Abstract
Engineering biomaterials mimicking the biofunctionality of the extracellular matrix (ECM) is important in instructing and eliciting cell response. The native ECM is highly dynamic and has been shown to support cellular attachment, migration, and differentiation. The advantage of synthesizing an ECM-based biomaterial is that it mimics the native cellular environment. However, the ECM has tissue-specific composition and patterned arrangement. In this study, we have employed biomimetic strategies to develop a novel collagen/chitosan template that is embedded with the native ECM of differentiating human marrow stromal cells (HMSCs) to facilitate osteoblast differentiation. The scaffold was characterized for substrate stiffness by magnetic resonance imaging and nanoindentation and by immunohistochemical analysis for the presence of key ECM proteins. Gene expression analysis showed that the ECM scaffold supported osteogenic differentiation of undifferentiated HMSCs as significant changes were observed in the expression levels of growth factors, transcription factors, proteases, receptors, and ECM proteins. Finally, we demonstrate that the scaffold had the ability to nucleate calcium phosphate polymorphs to form a mineralized matrix. The results from this study suggest that the three-dimensional native ECM scaffold directly controls cell behavior and supports the osteogenic differentiation of mesenchymal stem cells.
Introduction
Regeneration or repair of native tissues requires scaffolds, appropriate cell types, and cell signaling molecules. The purpose of the scaffold is to provide mechanical support and a microenvironment for cells. However, the desired cellular response is currently triggered by several approaches such as controlled release of signaling molecules, use of genetically engineered cells that constitutively express the desired factors, or coupling signaling molecules to the scaffolds.1,2 These approaches have significant drawbacks such as inconsistent release kinetics, unpredictable diffusion rates of released molecules, risk of oncogenic transformation of transfected cells, and loss of activity of coupled molecules.1,2 Often times, scaffolds are selected based on their ability to promote adhesion and proliferation of the desired cell types. Although several of the scaffolds used for tissue engineering applications are derived from extracellular matrix (ECM) proteins or polymers designed to mimic ECM proteins, they do not mimic the native ECM, and therefore, in vivo-like behavior of the embedded cells cannot be obtained. To overcome the aforementioned drawbacks, the cellular response needs to be triggered by engineering a scaffold mimicking the extracellular environment of native tissue.
In tissues, cells are surrounded by their ECM. The native ECM is a complex entity comprising of structural proteins (primarily collagen) that provide support, proteoglycans and hyaluronan that can bind to growth factors, and specialized multi-adhesive proteins that bind cells to the matrix.3–8 Although ECM proteins can be categorized, a majority of them are multifunctional and they can also transmit biochemical signals that influence cell migration, proliferation, and differentiation. The composition of the ECM is unique for each cell type and tissue. The ECM of mesenchymal cells contains collagen type I, III, IV, and V, fibronectin, laminin, and other structural proteins along with proteoglycans such as perlecan, syndecan, and biglycan.3–5 Published reports demonstrate that the ECM of mesenchymal stem cells (MSCs) helps maintain the “stemness” when undifferentiated mesenchymal cells were cultured on it.6 Recent studies have also shown that the ECM of MSCs cultured in osteogenic medium influence the differentiation of these stem cells toward an osteoblastic lineage.7,8 Therefore, it is reasonable to expect that engineered native ECM scaffolds will play a significant role in future tissue engineering applications.
In the present study, we have fabricated a three-dimensional (3D) biomimetic scaffold for bone tissue engineering that could trigger differentiation of marrow stem cells toward an osteogenic lineage. For skeletal tissue regeneration, human marrow stromal cells (HMSCs) serve as a good source for adult stem cells as they can differentiate into osteoblasts, chondrocytes, or adipocytes depending on the microenvironment.9 Further, these cells are relatively easy to isolate from patients and can be expanded in culture. Differentiating HMSCs embedded in a collagen and chitosan copolymer was used to prepare the ECM scaffold. These cells secreted a functional ECM after several days in osteogenic culture.
Our rationale for this study is that an osteogenic ECM-embedded collagen/chitosan scaffold would provide a cell-instructive structural framework that would induce the differentiation of mesenchymal cells (HMSCs) into osteogenic cells and promote nucleation of calcium phosphate polymorphs. We tested this hypothesis by analyzing the expression of osteoblast markers using quantitative real-time polymerase chain reaction (PCR) to assess osteogenic differentiation and performed in vitro mineralization assay to test the functionality of the scaffold with respect to mineralized matrix formation.
Materials and Methods
Cell culture
HMSCs were used throughout this study. HMSCs isolated from the iliac crest of normal adult donors were obtained from NIH-funded center for research resources, Tulane Center for the preparation and distribution of adult stem cells.10 Cells obtained from passage 3 were utilized for all experiments. Cells were cultured in minimum essential medium–alpha (Gibco) containing 20% fetal bovine serum (Gibco), 1% l-glutamine (Gibco), and 1% antibiotic–antimycotic cocktail (Gibco).
Fabrication of ECM scaffold
HMSCs (2×106 cells/mL) were embedded in a 1:1 copolymer matrix consisting of 1 mg/mL type I collagen and 1 mg/mL chitosan as previously described.11 A 1:1 matrix was selected based on our earlier study8 that indicated that this ratio was ideal for mesenchymal cell culture. This ratio provided the perfect mix of hydrogel stability combined with proliferation rate similar to that of type I collagen hydrogel. The cells were cultured for 48 h in growth media. Subsequently, differentiation media containing 100 μg/mL ascorbic acid, 10 mM β-glycerophosphate, and 10 mM dexamethasone was used for culturing the cells for 2 weeks. At the end of 2 weeks, the cells were treated with buffer 1 (10 mM sodium phosphate, 150 mM sodium chloride, and 0.5% Triton X-100) for 30 min at 37°C in a tissue culture incubator. The buffer was then changed to buffer 2 (25 mM ammonium hydroxide) and the scaffolds were incubated for 20 min at 37°C. Finally, the scaffolds were washed three times in Hanks balanced salt solution (HBSS) containing no calcium or magnesium. Scaffolds were then subjected to three freeze–thaw cycles in liquid nitrogen and in a 37°C cell culture incubator, respectively. Finally, the scaffolds were washed three times in HBSS and treated with DNAse (Gibco) at 37°C for 30 min to remove traces of DNA bound to the matrix. The scaffolds were then washed four times in HBSS and stored at 4°C in HBSS containing 5% antibiotic–antimycotic cocktail (Gibco). These scaffolds were used for further cell culture experiments. The flow chart in Figure 1 is a schematic representation of the procedure.
FIG. 1.
Model representing the experimental setup. It is a schematic representation that depicts the different steps involved in the generation of the ECM scaffold. ECM, extracellular matrix. Color images available online at www.liebertonline.com/tea
Immunohistochemical analysis
The ECM and control scaffolds were fixed in 4% paraformaldehyde, dehydrated, and embedded in paraffin. About 5 μm sections were cut using a microtome and mounted onto glass slides. The sections were deparaffinized in xylene and rehydrated by incubating in graded ethanol solutions. The sections were quenched by incubation in 3% hydrogen peroxide solution for 30 min. Immunohistochemical analysis was performed using the Vecta Stain ABC peroxidase kit (Vector Labs) according to the manufacturer's protocol and developed using the DAB kit (Vector Labs). The primary antibodies used were (a) mouse monoclonal anti-dentin matrix protein 1 (DMP1) antibody (1/2000) (a gift from Dr. Chunlin Qin, Baylor College of Dentistry, University of Texas), (b) rabbit polyclonal anti-fibronectin antibody (1/100; Sigma), (c) rabbit polyclonal anti-phosphorylated serine antibody (1/100; Sigma), (d) rabbit polyclonal anti-osteopontin antibody (1/100; a gift from Dr. Larry Fisher, NIH), and (e) rabbit polyclonal anti-bone sialoprotein (BSP) antibody (1/100; a gift from Dr. Larry Fisher, NIH). Immunohistochemistry was also performed using fluorescently tagged secondary antibodies.
Immunofluorescence and confocal microscopy
HMSCs were cultured either on control or ECM-embedded scaffolds as previously described for a period of 2 weeks. The scaffolds were then fixed in 4% paraformaldehyde, permeablized with PBS containing 0.5% Triton X-100, and stained for filamentous actin using phalloidin conjugated with TRITC (for red fluorescence). The samples were then imaged using a Zeiss LSM 510 confocal microscope. Z-stacks of the sections were taken at 1 μm intervals and reconstructed using the Zeiss imaging software. Scaffold sections immunostained with fluorescently tagged secondary antibodies were imaged using the Zeiss LSM 510 Meta confocal microscope.
Calcium phosphate nucleation on ECM-embedded scaffolds
Nucleation of calcium phosphate polymorphs was carried out using two different techniques on the ECM-embedded scaffolds and control scaffolds. In the first technique, nucleation was carried out in the presence of physiological concentrations of calcium and phosphate ions as previously described.12 Briefly, the scaffolds were placed into a channel connecting two halves of an electrolyte cell, one compartment containing calcium buffer (165 mM NaCl, 10 mM HEPES, and 2.5 mM CaCl2 [pH 7.4]) and the other phosphate buffer (165 mM NaCl and 10 mM HEPES KH2PO4 [pH 7.4]). A small electric current of 1 mA was passed through the system to facilitate movement of ions across the samples. The buffers were changed regularly (twice daily) to maintain a constant pH. The samples were subjected to nucleation for a period of 2 weeks. In the second technique, nucleation was carried out under high concentrations of calcium and phosphate. Briefly, the scaffolds were immersed in a 1 M calcium chloride solution for a period of 30 min. They were then washed extensively in water to remove any nonspecifically bound calcium and then immersed in a 1 M sodium phosphate solution for a period of 30 min. The samples were then washed in water and fixed in neutral buffered 4% paraformaldehyde solution.
Electron microscopy
Electron microscopy was performed on the ECM-embedded scaffolds before and after nucleation of calcium phosphate and also on scaffolds seeded with HMSCs. For scaffolds seeded with HMSCs, the samples were fixed 48 h postseeding in 4% paraformaldehyde. The scaffolds were dehydrated in graded ethanol solutions and then dried with hexamethyldisilazane. The samples were mounted onto scanning electron microscopy (SEM) grids, sputter coated, and examined under a field emission scanning electron microscope (JSM-6320F, LEO Gemini 1525 sFEG) or under a variable pressure Hitachi S-3000N SEM without sputter coating for samples mineralized under high concentrations of calcium and phosphate. For energy-dispersive X-ray (EDX) analysis, the samples were imaged and analyzed using a Hitachi S-3000N variable pressure SEM without coating.
Nanoindentation
All measurements were performed at room temperature using a calibrated TI-700 Ubi nanoindentation system (Hysitron, Inc.). The tip used was a standard cono-spherical diamond tip with a radius of ∼100 μm. A trapezoidal load pattern with loading time of 5 s, a hold time segment of 40 s (to allow the viscoelastic creep to dissipate prior to unloading), and an unloading time segment of 5 s was used, with a maximum load of 50/20 μN. Indents were made at intervals of about 20 μm apart and 12 indents were made across each sample starting at visually selected test sites. The reduced modulus was calculated using the Oliver Pharr method and the hardness was calculated using the following formula: H=Pmax/Ar, where Pmax=maximum load and Ar=residual indentation area.
Magnetic resonance imaging
The magnetic resonance imaging (MRI) measurements were performed on control collagen/chitosan scaffold and ECM-embedded scaffold using a Bruker 500 MHz (11.7 T) microimaging facility at UIC. The Bruker microimager uses 56 mm vertical bore magnet (Oxford Instruments) and a Bruker DRX-500 MHz Avance Spectrometer (Bruker Instruments) controlled by a Silicon Graphics SG12 and a Bruker imaging software Paravision 4.0. The system is equipped with a Bruker linear triple axis gradient system with a maximum magnetic field gradient strength of 200 G/cm and micro 5 imaging probes. All experiments were performed using commercial Bruker 10 mm RF saddle coils on this probe. T1 quantification was done using pulse sequence RAREVTR (RARE with variable repetition time).13 The experimental parameters were FOV=1.2×1.2 cm, matrix size=32×32, slice thickness=2.5 mm, TE=13 ms, and TR=104.6, 207.4, 317.6, 436.6, 565.9, 707.4, 863.7, 1038.1, 1235.5, 1462.9, 1731.0, 2057.8, 2476.1, 3058.2, 4022.2, and 7500.0 ms. The T2 maps were calculated using the pulse sequence multislice multiecho.14 The experimental parameters were FOV=1.2×1.2 cm, matrix size=128×128, slice thickness=2.5 mm, TR=2 s, and TE=32 linearly spaced echoes with a step of 10 ms. The apparent diffusion maps were calculated using the standard DTI pulse sequence.15,16 The experimental parameters were FOV=0.8×0.8 mm (control) and 1.2×1.2 cm (scaffold), matrix size=64×64, slice thickness=2.5 mm, TE=24 ms, TR=1.5 s, b-value=10, 20, 30, 50, 100, 300, 500, 700, 800, and 1000 ms.
Quantitative real-time RT-PCR
HMSCs (1×106 cells) were seeded onto either control scaffolds (1:1 collagen:chitosan scaffold) or onto the ECM scaffolds. The cells were cultured in growth medium for either 2 or 4 weeks with media changed once in 2 days. Triplicate scaffolds from each group were processed for RNA isolation using Trizol reagent (Invitrogen) as per the manufacturer's protocol and purified using the SA Biosciences RNA isolation kit. cDNA synthesis was performed using the First strand kit (SA Biosciences) as per the manufacturer's protocol. Quantitative real-time RT-PCR array was performed for the osteogenesis pathway (SA Biosciences). Changes in gene expression profile and the statistical significance were calculated using the data analysis program provided by the manufacturer.
Results
Identification of ECM components in the 3D scaffold
Immunohistochemical analysis of ECM-embedded scaffold sections showed the presence of various ECM components that are known to be present in the bone matrix. Apart from structural proteins, noncollagenous proteins (NCPs) are known to play a significant role both in the differentiation of mesenchymal cells and in the nucleation of hydroxyapatite. Immunohistochemical analysis revealed the presence of NCPs such as DMP1 (Fig. 2A), BSP (Fig. 2B), osteopontin (Fig. 2C), and thrombospondin (Fig. 2D). Figure 2E shows the presence of fibronectin and Figure 2F shows positive staining with anti-phosphorylated serine antibody. Figure 2G and H are rabbit and mouse secondary antibody controls. Sections from non–ECM-embedded control scaffolds (1:1 collagen:chitosan scaffolds) were stained with the same antibodies. No positive staining was observed with the control sections (data not shown). DAPI staining of the sections did not show any positive stain, indicating absence of cellular DNA (Fig. 2I). Immunohistochemistry was also performed with fluorescently tagged secondary antibodies so that the scaffolds could be imaged using a confocal microscope. Supplementary Fig. S1 (Supplementary Data are available online at www.liebertonline.com/tea) shows fluorescence labeling of the ECM scaffold for the aforementioned proteins. Additionally, proof of ECM deposition could also be witnessed by SEM analyses with evidence of matrix being deposited in the shape of a cell (Supplementary Fig. S2).
FIG. 2.
Immunohistochemical localization and SEM analysis of ECM components. (A–F) Representative images that show the presence of dentin matrix protein 1, bone sialoprotein, osteopontin, thrombospondin, fibronectin, and phosphoserine, respectively. (G, H) Rabbit and mouse secondary antibody controls, respectively. Scale bar: 50 μm for all images. (I) Representative image of DAPI-stained ECM scaffold showing absence of cellular DNA. Scale bar: 20 μm for this image. SEM, scanning electron microscopy. Color images available online at www.liebertonline.com/tea
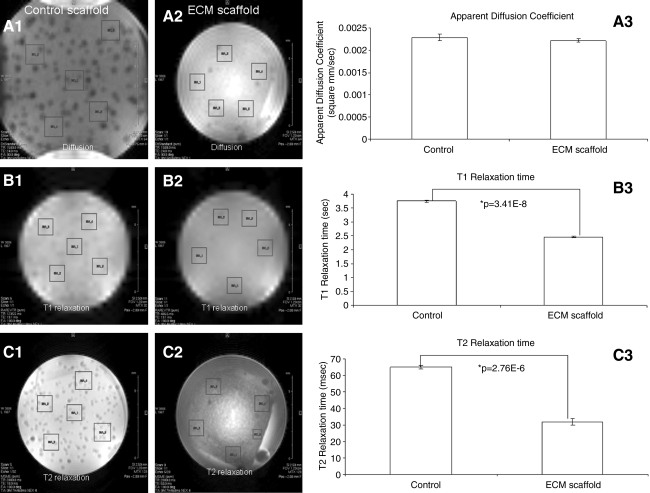
MRI of the scaffold
MRI has potential to detect early changes such as stiffness and porosity in growing tissue.17,18 These changes are reflected as changes in diffusion coefficient and relaxation times. Imaging techniques were used to estimate the apparent diffusion coefficients and T1 and T2 relaxation times in the ECM scaffold with respect to the control collagen/chitosan scaffold. Results in Figure 3 show diffusion and T1 and T2 relaxation maps for both the control and ECM scaffolds. Values were averaged over five randomly selected areas in the scaffold (boxed regions in the images). As can be seen from the graphical representation of the data, there was a small change in the diffusion coefficient between the control and the ECM scaffold (Fig. 3A3), but was not statistically significant. The values obtained were similar to the diffusion coefficient of free water proton.19 This indicates that the addition of several ECM proteins secreted by the cells onto the scaffold will not affect the diffusion of nutrients within the scaffold. However, there was a significant reduction (measured by Student's t-test) in both T1 (Fig. 3B3) and T2 (Fig. 3C3) relaxation times. For water proton, T1 and T2 relaxation times depend on the strength of dipolar couplings, on the orientation and distance between the nuclei, and on the motion. Additionally, scalar couplings also can affect T2 relaxation. As tissue stiffness changes, dipolar coupling between the spins is expected to be high and this change is reflected as a change in relaxation times. Although the T1 and T2 relaxation data do not provide an absolute value for the stiffness of the scaffold, they are indicative of the changes in stiffness.20 The relaxation times are inversely proportional to the stiffness of the material and a decrease in relaxation time indicates an increase in the stiffness of the scaffold. The T1 relaxation time decreased from 3.75 to 2.46 s and the T2 relaxation time decreased from 65 to 32 ms.
FIG. 3.
Magnetic resonance imaging. (A1, A2) Representative apparent diffusion coefficient (ADC) slices of control and ECM-embedded scaffolds, respectively. Boxed regions mark areas for which diffusion coefficients were calculated. (A3) Graph showing ADCs for the control and ECM scaffold. Data represent mean±SD of the values from the five boxed regions. (B1, B2) Representative T1 relaxation slices for control and ECM-embedded scaffolds, respectively. Boxed regions mark areas for which T1 relaxation times were calculated. (B3) Graph showing T1 relaxation times for the control and ECM scaffold. Data represent mean±SD of the values from the five boxed regions. (C1, C2) Representative T2 relaxation slices for control and ECM-embedded scaffolds, respectively. Boxed regions mark areas for which T2 relaxation times were calculated. (C3) Graph showing T2 relaxation times for the control and ECM scaffold. Data represent mean±SD of the values from the five boxed regions.
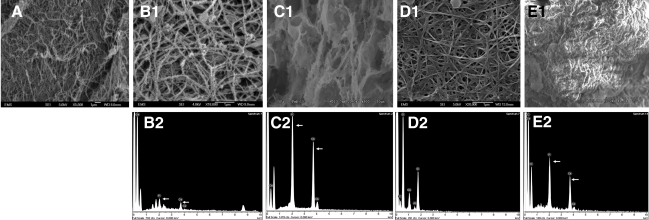
Nucleation of calcium phosphate polymorphs on the ECM scaffold
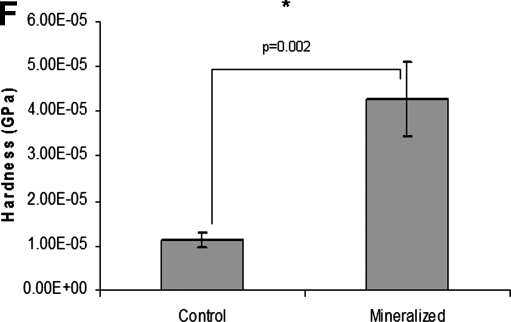
Having identified several of the extracellular components, we then proceeded to analyze whether the ECM scaffold had the ability to nucleate calcium phosphate. Figure 4A is a SEM image of the ECM-integrated scaffold before initiation of mineralization. Calcium phosphate deposition was not observed in this scaffold (data not shown). Results in Figure 4 are representative images that show the ECM scaffold facilitated calcium phosphate deposition both under physiological (Fig. 4B1, B2) as well as in the presence of high concentrations (Fig. 4C1, C2) of calcium and phosphate ions. Under physiological concentrations, the control scaffold did not initiate nucleation of calcium phosphate polymorphs (Fig. 4D1, D2). As the control scaffold was difficult to handle, it was fixed onto a cover glass. The silicon peak that can be seen in the EDX analysis is from the cover glass. Under high calcium and phosphate conditions, the fibrils in the ECM scaffold were highly mineralized (Fig. 4C1) and EDX analysis revealed a Ca/P ratio of 1.62 (n=5, s.e.m.=0.114), suggesting formation of hydroxyapatite as in native bone. Nucleation of calcium phosphate was observed in the control scaffold as well, possibly because of the ability of chitosan to bind metal ions (Fig. 4E1, E2). However, the mineral appeared morphologically different when compared with the ECM scaffold (Fig. 4E1). EDX analyses revealed a Ca/P ratio of 1.21 (n=6, s.e.m.=0.02), indicating the presence of amorphous calcium phosphate. Additionally, the hardness of ECM scaffolds subjected to in vitro nucleation in the presence of high concentrations of calcium and phosphate ions were compared with the control ECM scaffold. Results in Figure 4F show that the hardness of the scaffold subjected to in vitro mineralization increased approximately fourfold and was statistically significant with a p-value of 0.002 (Student's t-test).
FIG. 4.
SEM analysis of mineralized and nonmineralized ECM scaffolds and their mechanical properties. (A) SEM micrograph of the ECM-embedded scaffold before in vitro mineralization. (B1, B2) SEM micrograph of the ECM scaffold subjected to in vitro mineralization under physiological concentrations of calcium and phosphate ions and EDX analysis, respectively. (C1, C2) SEM micrograph of the ECM scaffold subjected to in vitro mineralization at high concentrations of calcium and phosphate ions and EDX analysis, respectively. (D1, D2) SEM micrograph of the control scaffold subjected to in vitro mineralization under physiological concentrations of calcium and phosphate ions and EDX analysis, respectively. (E1, E2) SEM micrograph of the control scaffold subjected to in vitro mineralization at high concentrations of calcium and phosphate ions and EDX analysis, respectively. (F) Graphical representation of the hardness data obtained by using a nanoindenter for the ECM scaffold before mineralization and after mineralization under high concentrations of calcium and phosphate ions. Data represent mean±s.e.m. EDX, energy-dispersive X-ray.
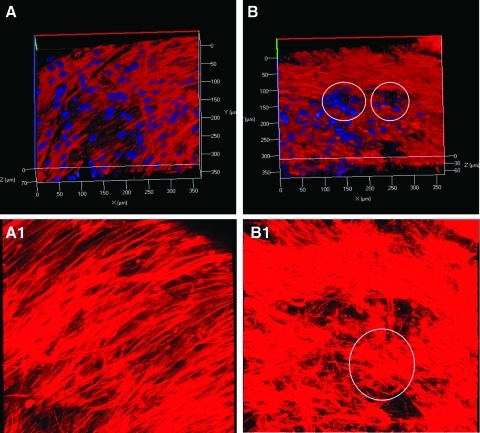
Clustering of HMSCs seeded on ECM scaffold
Confocal microscopy images were obtained for both control and ECM-embedded scaffolds to assess any possible changes in the orientation and morphology of the attached HMSCs. Figure 5 shows representative images of z-stacks from this experiment. Figure 5A and A1 are different 3D reconstructions of the z-stack images and show the distribution of HMSCs on control scaffold after 2 weeks of culture. Images depict homogenous distribution of the cells within the scaffold. Figure 5B and B1 are similar reconstructions to show the distribution of HMSCs on the ECM scaffold after 2 weeks. Comparing the images (5A to 5B and 5A1 to 5B1) it is evident that the HMSCs on the ECM scaffold are more clustered into nodule-like formations when compared with the control. The circled areas in Figure 5B and B1 represent such an arrangement in 3D. Clustering of cells and matrix synthesis is an indicator of osteogenic differentiation and mineralized nodule formation.21 This qualitative analysis points toward a differentiating mesenchymal cell population within the ECM scaffold.
FIG. 5.
Clustering of HMSCs on ECM scaffold. (A, A1) Representative z-stack projections of HMSCs cultured on control collagen/chitosan scaffold. (B, B1) Representative z-stack projections of HMSCs cultured on ECM-embedded collagen/chitosan scaffold. Circles depict clustering of HMSCs. HMSC, human marrow stromal cell. Color images available online at www.liebertonline.com/tea
ECM scaffold promotes cell–matrix and cell–cell interactions
SEM analysis was performed on the ECM scaffold seeded with HMSCs to observe cell–matrix and cell–cell interactions. Figure 6A shows a cell process embedded in the scaffold (white arrows). Figure 6A1 is a magnified image of the boxed area in Figure 6A and shows the branching of the tip of the cellular process used to adhere to the ECM scaffold. These representative images indicate that the ECM scaffold promotes cell–matrix interactions. Figure 6B shows two adjacent cells on the ECM scaffold (marked as 1 and 2). The boxed area in Figure 6B shows cellular processes that connect with the adjacent cell. This is more evident in the magnified image shown in Figure 6B1 (white arrows mark interacting processes). Cell–cell and cell–matrix interactions facilitate signal transduction and influence cellular behavior. Studies using coculture systems have shown that cell–cell interactions between osteoblasts and MSCs induce osteogenic differentiation of the MSCs.22 We believe that interaction between the differentiating cells within the scaffold could induce signal transduction, leading to the formation of a homogeneous population of differentiating cells.
FIG. 6.
ECM scaffold promotes cell–matrix and cell–cell interactions. (A, A1) Representative SEM images showing cell–matrix interactions. White arrows in A indicate cellular processes embedded in the scaffold. (A1) High-magnification image of the boxed area in A. (B, B1) Representative images showing cell–cell interactions on the ECM scaffold. Numbers 1 and 2 in B indicate two adjacent cells. (B1) Higher-magnification image of the boxed area in B. Arrows in B1 indicate points of cell–cell interaction.
ECM scaffold induces osteogenic differentiation of HMSCs in vitro
We tested the ability of the ECM-embedded scaffold to facilitate differentiation of HMSCs into osteogenic cells in the absence of external additives or growth factors. Osteoblast gene expression analysis was performed by quantitative real-time RT-PCR and compared with HMSCs cultured on control collagen/chitosan scaffolds. Table 1 shows the list of osteoblast-specific genes that showed a significant change in expression compared with control after 2 weeks in culture in the ECM scaffold. Several osteoblast-specific genes showed significant upregulation, indicating osteogenic differentiation. Table 2 shows the list of genes that showed significant difference in expression levels between 2 and 4 weeks of culture in the ECM scaffold. Table 2 contains fewer genes when compared with Table 1, as it only lists genes that changed significantly between 2 and 4 weeks of culture. Many genes present in Table 1 are not in Table 2. This is because they did not undergo any statistically significant change between 2 and 4 weeks of culture in the ECM scaffold. On the other hand, there were a few genes such as insulin-like growth factor 1 (IGF1), matrix metalloproteinase 8 (MMP8), and calcitonin receptor that were upregulated at 4 weeks, indicating a possible progression in differentiation. Downregulation of certain genes in Table 2 may indicate that they may be required only during early stages of differentiation. Table 3 lists the genes that were significantly regulated between 2 weeks of culture in the ECM scaffold compared with 4 weeks of culture in the control scaffold. Almost all of the genes expressed at 2 weeks (Table 1) showed significant regulation in this comparison (Table 3), indicating that osteoblast differentiation genes were significantly expressed at 2 weeks of culture in ECM scaffold even when compared with 4 weeks of culture in the control scaffold, although 4 weeks of culture in the control scaffold did show a certain degree of differentiation possibly because of the 3D culture conditions and also confluent population of cells leading to differentiation.
Table 1.
List of Genes That Are Significantly Regulated at the 2 Week Time Point When Human Marrow Stromal Cells Were Cultured on the Extracellular Matrix-Embedded Collagen/Chitosan Scaffold Compared with Control Collagen/Chitosan Scaffold
| Gene symbol | Description | Fold change | p-Value (t-test) |
|---|---|---|---|
| Growth factors | |||
| BMP6 | Bone morphogenetic protein 6 | 9.31 | 0.075 |
| EGF | Epidermal growth factor | 2.37 | 0.0494 |
| FGF2 | Fibroblast growth factor 2 | 4.75 | 0.00027 |
| GDF10 | Growth and differentiation factor 10 | 20.13 | 0.043 |
| IGF2 | Insulin-like growth factor 2 | 60.88 | 0.0217 |
| TGFB1 | Transforming growth factor β1 | 9.03 | 0.0011 |
| TGFB3 | Transforming growth factor β3 | 3.07 | 0.0040 |
| VEGFB | Vascular endothelial growth factor β | 4.63 | 0.0006 |
| PDGFA | Platelet-derived growth factor A | 7.4 | 0.00003 |
| Transcription factors | |||
| MSX1 | Msh homeobox 1 | 7.31 | 0.061 |
| NFKB1 | Nuclear factor kappa of B-cells 1 | 2.32 | 0.00065 |
| RUNX2 | Runt-related transcription factor 2 | 2.35 | 0.019 |
| SMAD2 | SMAD family member 2 | 1.56 | 0.05 |
| SMAD3 | SMAD family member 3 | 2.19 | 0.0056 |
| SMAD4 | SMAD family member 4 | 2.09 | 0.0025 |
| VDR | Vitamin-D receptor | 4.10 | 0.0006 |
| Phosphatases | |||
| MINPPI | Multiple Inositol polyphosphate histidine phosphatase 1 | 4.07 | 0.0012 |
| ALPL | Alkaline phosphatase | 9.8 | 0.0002 |
| ECM proteins | |||
| BGN | Biglycan | 14.52 | 0.00002 |
| COMP | Cartilage oligomeric matrix protein | 23.17 | 0.0006 |
| COL10A1 | Collagen type X, alpha 1 | 2.72 | 0.0009 |
| COL11A1 | Collagen type XI, alpha 1 | 4.14 | 0.0015 |
| COL12A1 | Collagen type XII, alpha 1 | 4.02 | 0.002 |
| COL14A1 | Collagen type XIV, alpha 1 | 3.56 | 0.006 |
| COL15A1 | Collagen type XV, alpha 1 | 5.44 | 0.002 |
| COL1A1 | Collagen type I, alpha 1 | 15.7 | 0.003 |
| COL1A2 | Collagen type I, alpha 2 | 8.63 | 0.0015 |
| COL3A1 | Collagen type III, alpha 1 | 10.67 | 0.0001 |
| COL4A3 | Collagen type IV, alpha 3 | 3.13 | 0.11 |
| COL5A1 | Collagen type V, alpha 1 | 9.36 | 0.002 |
| FN1 | Fibronectin 1 | 1.94 | 0.064 |
| COL2A1 | Collagen type II, alpha 1 | −4.42 | 0.057 |
| Proteases | |||
| BMP1 | Bone morphogenetic protein 1 | 6.42 | 0.0012 |
| MMP2 | Matrix metalloproteinase 2 | 5.10 | 0.0025 |
| Receptors | |||
| CDH11 | Cadherin 11 | 2.66 | 0.0055 |
| EGFR | Epidermal growth factor receptor | 2.03 | 0.067 |
| FGFR1 | Fibroblast growth factor receptor 1 | 2.83 | 0.002 |
| FGFR2 | Fibroblast growth factor receptor 2 | 4.15 | 0.00007 |
| FLT1 | Vascular endothelial growth factor receptor 1 | 11.48 | 0.013 |
| ICAM1 | Intercellular adhesion molecule 1 | 1.95 | 0.08 |
| IGF1R | Insulin-like growth factor 1 receptor | 2.66 | 0.0014 |
| SCARB1 | Scavenger receptor class B, member 1 | 3.11 | 0.003 |
| TGFBR2 | Transforming growth factor β receptor II | 2.38 | 0.0015 |
| ITGA3 | Integrin alpha 3 | 2.16 | 0.04 |
Data represent mean of triplicate experiments. p-Value was obtained from the Student's t-test.
Table 2.
List of Genes That Are Significantly Regulated Between the 2 and 4 Week Time Point When Human Marrow Stromal Cells Were Cultured on the Extracellular Matrix-Embedded Collagen/Chitosan Scaffold
| Gene symbol | Description | Fold change | p-Value |
|---|---|---|---|
| Growth factors | |||
| BMP3 | Bone morphogenetic protein 3 | 3.24 | 0.029 |
| BMP4 | Bone morphogenetic protein 4 | −2.56 | 0.05 |
| BMP5 | Bone morphogenetic protein 5 | 9.01 | 0.015 |
| IGF1 | Insulin-like growth factor 1 | 2.04 | 0.007 |
| GDF10 | Growth and differentiation factor 10 | 8.36 | 0.021 |
| TGFB3 | Transforming growth factor β3 | −2.50 | 0.032 |
| PDGFA | Platelet derived growth factor alpha | −1.80 | 0.014 |
| ECM proteins | |||
| BGLAP | Bone gamma-carboxyglutamate (gla) protein | −3.2 | 0.002 |
| COL2A1 | Collagen type II, alpha 1 | 4.94 | 0.042 |
| Proteases | |||
| BMP1 | Bone morphogenetic protein 1 | −1.69 | .046 |
| MMP8 | Matrix metalloproteinase 8 | 3.45 | 0.037 |
| Receptors | |||
| CALCR | Calcitonin receptor | 5.87 | 0.001 |
| FGFR1 | Fibroblast growth factor receptor 1 | −2.40 | 0.007 |
| FLT1 | Vascular endothelial growth factor receptor 1 | −2.45 | 0.045 |
The (−) sign before the fold change indicates downregulation. Data represent mean of triplicate experiments. p-Value was obtained from the Student's t-test.
Table 3.
List of Genes That Are Significantly Regulated When the 2-Week Culture of Human Marrow Stromal Cells Cultured on the Extracellular Matrix-Embedded Collagen/Chitosan Scaffold Were Compared with the 4-Week Culture of Human Marrow Stromal Cells Cultured in Control Collagen/Chitosan Scaffold
| Gene symbol | Description | Fold change | p-value [t-test] |
|---|---|---|---|
| Growth factors | |||
| BMP4 | Bone morphogenetic protein 4 | 4.58 | 0.038 |
| EGF | Epidermal growth factor | 2.13 | 0.056 |
| IGF2 | Insulin-like growth factor 2 | 10.04 | 0.03 |
| TGFB1 | Transforming growth factor β1 | 3.93 | 0.011 |
| VEGFB | Vascular endothelial growth factor β | 3.34 | 0.008 |
| PDGFA | Platelet-derived growth factor A | 3.18 | 0.003 |
| Transcription factors | |||
| NFKB1 | Nuclear factor kappa of B-cells 1 | 1.9 | 0.003 |
| RUNX2 | Runt-related transcription factor 2 | 2.3 | 0.021 |
| SMAD3 | SMAD family member 3 | 2.17 | 0.027 |
| VDR | Vitamin-D receptor | 2.73 | 0.015 |
| Phosphatases | |||
| ALPL | Alkaline phosphatase | 4.83 | 0.0007 |
| MINPP1 | Multiple inositol polyphosphate histidine phosphatase 1 | 2.03 | 0.009 |
| ECM proteins | |||
| BGLAP | Bone gamma-carboxyglutamate (gla) protein | 3.68 | 0.028 |
| BGN | Biglycan | 5.18 | 0.002 |
| COMP | Cartilage oligomeric matrix protein | 6.15 | 0.002 |
| COL10A1 | Collagen type X, alpha 1 | 4.31 | 0.011 |
| COL11A1 | Collagen type XI, alpha 1 | 4.10 | 0.002 |
| COL12A1 | Collagen type XII, alpha 1 | 2.3 | 0.027 |
| COL15A1 | Collagen type XV, alpha 1 | 4.63 | 0.027 |
| COL1A1 | Collagen type I, alpha 1 | 7.9 | 0.009 |
| COL1A2 | Collagen type I, alpha 2 | 4.3 | 0.022 |
| COL3A1 | Collagen type III, alpha 1 | 4.28 | 0.006 |
| COL5A1 | Collagen type V, alpha 1 | 4.93 | 0.008 |
| SERPINHI | Serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) | 3.76 | 0.036 |
| Proteases | |||
| BMP1 | Bone morphogenetic protein 1 | 2.53 | 0.054 |
| MMP10 | Matrix metalloproteinase 10 | −26.08 | 0.044 |
| Receptors | |||
| CDH11 | Cadherin 11 | 2.84 | 0.027 |
| FGFR1 | Fibroblast growth factor receptor 1 | 3.56 | 0.043 |
| FGFR2 | Fibroblast growth factor receptor 2 | 3.35 | 0.0008 |
| FLT1 | Vascular endothelial growth factor receptor 1 | 2.83 | 0.031 |
p-Value was obtained from the Student's t-test.
Absence of chondrogenic and adipogenic differentiation of HMSCs
HMSCs are multipotent and possess the ability to differentiate into multiple cell types such as chondrocytes and adipocytes besides osteoblasts. It is therefore important to show that the osteogenic ECM scaffold does not induce chondrogenic or adipogenic differentiation. Gene expression analyses revealed the absence of chondrogenic and adipogenic differentiation of the HMSCs cultured in the ECM scaffolds. Chondrogenic marker genes were analyzed by real-time PCR and did not show any significant changes with time in culture. Type II collagen expression (a hallmark for chondrocytes) was low to begin with (Ct value of 34.5). It reduced by 4.4-fold after 2 weeks in culture and returned to basal levels by 4 weeks. There was no significant change in the expression of chondrocyte-specific master transcription factor Sox9. Additionally, MMP2, an inhibitor of mesenchymal cell chondrogenic differentiation, was upregulated fivefold after 2 weeks of culture in the ECM scaffold.
With respect to adipogenic differentiation, one of the primary marker genes for adipocyte differentiation is CD36.23 We did not see any significant changes in CD36 gene regulation. Growth and differentiation factor 10 (GDF10) and vitamin D receptor, both negative regulators of adipogenesis,24,25 were upregulated. GDF10 was upregulated 20-fold and 8-fold after 2 and 4 weeks of culture in the ECM scaffold as opposed to the control scaffold. Vitamin D receptor was upregulated fourfold after 2 weeks of culture in the ECM scaffold, suggesting an absence of induction of the adipocyte differentiation pathway.
Discussion
Cells derive regulatory cues from the ECM and permit them to organize into functional tissues. ECM contains many macromolecules such as fibronectin, proteoglycans, collagens, laminin, and sequestered growth factors. It is primarily this molecular information that confers its bioactivity.18 The multitude of ECM proteins presented to cells in a given tissue is likely to be critical in determining how cells behave within the tissue.26 Therefore, it is necessary for biomaterials used in tissue engineering to provide cells with tissue-specific biological cues. The present study highlights the fabrication of ECM-embedded 3D scaffolds using collagen and chitosan as a template for osteogenic differentiation. Collagen was the material of choice as it is a major constituent of bone and dentin ECM and self-assembles to form a template for hydroxyapatite nucleation and growth.27 Several published studies have used type I collagen as a scaffolding matrix coupled with hydroxyapatite, growth factors, and other naturally occurring polymers such as chitosan for tissue engineering.11,28–30 Chitosan was selected, as it is a naturally occurring biopolymer that is obtained from deacylation of chitin from crustaceans.31 Further, chitosan has been shown to be biocompatible and biodegradable and is being used in a variety of tissue engineering applications.32,33 Additionally, chitosan also possesses antimicrobial characteristics,34 making it an attractive candidate for in vivo applications. We have previously published that a scaffold containing a blend of type I collagen with chitosan increased the mechanical properties of the matrix and that the cellular proliferation rate could be manipulated by varying the composition of the components in a 3D coculture system.11 Addition of chitosan to collagen hydrogels increases their mechanical strength by coating the collagen fibers,35 but reduces the proliferation rate of embedded cells.35 Additionally, collagen–chitosan hydrogel maintains its shape better when compared with plain collagen hydrogel that collapses with time in culture. A detailed characterization of collagen–chitosan hydrogel blends has been previously published by Dr. Desai's group.35
Immunohistochemical analysis of the ECM scaffold demonstrated the presence of key matrix molecules synthesized by osteoblasts. Among them were DMP1, osteopontin, thrombospondin, and BSP. We have published earlier that DMP1 is present in the ECM of bone and binds to type I collagen and functions as a hydroxyapatite nucleator.12,36 Both osteopontin and BSP are secreted NCPs that possess both collagen and calcium binding properties.37 Thrombospondin is an ECM protein important for bone remodeling and for differentiation of MSCs (possibly through notch signaling).38 The presence of phosphorylated ECM proteins was observed using the phosphorylated serine-specific antibody. Phosphoproteins play an important role during bone formation and perform dual functions, namely, serve as a nucleator or inhibitor of hydroxyapatite and also function as signaling molecules.39 Fibronectin, a key ECM cell-adhesive component was also identified. The presence of these ECM proteins suggests that the scaffold can provide the necessary microenvironment required for osteoblast differentiation.
The ideal scaffold for bone tissue engineering should have the ability to nucleate calcium phosphate polymorphs. In vitro nucleation assays were performed to test the ability of the fabricated ECM scaffold to nucleate calcium phosphate. The assays showed calcium phosphate deposition in the presence of both physiological concentrations as well as high concentrations of calcium and phosphate ions. It was interesting to note that the presence of ECM components within the scaffold triggered nucleation of hydroxyapatite as opposed to amorphous calcium phosphate in control scaffolds. Higher concentrations of calcium and phosphate were used to test the possibility of coating the ECM scaffold with calcium phosphate as they are known for their osteoconductive and osteoinductive properties. Substrate stiffness is known to affect cell spreading, growth rate, gene expression, and osteogenic differentiation of undifferentiated stem cells. In this study, MRI was used to determine the stiffness of the scaffold. Results indicated that the ECM scaffold demonstrated a significant increase in the stiffness when compared with the control collagen/chitosan scaffold. This change in stiffness of the scaffold can contribute toward cellular differentiation. Additionally, the diffusion constant of water in the ECM scaffold remained unaltered when compared with the control, suggesting that the deposition of the ECM will not affect the diffusion of nutrients.
One of the powerful advantages of using the ECM scaffolds for tissue engineering is that their cell-instructive ability can induce lineage-specific differentiation of precursor cells. In this study, we focused on the ability of the ECM scaffold to drive HMSCs toward an osteogenic lineage. Our results indicate that the ECM scaffold was able to induce differentiation by upregulation of several growth factors, transcription factors, ECM components, proteases, and receptors. More importantly, these changes were brought about without the need for growth factors or differentiating agents. Genes that were significantly regulated as a result of the instructive ECM matrix were grouped into the following categories:
Growth factors
Several growth factors were upregulated significantly when HMSCs were cultured on ECM-embedded scaffolds. In the TGFβ/BMP (transforming growth factor β/bone morphogenetic protein) superfamily of proteins that are critical for osteogenesis, BMP6, GDF10, and TGFβ1 and 3 were upregulated at 2 weeks. Additionally, TGFβ3 that was upregulated at 2 weeks was downregulated at 4 weeks (Table 2), suggesting that it may be an early event in the osteogenesis pathway. BMP6 has been shown to induce osteoblast differentiation by phosphorylating SMADs 1 and 540 and also is constitutively expressed at higher levels following fracture.41 TGFβ1 is expressed constitutively in bone and in higher levels immediately following fracture,41 suggesting that it is required for osteoblast differentiation and maintenance of bone. However, TGFβ3 is upregulated after fracture, suggesting its necessity in the formation of new bone and in the differentiation of mesenchymal cells at the fracture site.41 GDF10/BMP3b showed an increase of 20-fold at 2 weeks compared with control scaffold at the same time point (Table 1). GDF10/BMP3b is upregulated immediately following fracture and is constitutively expressed at a higher level throughout osteogenesis. GDF10 is therefore required throughout the process of osteogenesis.41 In our experiments, GDF10 was upregulated at both 2 and 4 weeks of culture (Tables 1 and 2). GDF10 is also a negative regulator of adipogenesis. Between 2 and 4 weeks in the ECM scaffold, BMP3 and BMP5 were upregulated and BMP4 was downregulated (Table 2). The fact that BMPs 3 and 5 were upregulated at 4 weeks suggests that these BMPs could be required at later stages of osteogenic differentiation. The upregulation of the aforementioned growth factors by HMSCs cultured in the ECM scaffold indicates that the scaffold favors the osteogenic differentiation of these cells.
Recently, IGF2 has been shown to induce the differentiation of marrow stromal cells by potentiating BMP9.42 Additionally, BMP9 has been found to be the most potent of BMPs in terms of osteogenic ability, surpassing even BMP2.43 In this context, it was interesting to note a 60-fold increase in IGF2 expression in HMSCs cultured for 2 weeks on ECM scaffold (Table 1).
All of the aforementioned growth factors directly affect the differentiation of mesenchymal cells. On the other hand, vascular endothelial growth factor (VEGF) and platelet-derived growth factor alpha (PDGFA) are mitogenic and angiogenic factors that induce the migration of endothelial cells to promote angiogenesis in vivo. Promotion of angiogenesis is an extremely important property to be considered if the scaffold has to be successful for in vivo bone regeneration. Additionally, upregulation of angiogenic factors also indicates osteogenic differentiation of these cells as opposed to chondrogenic differentiation that the HMSCs are capable of. VEGFB was upregulated fivefold after 2 weeks of culture in the ECM scaffold compared with the control scaffold (Table 1) and did not show any significant change after 4 weeks. PDGFA was upregulated sevenfold at 2 weeks (Table 1) and subsequently downregulated by twofold between 2 and 4 weeks (Table 2). Interestingly, BMP4, TGFβ1, epidermal growth factor (EGF), IGF2, GDF10, VEGFB, and PDGFA were all upregulated significantly when data from 2-week ECM scaffold culture and 4-week control culture (Table 3) was compared. This indicates that although 3D culture might induce differentiation of HMSCs to an extent, a 2-week culture in ECM scaffold significantly accelerates the induction of growth factors when compared with even 4 weeks of HMSC culture in the control scaffold. Another growth factor that was upregulated at 2 weeks was EGF (2.4-fold; Table 1). EGF upregulation was maintained at twofold when 2-week ECM scaffold culture was compared with 4-week control scaffold culture (Table 3), indicating that there was no change in EGF expression when the cells were grown in the control scaffold.
ECM components
HMSCs cultured on the ECM scaffold showed significant upregulation of several genes encoding ECM proteins. Almost all types of collagens were upregulated. Type I collagen was upregulated 16-fold, indicating osteogenic differentiation (Table 1). Proteoglycans such as biglycan (14.5-fold) and cartilage oligomeric matrix protein (COMP) (23-fold) were also significantly upregulated (Table 1). Published reports indicate that biglycan may modulate osteoblast differentiation44 and may also be involved in the nucleation of hydroxyapatite.45 COMP has been reported to be expressed by both embryonic and adult osteoblasts.46 Immunostaining and in situ hybridization in published reports have shown the presence of COMP in the bone collar, the newly formed bone near the growth plate and in the diaphysis of a 21-day human fetus.46
All the genes encoding ECM proteins that were upregulated as shown in Table 1 (except COMP) were significantly upregulated when 2-week ECM scaffold culture was compared with 4-week control scaffold culture (Table 3). The expression level of type II collagen gene in control HMSCs was very low (average Ct value of 34.5). With 2 weeks of culture in the ECM scaffold, the expression of type II collagen decreased 4.4 times (Table 1). The expression was returned to basal levels at 4 weeks (Table 2), suggesting that the pluripotent HMSCs are not undergoing chondrogenic differentiation.
Phosphatases
Alkaline phosphatase (ALP) and multiple inositol polyphosphate histidine phosphatase 1 (MINPP1) are key phosphatases necessary for bone formation. ALP was upregulated 10-fold and MINPP1 was upregulated fourfold in the 2-week ECM scaffold cultures compared with control cultures (Table 1). Phosphatases are known to cleave phosphates from several matrix proteins. The cleaved phosphate is responsible for the formation of calcium phosphate crystals. It is possible that the phosphorylated proteins present in the ECM could trigger downstream signaling events leading to upregulation of ALP in the ECM scaffolds. Upregulation of these genes suggests that the cells are possibly differentiating into osteoblasts.
Proteases
Functions of the proteases range from remodeling the ECM network to activating and deactivating different proteins by cleaving them at specific sites. After 2 weeks of culture in the ECM scaffold, two proteases were upregulated compared with the control scaffold. They are BMP1 (6.4-fold) and MMP2 (5-fold) (Table 1). Loss of MMP2 has been attributed to decreased mineralization and defects in the growth of osteoblasts and osteoclasts.47 MMP2 has also been implicated as a negative regulator of chondrogeneis.48 BMP1, also know as procollagen peptidase functions to cleave procollagen types I, II, and III. Upregulation of BMP1 might indicate processing of collagens and other matrix proteins such as DMP1. However, BMP1 was downregulated 1.7-fold at 4 weeks (Table 2); correspondingly, no significant change in collagen expression was observed at 4 weeks. MMP2 levels did not show a significant change between 2 and 4 weeks of culture on the ECM scaffolds. Additionally, MMP 8 was upregulated 3.5 times at 4 weeks (Table 2). Comparing the gene expression patterns at 2-week culture on the ECM scaffold with 4-week culture on the control scaffold, BMP1 remained upregulated 2.5-fold (Table 3). An interesting observation was the 26-fold downregulation of MMP10 (stromylysin 2) expression by HMSCs in the 2-week ECM scaffold culture compared with 4-week control scaffold culture (Table 3). Based on the observation that expression of osteoblast differentiation markers in the control scaffold lags behind the ECM scaffold, it can be hypothesized that MMP10 upregulation could be an early event during osteogenesis.
Transcription factors
Several transcription factors required for osteoblast differentiation were upregulated when HMSCs were cultured on ECM scaffolds. Runt-related transcription factor 2 (RUNX2) was upregulated 2.3-fold in the ECM scaffold at 2 weeks when compared with the control scaffold (Table 1). This fold change remained constant when the 2-week ECM scaffold data were compared with the 4-week control scaffold data, indicating that in the control scaffold, RUNX2 was not upregulated even after 4 weeks of culture. RUNX2 is an osteogenic-specific master transcription factor49,50 and RUNX2-deficient mice die at birth and are characterized by a complete absence of mineralized skeleton.51,52 The upregulation of RUNX2 is a clear indication of osteogenic differentiation.
SMADs 2, 3, and 4 were upregulated 1.5-, 2.2-, and 2-fold, respectively, after 2 weeks of culture on ECM scaffolds compared with control scaffolds (Table 1). Receptor-regulated SMADS 1, 2, 3, 5, and 8/9 are downstream transcription factors for TGFβ/BMP family of proteins. Of these, SMADs 2 and 3 are linked to TGFβ signaling cascade.53,54 Activation of TGF beta receptors leads to phosphorylation of receptor SMADS, which oligomerizes with SMAD4 and accumulate in the nucleus where they recognize gene regulatory regions and orchestrate transcription of osteogenic genes. Upregulation of SMADs in this study indicates that TGFβ signaling has been potentiated. Upregulation of SMAD4 might be due to an increase in the expression of SMADs 2 and 3. Other transcription factors that were upregulated at 2 weeks were MSX1 (msh homeobox 1) (7.3-fold), NFKB1 (nuclear factor kappa of B-cells 1) (2.3-fold), and vitamin-D receptor (4.1-fold) (Table 1). All these transcription factors play a significant role in osteogenesis.55,56 More specifically, vitamin D receptor has been shown to be a negative regulator of adipogenesis.25 Upregulation of this nuclear receptor is an indication of suppression of the adipogenic differentiation pathway. Interestingly, these transcription factors also remained upregulated when gene expression levels were compared for the 2-week ECM scaffold with the 4-week control scaffold (Table 3).
Receptors
Receptors are membrane proteins to which cytokines and growth factors bind to transduce their signal. The potency of a growth factor depends on the number of receptors present on the membrane surface and its recycling rate. Therefore, it is logical to assume that if there was an increase in the growth factor concentration, then the number of receptors specific to the growth factors would also increase in expression proportionately. However, if the recycling rate of the receptor is high, then a corresponding increase of the receptors is not necessary to compensate for an increase in growth factor expression. In our model system, after 2 weeks of culture on the ECM scaffold, significant increase in gene expression was observed with EGF receptor (twofold corresponding to its growth factor increase), FGF receptors 1 and 2 (2.8 and 4.1-fold respectively), VEGF receptor (11.5-fold), IGF1 receptor (2.7-fold), TGFβ receptor 2 (2.4-fold), integrin alpha 3 (2.2-fold), and intercellular adhesion molecule 1 (Table 1). Between 2 and 4 weeks of culture on the ECM scaffold, calcitonin receptor was upregulated sixfold (Table 2). When the 2-week ECM scaffold culture was compared with the 4-week control scaffold culture, cadherin 11, FGF receptors 1 and 2, and VEGF receptor were all upregulated approximately threefold.
Looking at the gene expression profile in its entirety, it is clear that HMSCs, when cultured on the ECM-embedded collagen/chitosan scaffolds, were able to differentiate toward an osteoblastic lineage. The expression of osteoblast differentiation factors were significantly higher in HMSCs cultured on the ECM scaffold for 2 weeks when compared with even 4 weeks of culture in the control scaffold. In general, our construct exhibited more robust changes in the expression of osteoblast-specific genes that regulate osteoblast differentiation than other constructs reported thus far.7,8 We believe that our construct has the potential to replace the existing surgical aids such as demineralized bone matrix (DBM) and freeze-dried bone. DBM has been shown to possess intact BMP2, but growth factor concentration and behavior varies between manufacturers and does not perform consistently even between batches from the same source.57 DBMs and freeze-dried allograft bone are also a source for possible HIV transmission. More importantly, the chemical and mechanical processing of these matrices induces many changes to the protein structure and native 3D arrangement of the ECM components. This might render many of the ECM molecules ineffective. On the other hand, the ECM scaffold would be able to provide intact cell-secreted ECM in a native 3D state.
Although we show significant upregulation of osteogenic markers with HMSCs cultured in the ECM scaffold, the results were obtained from a pool of HMSCs obtained from a single source. These results will have to be verified in HMSCs from other sources to make certain that the findings of this study are universal. Additionally, future studies would focus on the in vivo regenerative potential of the ECM scaffold for bone tissue engineering.
Conclusion
Based on the results obtained, the main highlights from this study can be summarized as follows:
Our approach for the development of ECM-embedded collagen/chitosan scaffold yields a 3D porous matrix that contains tissue-specific ECM components secreted by differentiating mesenchymal cells.
The ECM-embedded collagen–chitosan scaffold can function as a template for the nucleation of calcium phosphate polymorphs under physiological conditions.
The increased stiffness of the hydroxyapatite-coated ECM-embedded scaffolds would facilitate its use in load-bearing bone tissue engineering.
The ECM-embedded scaffold promotes cell–cell and cell–matrix interactions.
Analysis of the gene expression profile suggests that the ECM-embedded scaffold can induce the differentiation of HMSCs into osteogenic lineage without the need for external intervention by means of growth factors or differentiating agents.
Supplementary Material
Acknowledgments
This project was funded by the NIH grant DE 11657 (to A.G.), EB007537 (to R.L.M.), and the Brodie Endowment fund. The authors thank Kristina Jarosius of the UIC Research Resources Center Electron Microscopy Facility for her technical assistance.
Disclosure Statement
No competing financial interests exist.
References
- 1.Silva A.K. Richard C. Bessodes M. Scherman D. Merten O.W. Growth factor delivery approaches in hydrogels. Biomacromolecules. 2009;10:9. doi: 10.1021/bm801103c. [DOI] [PubMed] [Google Scholar]
- 2.Lee T.C. Ho J.T. Hung K.S. Chen W.F. Chung Y.H. Yang Y.L. Bone morphogenetic protein gene therapy using a fibrin scaffold for a rabbit spinal-fusion experiment. Neurosurgery. 2006;58:373. doi: 10.1227/01.NEU.0000199725.03186.F6. [DOI] [PubMed] [Google Scholar]
- 3.Hamilton R. Campbell F.R. Immunochemical localizationof extracellular materials in bone marrow of rats. Anat Rec. 2001;231:218. doi: 10.1002/ar.1092310210. [DOI] [PubMed] [Google Scholar]
- 4.Gordon M.Y. Extracellular matrix of the marrow microenvironment. Br J Haematol. 1988;7:1. doi: 10.1111/j.1365-2141.1988.tb02425.x. [DOI] [PubMed] [Google Scholar]
- 5.Hocking A.M. Shinomura T. McQuillan D.J. Leucine-rich repeat glycoproteins of the extracellular matrix. Matrix Biol. 1998;17:1. doi: 10.1016/s0945-053x(98)90121-4. [DOI] [PubMed] [Google Scholar]
- 6.Chen X.D. Dusevich V. Feng J.Q. Manolagas S.C. Jilka R.L. Extracellular matrix made by bone marrow cells facilitates expansion of marrow-derived mesenchymal progenitor cells and prevents their differentiation into osteoblasts. J Bone Miner Res. 2007;22:1943. doi: 10.1359/jbmr.070725. [DOI] [PubMed] [Google Scholar]
- 7.Thibault R.A. Scott Baggett L. Mikos A.G. Kasper F.K. Osteogenic differentiation of mesenchymal stem cells on pregenerated extracellular matrix scaffolds in the absence of osteogenic cell culture supplements. Tissue Eng Part A. 2010;16:431. doi: 10.1089/ten.tea.2009.0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pham Q.P. Kasper F.K. Scott Baggett L. Raphael R.M. Jansen J.A. Mikos A.G. The influence of an in vitro generated bone-like extracellular matrix on osteoblastic gene expression of marrow stromal cells. Biomaterials. 2008;29:2729. doi: 10.1016/j.biomaterials.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 9.Pittenger M.F. Mackay A.M. Beck S.C. Jaiswal R.K. Douglas R. Mosca J.D., et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 10.Sekiya I. Larson B.L. Smith J.R. Pochampally R. Cui J.G. Prockop D.J. Expansion of human adult stem cells from bone marrow stroma: conditions that maximize the yields of early progenitors and evaluate their quality. Stem Cells. 2002;20:530. doi: 10.1634/stemcells.20-6-530. [DOI] [PubMed] [Google Scholar]
- 11.Ravindran S. Song Y. George A. Development of three-dimensional biomimetic scaffold to study epithelial–mesenchymal interactions. Tissue Eng Part A. 2010;16:327. doi: 10.1089/ten.tea.2009.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He G. Dahl T. Veis A. George A. Nucleation of apatite crystals in vitro by self-assembled dentin matrix protein 1. Nat Mater. 2003;2:552. doi: 10.1038/nmat945. [DOI] [PubMed] [Google Scholar]
- 13.Hennig J. Nauerth A. Friedburg H. RARE Imaging: a fast imaging method for Clinical, M.R. Mag Reson Med. 1986;3:823. doi: 10.1002/mrm.1910030602. [DOI] [PubMed] [Google Scholar]
- 14.Pai A. Li X. Majumdar S. A comparative study at 3T of sequence dependence of T2 quantification in the knee. Mag Reson Imaging. 2008;26:1215. doi: 10.1016/j.mri.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stejskal E.O. Use of spin echoes in a pulsed magnetic-field gradient to study anisotropc, restricted diffusion and flow. J Chem Phys. 1965;43:3597. [Google Scholar]
- 16.Taylor D.G. Bushell M.C. The spatial mapping of translational diffusion coefficients by the NMR imaging technique. Phys Med Biol. 1985;30:345. doi: 10.1088/0031-9155/30/4/009. [DOI] [PubMed] [Google Scholar]
- 17.Xu H.H. Othman S.F. Magin R.L. Monitoring tissue engineering using magnetic resonance imaging. J Biosci Bioeng. 2008;106:515. doi: 10.1263/jbb.106.515. [DOI] [PubMed] [Google Scholar]
- 18.L1 W. Hong L. Hu L. Magin R.L. Magnetization transfer imaging provides a quantitative measure of chondrogenic differentiation and tissue development. Tissue Eng Part C Methods. 2010;16:1407. doi: 10.1089/ten.tec.2009.0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietrich O. Biffar A. Baur-Melnyk Reiser M.F. Technical aspects of MR diffusion imaging of the body. Eur J Radiol. 2010;76:314. doi: 10.1016/j.ejrad.2010.02.018. [DOI] [PubMed] [Google Scholar]
- 20.June R.K. Fyhrie D.P. Molecular N.M. R T2 values can predict cartilage stress relaxation. Biochem Biophys Res Commun. 2008;377:57. doi: 10.1016/j.bbrc.2008.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woll N.L. Heaney J.D. Bronson S.K. Osteogenic nodule formation from single embryonic stem cell-derived progenitors. Stem Cells Dev. 2006;15:865. doi: 10.1089/scd.2006.15.865. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y. Volloch V. Pindrus M.A. Blasioli D.J. Chen J. Kaplan D.L. Murine osteoblasts regulate mesenchymal stem cells via WNT and cadherin pathways: mechanism depends on cell-cell contact mode. J Tissue Eng Regen Med. 2007;1:39. doi: 10.1002/term.6. [DOI] [PubMed] [Google Scholar]
- 23.Coburn C.T. Hajri T. Ibrahimi A. Abumrad N.A. Role of CD36 in membrane transport and utilization of long-chain fatty acids by different tissues. J Mol Neurosci. 2001;16:117. doi: 10.1385/JMN:16:2-3:117. [DOI] [PubMed] [Google Scholar]
- 24.Hino J. Miyazawa T. Miyazato M. Kangawa K. Bone morphogenetic protein-3b (BMP-3b) is expressed in adipocytes and inhibits adipogenesis as a unique complex. Int J Obes. doi: 10.1038/ijo.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood R.J. Vitamin D and adipogenesis: new molecular insights. Nutr Rev. 2008;66:40. doi: 10.1111/j.1753-4887.2007.00004.x. [DOI] [PubMed] [Google Scholar]
- 26.Place E.S. Evans N.D. Stevens M.M. Complexity in biomaterials for tissue engineering. Nat Mater. 2009;8:457. doi: 10.1038/nmat2441. [DOI] [PubMed] [Google Scholar]
- 27.George A. Ravindran S. Protein templates in hard tissue engineering. Nano Today. 2010;5:255. doi: 10.1016/j.nantod.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pek Y.S. Gao S. Arshad M.S. Leck K.J. Ying J.Y. Porous collagen-apatite nanocomposite foams as bone regeneration scaffolds. Biomaterials. 2008;29:4300. doi: 10.1016/j.biomaterials.2008.07.030. [DOI] [PubMed] [Google Scholar]
- 29.Geiger M. Li R.H. Friess W. Collagen sponges for bone regeneration with rhBMP-2. Adv Drug Deliv Rev. 2003;55:1613. doi: 10.1016/j.addr.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y. Jayarama Reddy V. Wong S.Y. Li X. Su B. Ramakrishna S., et al. Enhanced biomineralization in osteoblasts on a novel electrospun biocomposite nanofibrous substrate of hydroxyapatite/collagen/chitosan. Tissue Eng Part A. 2010;16:1949. doi: 10.1089/ten.TEA.2009.0221. [DOI] [PubMed] [Google Scholar]
- 31.Chandy T. Sharma C. Chitosan as a biomaterial. Biomater Art Cells Art Org. 1990;18:1. doi: 10.3109/10731199009117286. [DOI] [PubMed] [Google Scholar]
- 32.Xia W. Liu W. Cui L. Liu Y. Zhong W. Liu D., et al. Tissue engineering of cartilage with the use of chitosan-gelatin complex scaffolds. J Biomed Mater Res B Appl Biomater. 2004;71:373. doi: 10.1002/jbm.b.30087. [DOI] [PubMed] [Google Scholar]
- 33.Wu X. Black L. Santacana-Laffitte G. Patrick C.W. Preparation and assessment of glutaraldehyde-crosslinked collagen-chitosan hydrogels for adipose tissue engineering. J Biomed Mater Res A. 2007;81:59. doi: 10.1002/jbm.a.31003. [DOI] [PubMed] [Google Scholar]
- 34.Kim K.W. Thomas R.L. Lee C. Park H.J. Antimicrobial activity of native chitosan, degraded chitosan, and O-carboxymethylated chitosan. J Food Prot. 2003;66:1495. doi: 10.4315/0362-028x-66.8.1495. [DOI] [PubMed] [Google Scholar]
- 35.Tan W. Krishnaraj R. Desai T.A. Evaluation of nanostructured composite collagen—chitosan matrices for tissue engineering. Tissue Eng. 2001;7:203. doi: 10.1089/107632701300062831. [DOI] [PubMed] [Google Scholar]
- 36.He G. George A. Dentin matrix protein 1 immobilized on type I collagen fibrils facilitates apatite deposition in vitro. J Biol Chem. 2004;279:11649. doi: 10.1074/jbc.M309296200. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y. Bal B.S. Gorski J.P. Calcium and collagen binding properties of osteopontin, bone sialoprotein, and bone acidic glycoprotein-75 from bone. J Biol Chem. 1992;267:24871. [PubMed] [Google Scholar]
- 38.Delany A.M. Hankenson K.D. Thrombospondin-2 and SPARC/osteonectin are critical regulators of bone remodeling. J Cell Commun Signal. 2009;3:227. doi: 10.1007/s12079-009-0076-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.George A. Veis A. Phosphorylated proteins and control over apatite nucleation, crystal growth, and inhibition. Chem Rev. 2008;108:4670. doi: 10.1021/cr0782729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebisawa T. Tada K. Kitajima I. Tojo K. Sampath T.K. Kawabata M. Miyazono K. Imamura T. Characterization of bone morphogenetic protein-6 signaling pathways in osteoblast differentiation. J Cell Sci. 1999;112:3519. doi: 10.1242/jcs.112.20.3519. [DOI] [PubMed] [Google Scholar]
- 41.Cho T.J. Gerstenfeld L.C. Einhorn T.A. Differential temporal expression of members of the transforming growth factor beta superfamily during murine fracture healing. J Bone Miner Res. 2002;17:513. doi: 10.1359/jbmr.2002.17.3.513. [DOI] [PubMed] [Google Scholar]
- 42.Chen L. Jiang W. Huang J. He B.C. Zuo G.W. Zhang W., et al. Insulin-like growth factor 2 (IGF2) potentiates BMP9-induced osteogenic differentiation and bone formation. J Bone Miner Res. 2010;25:2447. doi: 10.1002/jbmr.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu H.H. Song W.X. Luo X. Manning D. Luo J. Deng Z.L., et al. Distinct roles of bone morphogenetic proteins in osteogenic differentiation of mesenchymal stem cells. J Orthop Res. 2007;25:665. doi: 10.1002/jor.20359. [DOI] [PubMed] [Google Scholar]
- 44.Parisuthiman D. Mochida Y. Duarte W.R. Yamauchi M. Biglycan modulates osteoblast differentiation and matrix mineralization. J Bone Miner Res. 2005;20:1878. doi: 10.1359/JBMR.050612. [DOI] [PubMed] [Google Scholar]
- 45.Boskey A.L. Spevak L. Doty S.B. Rosenberg L. Effects of bone CS-proteoglycans, DS-decorin, and DS-biglycan on hydroxyapatite formation in a gelatin gel. Calcif Tissue Int. 1997;61:298. doi: 10.1007/s002239900339. [DOI] [PubMed] [Google Scholar]
- 46.Di Cesare P.E. Fang C. Leslie M.P. Tulli H. Perris R. Carlson C.S. Expression of cartilage oligomeric matrix protein (COMP) by embryonic and adult osteoblasts. J Orthop Res. 2000;18:713. doi: 10.1002/jor.1100180506. [DOI] [PubMed] [Google Scholar]
- 47.Mosig R.A. Dowling O. DiFeo A. Ramirez M.C. Parker I.C. Abe E. et al. Loss of MMP-2 disrupts skeletal and craniofacial development and results in decreased bone mineralization, joint erosion and defects in osteoblast and osteoclast growth. Hum Mol Genet. 2007;16:1113. doi: 10.1093/hmg/ddm060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin E.J. Choi Y.A. Kyun Park E. Bang O.S. Kang S.S. MMP-2 functions as a negative regulator of chondrogenic cell condensation via down-regulation of the FAK-integrin beta1 interaction. Dev Biol. 2007;308:474. doi: 10.1016/j.ydbio.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 49.Banerjee C. McCabe L.R. Choi J.Y. Hiebert S.W. Stein J.L. Stein G.S., et al. Runt homology domain proteins in osteoblast differentiation: AML-3/CBFA1 is a major component of a bone specific complex. J Cell Biochem. 1997;66:1. doi: 10.1002/(sici)1097-4644(19970701)66:1<1::aid-jcb1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 50.Ducy P. Zhang R. Geoffroy V. Ridall A.L. Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 51.Choi J.Y. Pratap J. Javed A. Zaidi S.K. Xing L. Balint E., et al. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc Natl Acad Sci U S A. 2001;98:8650. doi: 10.1073/pnas.151236498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Komori T. Yagi H. Nomura S. Yamaguchi A. Sasaki K. Deguchi K., et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 53.Macías-Silva M. Abdollah S. Hoodless P.A. Pirone R. Attisano L. Wrana J.L. MADR2 is a substrate of the TGFbeta receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87:1215. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 54.Massagué J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Z. Song Y. Zhang X. Tang J. Chen J. Chen Y. Msx1/Bmp4 genetic pathway regulates mammalian alveolar bone formation via induction of Dlx5 and Cbfa1. Mech Dev. 2003;120:1469. doi: 10.1016/j.mod.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 56.Dziedzic-Goclawska A. Toverud S.U. Kaminski A. Boass A. Yamauchi M. Decreased heterotopic osteogenesis in vitamin-D-deficient, but normocalcemic guinea pigs. Bone Miner. 1992;19:127. doi: 10.1016/0169-6009(92)90921-y. [DOI] [PubMed] [Google Scholar]
- 57.Bae H. Zhao L. Zhu D. Kanim L.E. Wang J.C. Delamarter R.B. Variability across ten production lots of a single demineralized bone matrix product. J Bone Joint Surg Am. 2010;92:427. doi: 10.2106/JBJS.H.01400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.