [Google Scholar]
- 2.Bisaria VS, Ghose TK. Biodegradation of cellulosic materials: substrate, microorganisms, enzymes and products. Enzyme Microb Technol. 1981;3:90–104. [Google Scholar]
- 3.Stephanopoulous G. Challenges in engineering microbes for biofuels production. Science. 2007;315:801–804. doi: 10.1126/science.1139612. [DOI] [PubMed] [Google Scholar]
- 4.Rouvinen J, Bergfors T, Teeri T, Knowles JKC, Jones TA. Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science. 1990;249:380–386. doi: 10.1126/science.2377893. [DOI] [PubMed] [Google Scholar]
- 5.Divne C, Stahlberg J, Reinikainen T, Ruohonen L, Pettersson G, Knowles JK, Teeri TT, Jones TA. The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science. 1994;265:524–528. doi: 10.1126/science.8036495. [DOI] [PubMed] [Google Scholar]
- 6.Kleywegt GJ, Zou JY, Divne C, Davies GJ, Sinning I, Ståhlberg J, Reinikainen T, Srisodsuk M, Teeri TT, Jones TA. The crystal structure of the catalytic core domain of endoglucanase I from Trichoderma reesei at 3.6 Å resolution, and a comparison with related enzymes. J Mol Biol. 1997;272:383–397. doi: 10.1006/jmbi.1997.1243. [DOI] [PubMed] [Google Scholar]
- 7.Sandgren M, Ståhlberg J, Mitchinson C. Structural and biochemical studies of GH family 12 cellulases: improved thermal stability, and ligand complexes. Prog Biophys Mol Biol. 2005;89:246–291. doi: 10.1016/j.pbiomolbio.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Suominen PL, Mäntylä AL, Karhunen T, Hakola S, Nevalainen H. High frequency one-step gene replacement in Trichoderma reesei. II. Effects of deletions of individual cellulase genes. Mol Gen Genet. 1993;241:523–530. doi: 10.1007/BF00279894. [DOI] [PubMed] [Google Scholar]
- 9.Teeri TT, Lehtovaara P, Kauppinen S, Salovuori I, Knowles J. Homologous domains in Trichoderma reesei cellulolytic enzymes: gene sequence and expression of cellobiohydrolase II. Gene. 1987;51:43–52. doi: 10.1016/0378-1119(87)90472-0. [DOI] [PubMed] [Google Scholar]
- 10.Kraulis PJ, Clore GM, Nilges M, Jones TA, Pettersson G, Knowles J, Gronenborn AM. Determination of the three-dimensional solution structure of the C-terminal domain of cellobiohydrolase I from Trichoderma reesei. A study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing. Biochemistry. 1989;28:7241–7257. doi: 10.1021/bi00444a016. [DOI] [PubMed] [Google Scholar]
- 11.Van Petegem F, Vandenberghe I, Bhat MK, Van Beeumen J. Atomic resolution structure of the major endoglucanase from Thermoascus aurantiacus. Biochem Biophys Res Commun. 2002;296:161–166. doi: 10.1016/s0006-291x(02)00775-1. [DOI] [PubMed] [Google Scholar]
- 12.Pereira JH, Chen Z, McAndrew RP, Sapra R, Chhabra SR, Sale KL, Simmons BA, Adams PD. Biochemical characterization and crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima. J Struct Biol. 2010;172:372–379. doi: 10.1016/j.jsb.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 13.Sakon J, Adney WS, Himmel ME, Thomas SR, Karplus PA. Crystal structure of thermostable family 5 endocellulase E1 from Acidothermus cellulolyticus in complex with cellotetraose. Biochemistry. 1996;35:10648–10660. doi: 10.1021/bi9604439. [DOI] [PubMed] [Google Scholar]
- 14.Hui JPM, White TC, Thibault P. Identification of glycan structure and glycosylation sites in cellobiohydrolase II and endoglucanases I and II from Trichoderma reesei. Glycobiology. 2002;12:837–849. doi: 10.1093/glycob/cwf089. [DOI] [PubMed] [Google Scholar]
- 15.Davies GJ, Mackenzie L, Varrot A, Dauter M, Brzozowski AM, Schülein M, Withers SG. Snapshots along an enzymatic reaction coordinate: analysis of a retaining β-glycoside hydrolase. Biochemistry. 1998;37:11707–11713. doi: 10.1021/bi981315i. [DOI] [PubMed] [Google Scholar]
- 16.Medve J, Karlsson J, Lee D, Tjerneld F. Hydrolysis of microcrystalline cellulose by cellobiohydrolase I and endoglucanase II from Trichoderma reesei: adsorption, sugar production pattern, and synergism of the enzymes. Biotechnol Bioeng. 1998;59:621–634. [PubMed] [Google Scholar]
- 17.Wang Q, Graham RW, Trimbur D, Warren RAJ, Withers SG. Changing enzymic reaction mechanisms by mutagenesis: conversion of a retaining glucosidase to an inverting enzyme. J Am Chem Soc. 1994;116:11594–11595. [Google Scholar]
- 18.Shaw A, Bott R, Vonrhein C, Bricogne G, Power S, Day AG. A novel combination of two classic catalytic schemes. J Mol Biol. 2002;320:303–309. doi: 10.1016/S0022-2836(02)00387-X. [DOI] [PubMed] [Google Scholar]
- 19.Parry NJ, Beever DE, Owen E, Vandenberghe I, Van Beeumen J, Bhat M. Biochemical characterization and mechanism of action of a thermostable beta-glucosidase purified from Thermoascus aurantiacus. Biochem J. 2001;353:117–127. [PMC free article] [PubMed] [Google Scholar]
- 20.Rosgaard L, Pedersen S, Langston J, Akerhielm D, Cherry JR, Meyer AS. Evaluation of minimal Trichoderma reesei cellulase mixtures on differently pretreated barley straw substrates. Biotechnol Prog. 2007;23:1270–1276. doi: 10.1021/bp070329p. [DOI] [PubMed] [Google Scholar]
- 21.Nakazawa H, Okada K, Kobayashi R, Kubota T, Onodera T, Ochiai N, Omata N, Ogasawara W, Okada H, Morikawa Y. Characterization of the catalytic domains of Trichoderma reesei endoglucanase I, II, and III expressed in Escherichia coli. Appl Microbiol Biotechnol. 2008;81:681–689. doi: 10.1007/s00253-008-1667-z. [DOI] [PubMed] [Google Scholar]
- 22.Qin Y, Wei X, Song X, Qu Y. Engineering endoglucanase II from Trichoderma reesei to improve the catalytic efficiency at a higher pH optimum. J Biotechnol. 2008;135:190–195. doi: 10.1016/j.jbiotec.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 23.Canutescu AA, Shelenkov AA, Dunbrack RL. A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. Iterative model building, structure refinement and density modification with the Phenix autobuild wizard. Acta Crystallogr Sect D. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


