Abstract
Here we describe the role of charged amino acids at the catalytic sites of Escherichia coli ATP synthase. There are four positively charged and four negatively charged residues in the vicinity of of E. coli ATP synthase catalytic sites. Positive charges are contributed by three arginine and one lysine, while negative charges are contributed by two aspartic acid and two glutamic acid residues. Replacement of arginine with a neutral amino acid has been shown to abrogate phosphate binding, while restoration of phosphate binding has been accomplished by insertion of arginine at the same or a nearby location. The number and position of positive charges plays a critical role in the proper and efficient binding of phosphate. However, a cluster of many positive charges inhibits phosphate binding. Moreover, the presence of negatively charged residues seems a requisite for the proper orientation and functioning of positively charged residues in the catalytic sites. This implies that electrostatic interactions between amino acids are an important constituent of initial phosphate binding in the catalytic sites. Significant loss of function in growth and ATPase activity assays in mutants generated through charge modulations has demonstrated that precise location and stereochemical interactions are of paramount importance.
1. Introduction
A typical 70 kg human generates approximately 2.0 million kg of ATP, the cell's energy currency, in a 75-year lifespan by converting food into useable energy by oxidation. ATP is generated by ATP synthase from ADP and inorganic phosphate (Pi) [1, 2]. ATP synthase is not only the essential means of cellular energy production in animals but also in plants and almost all microorganisms. ATP synthase is the final enzyme in the oxidative phosphorylation pathway and is responsible for ATP synthesis by oxidative or photophosphorylation in the membranes of bacteria, mitochondria, and chloroplasts. It is the smallest known biological nanomotor. In order to synthesize ATP, a mechanical rotation mechanism is used where subunits rotate at approximately 100 times per second. Basic [3] functional aspects of ATP synthase remain the same in both prokaryotes and eukaryotes [4].
Membrane bound F1Fo ATP synthase enzyme is structurally identical and highly conserved among different species. ATP hydrolysis and synthesis occur in the F1 sector, whereas proton transport occurs through the membrane embedded Fo [2, 5]. ATP synthesis is the result of proton gradient-driven clockwise rotation of γ (as viewed from the outer membrane), while ATP hydrolysis occurs from anticlockwise rotation of γ-sub unit. Detailed reviews of ATP synthase structure and function may be found in [6–16].
A number of diseases such as Leigh syndrome, ataxia, Batten's diseases, Alzheimer's, angiogenesis, hypertension, cancer, heart disease, mitochondrial diseases, immune deficiency, cystic fibrosis, diabetes, ulcers, and tuberculosis that affect both human and animals have been associated with ATP synthase ([1, 17] and references therein). The presence of ATP synthase on the surfaces of multiple cell types, and its involvement in a number of cellular processes, makes this enzyme an attractive molecular target in the development of treatments for numerous diseases. [18–21]. One particular way in which ATP synthase can be used as a therapeutic target is to inhibit it and thereby deprive abnormal cells of required energy leading to cell death [1, 17, 21, 22].
2. Inhibition of ATP Synthase
A wide range of natural and synthetic products are known to bind and inhibit ATP synthase [1, 17, 23, 24]. Biochemical and structural studies of ATP synthase have so far revealed about ten different inhibitor binding sites. A detailed list of known inhibitors and their actions on ATP synthase are discussed in reference [1, 17]. The inhibitory effects and the extent of inhibition on a molar scale are variable among different inhibitors. Some inhibitors prevent synthesis of ATP but not hydrolysis, or vice versa, while some are known to inhibit both synthesis and hydrolysis equally. Well-known inhibitors of ATP synthase are sodium azide (NaN3), aluminum fluoride (AlFx), scandium fluoride (ScFx), beryllium fluoride (BeFx), dicyclohexylcarbodiimide (DCCD), and 7-chloro- 4-nitrobenzo-2-oxa-1, 3-diazole (NBD-Cl) [11, 24–32]. Less well-known inhibitors of ATP synthase are peptides such as melittin, melittin-related peptide (MRP), ascaphin, aurein, caerin, dermaseptin, magainin II, and polyphenols such as resveratrol, piceatannol, quercetin, morin, and epicatechin [1, 18, 19, 21, 33–35].
The polyphenol piceatannol is one of the most portent inhibitors of ATP synthase [19, 22]. The binding site for polyphenols is at the interface of α-, β-, and γ-subunits of the F1 sector. X-ray structure shows that the following polyphenol binding pocket residues γGln274, γThr-277, βAla-264, βVal-265, γAla-270, γThr-273, γGlu-278, γGly-282, and αGlu-284, are highly conserved among different species and are within 4 Å of the bound polyphenol compounds. Consequently, piceatannol and other inhibitory polyphenols can form both hydrophobic and nonpolar interactions with the above residues [22, 36, 37]. We hypothesize that molecular modulation of both polyphenol-binding pocket residues and polyphenol structures may synergistically affect ATP synthase activity and provide additional clues to catalytic site function.
The βDELSEED-loop of E. coli ATP synthase is known to be the binding site for several basic amphiphilic α-helical peptide inhibitors of ATP synthase. Examples are melittin, melittin-related peptide (MRP), bacterial/chloroplast ATP synthase ε-subunit, and SynA2 (the synthetic derivative of cytochrome oxidase). The α-helical basic peptide, melittin, is composed of 26 residues and is the primary component of honey bee venom (Apis mellifera). MRP is a 23-residue long peptide derived from frog skin (Rana tagoi). Both melittin and MRP are potent inhibitors of ATPase activity of E. coli ATP synthase [1, 21, 38, 39].
Most ATP synthase inhibitory peptides studied so far are from anuran (frogs) sources. These antimicrobial peptides (AMPs) are cationic, between 10 and 50 residues in length, and frequently include a C-terminal amide group [1, 40–42]. Previous mode of action studies indicate that AMPs appear to interact with negatively charged phospholipids and then insert into the bacterial cell membrane or that they may also move across the cell membrane by passive transport and there disrupt a number of cellular processes [43].
Lately, it was observed that some of the antimicrobial effects of amphibian AMPs may be through their inhibitory effects on ATP synthase [1, 21]. Melittin and other peptide inhibitors inhibit ATPase activity in a reversible and noncompetitive fashion [38, 39, 44–47]. It is hypothesized that relatively short antibacterial or anticancer cationic peptides of approximately 10–30 amino acid residues with α-helical secondary structure may inhibit ATP synthase through their binding to the βDELSEED loop. For example, lysine-induced three positive charges of dermaseptin or four positive charges of magainin II interact with the five negative charges of βAsp-380, βGlu-381, βGlu-384, βGlu-385, or βAsp-386 and result in the inhibition of ATPase activity.
Of nearly 60 anuran-derived potential antimicrobial/anticancer peptides, only 13 have been tested for their inhibitory effects on ATP synthase [1, 21]. It was shown that MRP (melittin-related peptide) and MRP-amide strongly inhibited the ATPase activity of ATP synthase and that the presence of an amide group at the c-terminus of MRP caused a ~16% increase in inhibition of ATP synthase ATPase activity. Whether or not negative charges in the βDELSEED motif play any role in the structural stability of the catalytic sites through electrostatic interactions with site residues remains to be seen.
3. Structural and Functional Aspects of Charged Residues in the Catalytic Sites of ATP Synthase
Recent studies have illuminated the role of charged residues in Pi binding at the catalytic sites. Binding of inorganic phosphate (Pi) is an important step in the ATP synthase mechanism which has been extensively studied by biochemical approaches and may be directly coupled to subunit rotation [2, 11, 25, 48–53]. ATP synthase is the terminal enzyme of oxidative phosphorylation and photophosphorylation that synthesizes ATP from ADP and Pi. The energy for ATP synthesis comes from the transmembrane movement of protons down an electrochemical gradient that is generated by substrate oxidation or by light capture. As protons move through the interface between the a and c subunits in the membrane-bound Fo-sector of the enzyme, the free energy is transduced into mechanical rotation of a group of subunits (γεc10-14) which comprise the “rotor"”. The helical coiled coil domain of the γ-subunit projects into the central region of the α 3 β 3 hexagon in the membrane extrinsic F1-sector. The α 3 β 3 hexagon contains three catalytic sites at α/β interfaces. The “Stator” subunits b2 and δ function to prevent co-rotation of α 3 β 3 with the rotor [6, 54–58]. In this paper we present a detailed description of the catalytic site charged amino acids, their role in Pi binding, their effects on the spatial orientation, and effect of their modulation on one another.
Figure 1 represents the simplest form of ~530 kDa Escherichia coli ATP synthase containing eight different subunits, namely, α 3 β 3 γδεab2c10-15, divided into two sectors F1 and Fo. F1 corresponds to α 3 β 3 γδε and Fo to ab2c. Overall, F1Fo-ATP synthase is structurally and functionally similar among sources with only a few exceptions such as in chloroplasts, where there are two isoforms, and in mitochondria, where there are 7–9 additional subunits. ATP hydrolysis and synthesis occur on three catalytic sites in the F1 sector. The γ-subunit is comprised of three α-helices. Two of these helices form a coiled coil that extend into the central space of the α 3 β 3 hexagon. In recent nomenclature, the rotor consists of γεcn, and the stator consists of α 3 β 3 δab2. Current understanding of the F1Fo ATP synthase structure, function, catalytic mechanism, and its role in human health and disease has been thoroughly reviewed by Senior's group and others [1, 2, 7, 8, 17, 20, 59].
Figure 1.
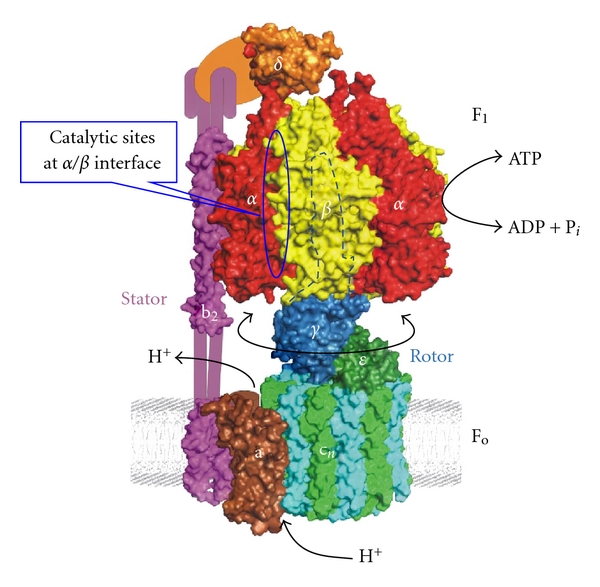
Escherichia coli ATP synthase structure: E. coli ATP synthase enzyme is composed of two sectors, water soluble F1 and membrane bound Fo. Catalytic activity occurs at the interface of αβ/subunits of F1 sector which consists of five subunits (α3β3γδε) and proton conduction occurs at the Fo sector consisting of three subunits (ab2c). One of the catalytic binding sites is identified with circle at the interface of α/β subunits. This model of E. coli ATP synthase is reproduced from Weber [6] with permission; copyright Elsevier.
Based on the binding of ATP, ADP, and Pi, the three catalytic sites located on the F1 sector of ATP synthase are designated βTP, βDP, and βE by X-ray crystallographers [60, 61]. βE is the empty site into which Pi (inorganic phosphate) must initially bind for initiation of ATP synthesis. It has been proposed that the synthesis reaction in the three catalytic sites do not occur independently but in a consecutive manner [51]. In this “binding change mechanism”, the three catalytic sites have different affinities for nucleotides at any given moment. Each catalytic site undergoes conformational transitions that lead to the following sequence: substrate binding (ADP and Pi) → ADP phosphorylation → ATP release. Experimental observations of rotation verified the predication made by Boyer [4, 51, 62] that catalysis requires the sequential participation of the three catalytic sites, with changing affinity for substrates and products, as it proceeds through the recurring mechanism, hence the term “binding change mechanism.” Proton motive force is converted by Fo to a mechanical rotation of the rotor shaft, which drives conformational changes of the catalytic domains of F1 causing synthesis of ATP by phosphorylation of ADP. Conformational changes in the catalytic sites are connected to γ-subunit rotation. γ-Subunit rotation in isolated α 3 β 3 γ subcomplex has been observed directly by Yoshida and Kinosita with colleagues in Japan and subsequently by several other labs [12, 63–68]. The focus of this paper, the role of charged residues at the catalytic sites of E. coli ATP synthase, is a fundamental issue, because catalytic site charged residues are also relevant to many other ATPases, GTPases, and their potential application to nanotechnology and nanomedicine [1, 11, 18, 69].
4. Catalytic and Motor Function of ATP Synthase
Determination of catalytic site Pi-binding residues has allowed a better understanding of the reaction mechanism of ATP synthesis and hydrolysis, and their relationship to the mechanical rotation of the γ-subunit. Characterization of catalytic site-charged residues can provide essential understanding in the following possible applications (1) development of effective modulator (inhibitory/stimulatory) molecules of ATP synthase catalytic function and (2) development of catalytic site mutants for biotechnological applications.
The analysis of charged catalytic sites residues involved in Pi binding has also helped answer the primary question of how the enzyme binds ADP and Pi rather than ATP at the catalytic sites? This is an often overlooked but crucial question in the mechanism of ATP synthesis. In active cells, the cytoplasmic concentrations of ATP and Pi are approximately in the 2–5 mM range, whereas that of ADP is at least 10–50-fold lower. Equilibrium-binding assays have established that both ADP and ATP bind to catalytic sites of purified F1 and detergent solubilized F1 Fo with relatively similar binding affinities [71–74]. Obviously, a specific mechanism must have evolved for selectively binding ADP into catalytic sites while simultaneously preventing ATP binding during proton driven rotation and ATP synthesis. One hypothesis is that during ATP synthesis, the proton gradient-driven rotation of subunits impels an empty catalytic site to bind Pi tightly, thus stereochemically precluding ATP binding and, therefore, selectively favoring ADP binding [7]. A second fundamental question is how does subunit rotation affect Pi binding [49–51]? It was shown that Pi binding appears to be “energy linked”, which entails a linkage to subunit rotation [56, 75, 76]. Therefore, for formulating a mechanism of ATP synthesis, it is of paramount importance to understand the features that determine Pi binding. Moreover, in the near future, it may be possible to use molecular features of Pi binding, derived from mutational and biochemical studies, in the development of potent and novel molecular modulators of ATP synthase.
5. Characterization of Charged Residues at the Catalytic Sites
X-ray structural studies and mutagenic analyses of F1 sector, the catalytic segment of ATP synthase, have so far identified four basic residues critical for catalysis, namely, αβLys-155, βArg-182, Arg-376, and βArg-246. Figure 2 shows the spatial orientation of these amino acids in close proximity to bound phosphate analog AlF4 −. E. coli residue numbers are used throughout. Figure 2 also shows the spatial relationship between positive and negative charged residues.
Figure 2.
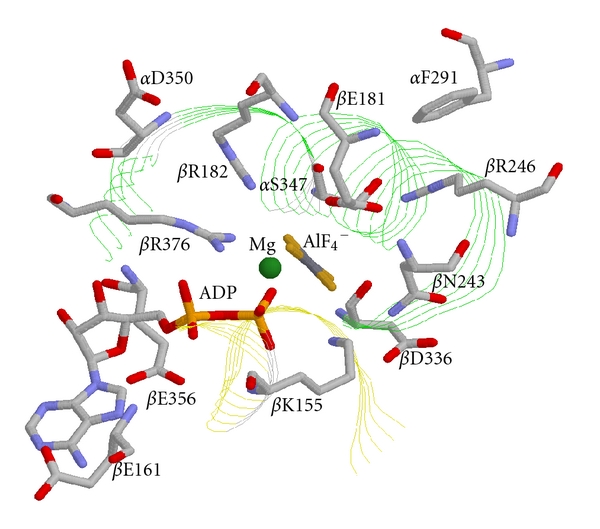
Catalytic sites X-ray structure of ATP synthase showing spatial relationship of charged α and β-subunit residues. The βDP site in the AlF4 −-inhibited enzyme structure is taken from [61]. E. coli residue numbering is used. Four positively and four negatively charged residue in close proximity to the bound phosphate analog AlF4 − are identified. Rasmol software [70] was used to generate this figure using PDB file 1H8E [61].
Initial studies employed the MgATP- and MgADP-binding parameters in mutant enzymes βK155Q, βR182K, βR182Q, βR246A, βR246Q, βR246K, αR376K, and αR376Q. These studies used fluorimetric analysis with introduced βTrp-331 as a specific catalytic site probe, while analysis of the transition state formation was done using MgADP-fluoroaluminate and MgADP-fluoroscandium as transition state analogs [77–81]. Absent from these analyses was a direct measurement of Pi binding in the mutant enzymes. The above residues were clearly demonstrated to be involved in Pi binding with the subsequent application of Pi protection against NBD-Cl inhibition assays [11, 48, 82–85].
The βLys-155 is part of the Walker A sequence in the catalytic sites of ATP synthase. X-ray structures of native F1 with bound MgAMPPNP and MgADP [5], of MgADP-BeFx inhibited F1 [87], of MgADP-AlF4- inhibited F1 representing the transition state [61], and of MgADP-AlF3 inhibited F1 representing the late transition state/early ground state [88] all show the ε-amino group of βLys-155 very close (≤3 Å) to the γ-phosphate position. The βK155Q mutant lacks ATP synthesis and has very low F1-ATPase activity (Table 1). Previous work had shown that βLys-155 plays a major role in binding MgATP, particularly at catalytic sites of high and medium nucleotide affinity, but not in binding MgADP [77]. βLys-155 is also critical for transition state formation [79, 80]. The earlier hypothesis that βLys-155 was important for Pi binding in ATP synthesis [2] was experimentally confirmed by Pi protection against NBD-Cl inhibition where Pi binding in the βE catalytic site is abolished in βK155Q. Therefore, residue βLys-155 is involved at all stages of ATP synthesis from Pi binding, to the transition state, to MgATP formation [84].
Table 1.
ATPase activity of E. coli membrane bound or purified F1 enzymes.
| Mutation a | ATPase activity μmol/min/mg |
|---|---|
| Wild-type | 28.0 (42.0) |
| Null | 0.0013 |
| βK155Q | (0.023) |
| βR182K | (0.250) |
| βR182Q | (0.020) |
| αR376K | (0.120) |
| αR376Q | (0.025) |
| αF291D | 0.07 |
| αF291E | 0.09 |
| βN243A | 0.95 |
| βN243D | 0.033 |
| βR246A | 0.050 (0.25) |
| βR246K | (0.27) |
| βR246Q | (0.27) |
| βN243R | 0.023 |
| βN243R/βR246A | 0.016 |
| αF291R | 0.035 |
| αF291R/βR246A | 0.52 |
| αF291R/βN243R | 0.028 |
aWild-type, pBWU13.4/DK8; Null, pUC118/DK8. All mutants were expressed with the βY331W mutation also present, which does not significantly affect growth. Data are means of four to six experiments each. bMeasured at 37°C and expressed as μmol ATP hydrolyzed/min/mg membrane protein. Each individual experimental point is itself the mean of duplicate assay tubes. Data in parentheses is from purified F1 ATP synthase. Data taken from [48, 82–84, 86].
βArg-182 is another important positively charged phosphate-binding residue in the catalytic sites of ATP synthase [84]. Mutants βR182Q and βR182K lack ATP synthesis activity and have low F1-ATPase activity (Table 1). Residue βArg-182 had been shown to be involved in MgATP binding at the site of highest affinity but not in MgADP binding. Transition state formation is abolished by βR182Q but retained in βR182K [78]. In this regard, it should be noted that βR182K F1 does have somewhat higher ATPase activity (Table 1). βArg-182 was also hypothesized to be required for Pi binding in ATP synthesis [2], and this confirmed that both βR182Q and βR182K mutations abolished Pi binding in the βE site. Therefore, residue βArg-182 is also involved in all stages of ATP synthesis from Pi binding through ATP formation [84].
The αArg-376 residue of E. coli ATP synthase has been described as the “arginine finger” based on G-protein literature and was thought to be a required ligand for the catalytic transition state. Nonetheless, this residue was not shown to be involved in MgATP or MgADP binding despite its apparent proximity to the γ-phosphate of MgAMPPNP in X-ray structures [65, 81]. Movement of this residue in and out of the catalytic site was inferred and was postulated to produce the rate acceleration (“positive catalytic cooperativity”) linked to subunit rotation and full (“tri-site”) catalytic site occupancy that is a hallmark of the mechanism [2]. Significant spatial displacements of residue αArg-376 have been noted in X-ray structures representing different reaction intermediates [5, 33, 61, 75, 87, 88]. Consequently, it was hypothesized that conformational freedom of this residue likely contributes to its importance in catalysis [87]. The previously hypothesized importance of this residue in catalysis [2] was confirmed by Pi protection against NBD-Cl inhibition in which Pi failed to protect αR376Q F1 from NBD-Cl inhibition. However, just as the αR376K mutant was able to form the transition state [81], it was also able to support Pi binding. It is nevertheless strongly impaired in both ATP synthesis and hydrolysis, which suggests that this residue has other required function(s) such as in conformational movements or in H-bonding to other side chains that are specific to Arg and not supported by Lys [11, 84].
βArg-246 is the fourth positively charged residue within the Pi-binding subdomain of catalytic sites identified in the X-ray crystallographic structure (Figure 2) that is involved in Pi binding. βArg-246 is equivalent to βArg-260 in mitochondrial F1 and is conserved among all species. Early random mutagenesis experiments revealed that mutations βR246H and βR246C impaired oxidative phosphorylation drastically and reduced ATPase activity in purified F1 to ~1% of wild type [89, 90]. Further work showed that these mutations caused the unisite catalysis parameter KdPi to change by 4 orders of magnitude, whereas the KdADP was largely unaltered by βR246C, and the ATP hydrolysis reaction equilibrium constant changed to favor ATP over ADP plus Pi [91]. Computer simulations have drawn attention to βArg-246, where movement of the residue during rotation, conformational change of the sites [92], and a role in binding Pi in the transition state were predicted [93, 94]. Site-directed mutagenesis of βR246 to Gln, Lys, and Ala was used to examine the effects of each mutation on function. A variety of inhibitors and ligands known to bind or react in the catalytic sites close to the Pi-binding subdomain were utilized in combination with the mutant enzymes to establish the role of the βArg-246 side chain. Substitutions of the Arg side chain to Gln (removes charge and preserves bulk), to Lys (preserves positive charge), and to Ala (removes side chain and charge) were all examined. All three substitutions severely impeded growth by oxidative phosphorylation and reduced ATPase activity of purified F1 to ~1% of wild type. Finally, as shown in Figure 3 Pi protection against NBD-Cl inhibition clearly demonstrated that βArg-246 residue side chain is an important constituent in binding Pi and in forming the transition state [48].
Figure 3.

Sequence alignment of α and β-subunit residues. α and β-subunit amino acids from different species are aligned. Conserved positively charged residues are shown in blue color, and negatively charged residues are identified by red color. E. coli starting residue for α-subunit is αD336 and for β-subunit is βK155.
Pi binding assays using Pi protection against NBD-Cl were devoid of any nucleotide and enzymes were prepared so as to have all three catalytic sites essentially empty. Therefore, the sites were in βE conformation. In this conformation αArg-376 and βArg-246 lie 2.6 and 4.0 Å from βArg-182, whereas βLys-155 lies 9.5, 7.3, and 6.3 Å from αArg-376, βArg-182, and βArg-246, respectively [88]. In essence, the X-ray structure [87] showed that bound MgADP-BeFx mimicked bound MgATP. In assays of F1-ATPase, it was shown that wild type and αR376Q were fully inhibited by MgADP-BeFx, whereas βK155Q and βR182Q were fully-resistant (Z. Ahmad, and A. E. Senior, unpublished work). These results supported the hypothesis that βLys-155 and βArg-182 are MgATP ligands, but that αArg-376 is not, and that the involvement of stringent stereochemical orientation factors plays a role in determining the functional interactions of αArg-376 [11, 84].
The four positively charged residues form a tetrahedral structure with βLys-155 at the apex and αArg-376, βArg-182, and βArg-246 on the base [11, 25]. A potential Pi-binding pocket can readily be envisaged at the center of this tetrahedron (see Figure 2). In ATP synthesis, the βE site will change to the βADP + Pi (“half-closed”) site in association with γ-rotation [2, 61]. The X-ray structure of this conformation [61] shows that the residues αArg-376, βLys-155, and βArg-182 are each located ≤3.0 Å from the nearest oxygen atom of bound SO4 2-) anion (modeling Pi), whereas βArg-246 is 4.5 Å from the sulfate. Thus, as the reaction proceeds, the three residues αArg-376, βLys-155 and βArg-182 close around the Pi and move it away from βArg-246 toward the site of transition state formation [11, 12, 48, 95].
The above results supported the following proposed molecular mechanism for ATP synthesis [2]. Initially, substrate Pi binds in the βE catalytic site using four basic residues as ligands, namely, αArg-376, βArg-182, βLys-155, and βArg-246 [11, 25, 48, 82–84, 86]. After binding of MgADP (in which these four residues are not involved), the catalytic transition state forms using αArg-376, βArg-182, and βLys-155 as direct ligands. Upon formation of MgATP, αArg-376 withdraws and no longer interacts, whereas βLys-155 and βArg-182 are still bound to the γ-phosphate. MgATP is released to the medium with the breaking of these bonds.
Historically, many attempts to measure Pi binding in purified E. coli F1 using [32P] Pi [50] or by competition with ATP or AMP-PNP in fluorescence assays of nucleotide binding [72, 96] failed to detect appreciable Pi binding at physiological Pi concentration. An assay devised by Perez et al. [97] in which the protection afforded by Pi against the inhibition of ATPase activity was induced by covalent reaction with 7-chloro-4-nitrobenzo-2-oxa-1, 3,-diazole (NBD-Cl) provided a measure of Pi binding. Orriss et al. [98] showed by X-ray crystallography that the covalent adduct formed by NBD-Cl is specifically in the βE catalytic site. Hence, protection afforded by Pi indicates that binding of Pi occurs at the βE catalytic site. By modifying the above assay for use with E. coli purified F1 or membrane bound F1 Fo, further studies have to date investigated the relationship between Pi binding and catalysis for eight residues, namely αPhe-291, αSer-347, αGly-351, αArg-376, βLys-155, βArg-182, βAsn-243, and βArg-246. It was shown that the five residues αSer-347, αArg-376, βLys-155, βArg-182, and βArg-246 were grouped in a tetrahedral relationship,and are involved in Pi binding. The other three residues αPhe-291, αGly-351, and βAsn-243 are not involved in Pi binding [11, 25, 48, 82–84, 86]. In consequence, the presence of positively charged residues in the catalytic site explains the preferential binding of ADP over ATP.
It may be noted that [32P]Pi binding was detected with a Kd(Pi) in the range of 0.1 mM using an alternative, pressure ultrafiltration method, and this result is consistent with data obtained from the NBD-Cl protection assay [99]. It is apparent that Pi dissociates more rapidly from E. coli F1 than it does from mitochondrial F1, undesirably, rendering the convenient centrifuge assay incompatible with the E. coli enzyme.
The story goes on with the presence of many charged/uncharged residues in close proximity to the Pi binding subdomain in the catalytic sites. These residues have been shown to exert either a direct or an indirect role in Pi binding. One of these residues is the charged αAsp-350 of the VISIT-DG sequence. αAsp-350 is part of the α-subunit VISIT-DG sequence, which is a highly conserved motif in this enzyme [25]. VISIT-DG sequence residues are of special interest in general and negatively charged αAsp-350 in particular, because of the close proximity to the known positively charged phosphate-binding residues. αAsp-350 is ~3 Å from αArg-376. It would be imperative to understand three specific aspects of αAsp-350 residue. First, is residue Asp-350 directly or indirectly involved in phosphate binding through αArg-376? Second, is αAsp-350 important for function through its role in maintaining the structural integrity of the Pi binding subdomain but not involved in Pi binding per se? Third, is the carboxyl side chain of αAsp-350 involved in the transition state at the catalytic site? Our hypothesis is that αAsp-350 interacts electrostatically with αR376. Such an interaction may provide proper orientation of αArg-376 side chain towards Pi.
6. Modulation of Charge in the Catalytic Sites
Understanding the role of charged residues and the effects of modifying them is important in understanding the molecular mechanism of ATP synthesis. NBD-Cl inactivation assay described earlier have shown that positively charged residues are functionally essential for Pi binding in the βE catalytic site of E. coli ATP synthase [11, 48, 83, 84]. The introduction of charged residues in place of uncharged residues in the vicinity of catalytically important residues has been shown to affect Pi binding by resulting in a loss or gain of ATPase activity [82, 83, 86]. Earlier work [82, 83] indicated that the introduction of negative charge in the Pi-binding pocket in the form of βN243D close to βArg-246 prevented Pi binding (Figure 5). Also, introduction of positive charge in the form of βN243R restored Pi binding in βR246A mutants. Similarly, the introduction of negative or positive charge in the form of αF291D/E/R with βR246 or βR246A resulted in a loss or gain of Pi binding (Figures 4 and 5) [83, 86]. These results suggested that modulation of charge in the Pi binding site could be used to understand the molecular mechanism of Pi binding. It is established that Arg residues occur particularly commonly in the Pi binding sites of proteins [100]. Therefore, varying the number of Arg residues in the Pi binding site of ATP synthase can be an instructive approach.
Figure 5.
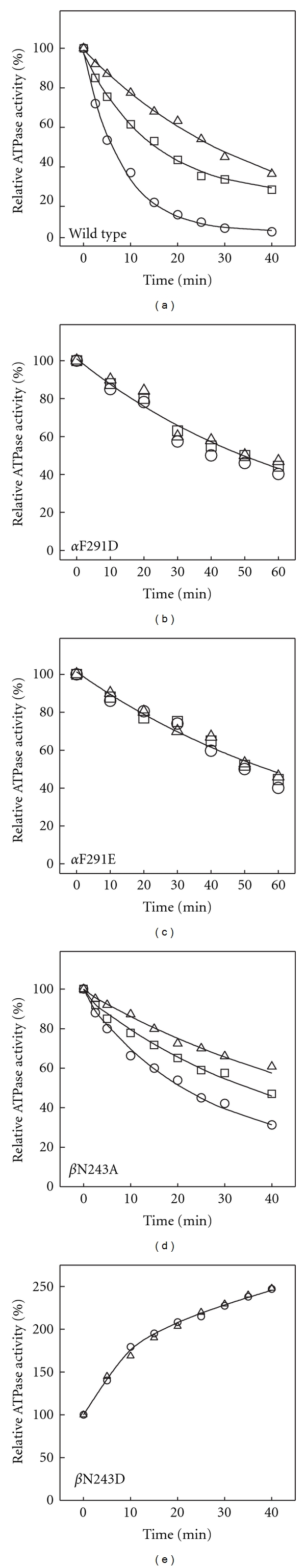
Loss of Pi protection from inactivation by NBD-Cl inhibition with inserted negative charge. Membranes were preincubated with Pi at zero, 2.5 or 10 mM concentration as shown, for 60 min at 23°C. Then, NBD-Cl (125 μM) was added and aliquots withdrawn for assay at time intervals as shown. ATPase activity remaining is plotted against time of incubation with NBD-Cl. ○, no Pi added; □, 2.5 mM Pi; Δ, 10 mM Pi. Data taken from [82, 86].
Figure 4.
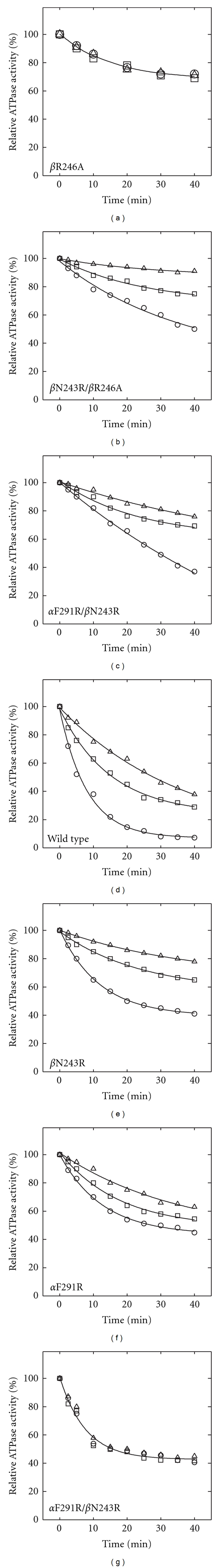
Compensatory effect inserted arginine residue. Membranes were preincubated with Pi at zero, 2.5 or 10 mM concentration as shown, for 60 min at 23°C. Then, NBD-Cl (125 μM) was added and aliquots withdrawn for assay at time intervals as shown. ATPase activity remaining is plotted against time of incubation with NBD-Cl. ○, no Pi added; □, 2.5 mM Pi; Δ, 10 mM Pi. Data taken from [83].
Residue βAsn-243 lies 3.2 Å from βArg-246 in both AlF3 and SO4 2−)-containing catalytic sites (nearest atom distances quoted) [83]. One experimental approach used was to introduce the mutation βN243R in a wild-type background (with βArg-246) and in presence of the βR246A mutation. Residue αPhe-291, located at the end of the Pi-binding pocket across the catalytic α/β interface, with its side-chain pointing toward the bound Pi analogs, is also a suitable location for introducing a new Arg. This residue lies at a distance of 3.2 Å from βArg-246 in the AlF3-containing catalytic site and 7.5 Å in the SO4 2−-containing catalytic site [61, 83, 88]. Arginine was introduced in the form of an αF291R mutation in the wild-type background and in the presence of the βR246A mutation. The actual distances of residues βArg-246, βAsn-243 and αPhe-291 were obtained from bound AlF3 and SO4 2− as determined by X-ray crystallography [61, 88], while speculative distances (in brackets) were calculated for mutant residues βAla-246, βArg-243 and αArg-291 using the “Deep View Swiss-Pdb Viewer” [101]. Apparently, mutations placed extra positive charge relatively close to Pi, and the βAla-246 mutation left a relatively large space into which a new Arg fits nicely.
7. Synergistic Stereochemical Interactions at the Catalytic Sites
The introduction of one or two extra positively charged Arg residues in the wild-type background at either β-243 or α-291, or both, has proven to inhibit Pi binding (Figure 4). Introduction of a new Arg at β-243 or α-291 in the βR246A background provided a significant compensatory effect on ATPase. ATP-driven proton pumping was also reinstated in the case of the αF291R/βR246A mutant. But these new arginines did not restore function to full normal [83].
The βR246A mutant did not show Pi binding, but both βN243R and αF291R mutations “rescued” Pi binding in combination with βAla-246 (Figure 4). Since neither βArg-243 nor αPhe-291 could be expected to assume the exact same stereochemical interactions that βArg-246 achieves, electrostatic interaction appears to be a significant factor. Presence of at least one positive charge at this general location is a requisite determinant of initial Pi binding in the catalytic site βE. βN243R or αF291R in the wild-type background (representing one extra positive charge) did not prevent Pi binding, but the combination of αF291R/βN243R (two extra charges) abrogated Pi binding (Figure 4). Presumably the local concentration of charge in the latter becomes too disruptive and distorts the Pi-binding site [83].
A similar pattern of effects has been reported when transition state stabilization was assessed by assaying inhibition of ATPase activity by the transition state analogs MgADP-fluoroaluminate and MgADP-fluoroscandium. Previously, it was shown that [48] that both inhibitors are potent against wild-type ATP synthase but that each inhibit βR246A mutant only to small extent, which indicates that βArg-246 is intimately involved in transition state stabilization. It was found that either mutant residue βArg-243 or αArg-291 partly “rescued” transition state stabilization when present with βAla-246 [83]. Raising the number of positively charged residues to two (βN243R and αF291R mutants in wild-type background) had an adverse effect as reflected by a lesser inhibition of ATPase activity. Raising the number of local positive charges to three reduced transition state stabilization right back to where it was in βR246A. Interestingly, even in the best cases among the mutants (βN243R/βR246A or αF291R/βR246A) transition state stabilization was incomplete as compared to wild-type, which suggests the importance of stereochemical interactions [83].
In summary, all the results showed that Pi binding is notably affected by the local positive charges in catalytic site βE of ATP synthase. Positive charge in the vicinity of the natural βArg-246 is important because its removal in βR246A mutant can be compensated partially by introduction of one Arg at either β-243 or α-291. Thus, electrostatic interaction is an important determinant of Pi binding. The presence of two arginines by introduction of either βArg-243 or αArg-291 in the presence of βArg-246 does not prevent Pi binding, but the presence of all three arginines eliminates Pi binding. Effects on transition state stabilization followed a parallel pattern, but the restoration of Pi binding in βE catalytic sites by charge compensation was not sufficient by itself to restore full function [83].
Acknowledgment
This work was supported by the National Institutes of Health Grant no. GM085771 to Z. Ahmad.
References
- 1.Ahmad Z, Laughlin TF. Medicinal chemistry of ATP synthase: a potential drug target of dietary polyphenols and amphibian antimicrobial peptides. Current Medicinal Chemistry. 2010;17(25):2822–2836. doi: 10.2174/092986710791859270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Senior AE, Nadanaciva S, Weber J. The molecular mechanism of ATP synthesis by FF-ATP synthase. Biochimica et Biophysica Acta. 2002;1553(3):188–211. doi: 10.1016/s0005-2728(02)00185-8. [DOI] [PubMed] [Google Scholar]
- 3.Frasch WD. The participation of metals in the mechanism of the F-ATPase. Biochimica et Biophysica Acta. 2000;1458(2-3):310–325. doi: 10.1016/s0005-2728(00)00083-9. [DOI] [PubMed] [Google Scholar]
- 4.Boyer PD. The ATP synthase—a splendid molecular machine. Annual Review of Biochemistry. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 5.Abrahams JP, Leslie AGW, Lutter R, Walker JE. Structure at 2.8 Å resolution of F-ATPase from bovine heart mitochondria. Nature. 1994;370(6491):621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 6.Weber J. ATP synthase: subunit-subunit interactions in the stator stalk. Biochimica et Biophysica Acta. 2006;1757(9-10):1162–1170. doi: 10.1016/j.bbabio.2006.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weber J, Senior AE. ATP synthase: what we know about ATP hydrolysis and what we do not know about ATP synthesis. Biochimica et Biophysica Acta. 2000;1458(2-3):300–309. doi: 10.1016/s0005-2728(00)00082-7. [DOI] [PubMed] [Google Scholar]
- 8.Senior AE, Nadanaciva S, Weber J. Rate acceleration of ATP hydrolysis by FF(o)-ATP synthase. Journal of Experimental Biology. 2000;203(1):35–40. doi: 10.1242/jeb.203.1.35. [DOI] [PubMed] [Google Scholar]
- 9.Nakamoto RK, Ketchum CJ, al-Shawi MK. Rotational coupling in the FF ATP synthase. Annual Review of Biophysics and Biomolecular Structure. 1999;28:205–234. doi: 10.1146/annurev.biophys.28.1.205. [DOI] [PubMed] [Google Scholar]
- 10.Pedersen PL. Transport ATPases into the year 2008: a brief overview related to types, structures, functions and roles in health and disease. Journal of Bioenergetics and Biomembranes. 2007;39(5-6):349–355. doi: 10.1007/s10863-007-9123-9. [DOI] [PubMed] [Google Scholar]
- 11.Ahmad Z, Senior AE. Identification of phosphate binding residues of Escherichia coli ATP synthase. Journal of Bioenergetics and Biomembranes. 2005;37(6):437–440. doi: 10.1007/s10863-005-9486-8. [DOI] [PubMed] [Google Scholar]
- 12.Senior AE. ATP synthase: motoring to the finish line. Cell. 2007;130(2):220–221. doi: 10.1016/j.cell.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Khan S. Rotary chemiosmotic machines. Biochimica et Biophysica Acta. 1997;1322(2-3):86–105. doi: 10.1016/s0005-2728(97)00075-3. [DOI] [PubMed] [Google Scholar]
- 14.Noji H, Yoshida M. The rotary machine in the cell, ATP synthase. Journal of Biological Chemistry. 2001;276(3):1665–1668. doi: 10.1074/jbc.R000021200. [DOI] [PubMed] [Google Scholar]
- 15.Ren H, Allison WS. On what makes the γ subunit spin during ATP hydrolysis by F. Biochimica et Biophysica Acta. 2000;1458(2-3):221–233. doi: 10.1016/s0005-2728(00)00075-x. [DOI] [PubMed] [Google Scholar]
- 16.Weber J, Senior AE. ATP synthesis driven by proton transport in FF-ATP synthase. FEBS Letters. 2003;545(1):61–70. doi: 10.1016/s0014-5793(03)00394-6. [DOI] [PubMed] [Google Scholar]
- 17.Hong S, Pedersen PL. ATP synthase and the actions of inhibitors utilized to study its roles in human health, disease, and other scientific areas. Microbiology and Molecular Biology Reviews. 2008;72(4):590–641. doi: 10.1128/MMBR.00016-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chinnam N, Dadi PK, Sabri SA, Ahmad M, Kabir MA, Ahmad Z. Dietary bioflavonoids inhibit Escherichia coli ATP synthase in a differential manner. International Journal of Biological Macromolecules. 2010;46(5):478–486. doi: 10.1016/j.ijbiomac.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dadi PK, Ahmad M, Ahmad Z. Inhibition of ATPase activity of Escherichia coli ATP synthase by polyphenols. International Journal of Biological Macromolecules. 2009;45(1):72–79. doi: 10.1016/j.ijbiomac.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Pedersen PL. The cancer cell’s “power plants” as promising therapeutic targets: an overview. Journal of Bioenergetics and Biomembranes. 2007;39(1):1–12. doi: 10.1007/s10863-007-9070-5. [DOI] [PubMed] [Google Scholar]
- 21.Laughlin TF, Ahmad Z. Inhibition of Escherichia coli ATP synthase by amphibian antimicrobial peptides. International Journal of Biological Macromolecules. 2010;46(3):367–374. doi: 10.1016/j.ijbiomac.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gledhill JR, Montgomery MG, Leslie AG, Walker JE. Mechanism of inhibition of bovine F-ATPase by resveratrol and related polyphenols. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(34):13632–13637. doi: 10.1073/pnas.0706290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng J, Ramirez VD. Inhibition of mitochondrial proton F0F1-ATPase/ATP synthase by polyphenolic phytochemicals. British Journal of Pharmacology. 2000;130(5):1115–1123. doi: 10.1038/sj.bjp.0703397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber J, Senior AE. Effects of the inhibitors azide, dicyclohexylcarbodiimide, and aurovertin on nucleotide binding to the three F-ATPase catalytic sites measured using specific tryptophan probes. Journal of Biological Chemistry. 1998;273(50):33210–33215. doi: 10.1074/jbc.273.50.33210. [DOI] [PubMed] [Google Scholar]
- 25.Li W, Brudecki LE, Senior AE, Ahmad Z. Role of α-subunit VISIT-DG sequence residues Ser-347 and Gly-351 in the catalytic sites of Escherichia coli ATP synthase. Journal of Biological Chemistry. 2009;284(16):10747–10754. doi: 10.1074/jbc.M809209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowler MW, Montgomery MG, Leslie AG, Walker JE. How azide inhibits ATP hydrolysis by the F-ATPases. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(23):8646–8649. doi: 10.1073/pnas.0602915103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frasch AC, Cazzulo JJ, Stoppani AO. Solubilization and some properties of the Mg-activated adenosine triphosphatase from Trypanosoma cruzi. Comparative Biochemistry and Physiology B. 1978;61(2):207–212. doi: 10.1016/0305-0491(78)90162-1. [DOI] [PubMed] [Google Scholar]
- 28.Hermolin J, Fillingame RH. H-ATPase activity of Escherichia coli FF is blocked after reaction of dicyclohexylcarbodiimide with a single proteolipid (subunit c) of the F complex. Journal of Biological Chemistry. 1989;264(7):3896–3903. [PubMed] [Google Scholar]
- 29.Tommasino M, Capaldi RA. Effect of dicyclohexylcarbodiimide on unisite and multisite catalytic activities of the adenosinetriphosphatase of Escherichia coli . Biochemistry. 1985;24(15):3972–3976. doi: 10.1021/bi00336a026. [DOI] [PubMed] [Google Scholar]
- 30.Yarlett N, Lloyd D. Effects of inhibitors on mitochondrial adenosine triphosphatase of Crithidia fasciculata: an unusual pattern of specificities. Molecular and Biochemical Parasitology. 1981;3(1):13–17. doi: 10.1016/0166-6851(81)90073-6. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida M, Allison WS, Esch FS, Futai M. The specificity of carboxyl group modification during the inactivation of the Escherichia coli F-ATPase with dicyclohexyl[C]carbodiimide. Journal of Biological Chemistry. 1982;257(17):10033–10037. [PubMed] [Google Scholar]
- 32.Zharova TV, Vinogradov AD. Energy-dependent Transformation of F0.F1-ATPase in Paracoccus denitrificans Plasma Membranes. Journal of Biological Chemistry. 2004;279(13):12319–12324. doi: 10.1074/jbc.M311397200. [DOI] [PubMed] [Google Scholar]
- 33.Abrahams JP, Buchanan SK, Van Raaij MJ, Fearnley IM, Leslie AG, Walker JE. The structure of bovine F-ATPase complexed with the peptide antibiotic efrapeptin. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(18):9420–9424. doi: 10.1073/pnas.93.18.9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gledhill JR, Montgomery MG, Leslie AG, Walker JE. How the regulatory protein, IF, inhibits F-ATPase from bovine mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(40):15671–15676. doi: 10.1073/pnas.0707326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lardy H, Reed P, Lin CH. Antibiotic inhibitors of mitochondrial ATP synthesis. Federation Proceedings. 1975;34(8):1707–1710. [PubMed] [Google Scholar]
- 36.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385(6617):602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 37.Walker EH, Pacold ME, Perisic O, et al. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Molecular Cell. 2000;6(4):909–919. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- 38.Bullough DA, Ceccarelli EA, Roise D, Allison WS. Inhibition of the bovine-heart mitochondrial F-ATPase by cationic dyes and amphipathic peptides. Biochimica et Biophysica Acta. 1989;975(3):377–383. doi: 10.1016/s0005-2728(89)80346-9. [DOI] [PubMed] [Google Scholar]
- 39.Gledhill JR, Walker JE. Inhibition in F-ATPase from bovine heart mitochondria. Biochemical Journal. 2005;386(3):591–598. doi: 10.1042/BJ20041513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z, Wang G. APD: the antimicrobial peptide database. Nucleic Acids Research. 2004;32:D590–D592. doi: 10.1093/nar/gkh025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zasloff M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(15):5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415(6870):389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 43.Jakubke H-D, Sewald N. Peptides from A to Z: A Concise Encyclopedia. Weinheim, Germany: Wiley-VCH; 2008. [Google Scholar]
- 44.Iwadate M, Asakura T, Williamson MP. The structure of the melittin tetramer at different temperatures: an NOE-based calculation with chemical shift refinement. European Journal of Biochemistry. 1998;257(2):479–487. doi: 10.1046/j.1432-1327.1998.2570479.x. [DOI] [PubMed] [Google Scholar]
- 45.Roise D, Horvath SJ, Tomich JM, Richards JH, Schatz G. A chemically synthesized pre-sequence of an imported mitochondrial protein can form an amphiphilic helix and perturb natural and artificial phospholipid bilayers. The EMBO journal. 1986;5(6):1327–1334. doi: 10.1002/j.1460-2075.1986.tb04363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roise D, Theiler F, Horvath SJ, et al. Amphiphilicity is essential for mitochondrial presequence function. The EMBO Journal. 1988;7(3):649–653. doi: 10.1002/j.1460-2075.1988.tb02859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terwilliger TC, Eisenberg D. The structure of melittin. I. Structure determination and partial refinement. Journal of Biological Chemistry. 1982;257(11):6010–6015. doi: 10.2210/pdb1mlt/pdb. [DOI] [PubMed] [Google Scholar]
- 48.Ahmad Z, Senior AE. Mutagenesis of residue βArg-246 in the phosphate-binding subdomain of catalytic sites of Escherichia coli F-ATPase. Journal of Biological Chemistry. 2004;279(30):31505–31513. doi: 10.1074/jbc.M404621200. [DOI] [PubMed] [Google Scholar]
- 49.Al-Shawi MK, Ketchum CJ, Nakamoto RK. The Escherichia coli F0F1 γM23K uncoupling mutant has a higher K0.5 for Pi. Transition state analysis of this mutant and others reveals that synthesis and hydrolysis utilize the same kinetic pathway. Biochemistry. 1997;36(42):12961–12969. doi: 10.1021/bi971478r. [DOI] [PubMed] [Google Scholar]
- 50.al-Shawi MK, Senior AE. Effects of Dimethyl Sulfoxide on catalysis in Escherichia coli F-ATPaSe. Biochemistry. 1992;31(3):886–891. doi: 10.1021/bi00118a034. [DOI] [PubMed] [Google Scholar]
- 51.Boyer PD. A perspective of the binding change mechanism for ATP synthesis. The FASEB Journal. 1989;3(10):2164–2178. doi: 10.1096/fasebj.3.10.2526771. [DOI] [PubMed] [Google Scholar]
- 52.Masaike T, Muneyuki E, Noji H, Kinosita K, Yoshida M. F-ATPase changes its conformations upon phosphate release. Journal of Biological Chemistry. 2002;277(24):21643–21649. doi: 10.1074/jbc.M110297200. [DOI] [PubMed] [Google Scholar]
- 53.Rosing J, Kayalar C, Boyer PD. Evidence for energy dependent change in phosphate binding for mitochondrial oxidative phosphorylation based on measurements of medium and intermediate phosphate water exchanges. Journal of Biological Chemistry. 1977;252(8):2478–2485. [PubMed] [Google Scholar]
- 54.Devenish RJ, Prescott M, Roucou X, Nagley P. Insights into ATP synthase assembly and function through the molecular genetic manipulation of subunits of the yeast mitochondrial enzyme complex. Biochimica et Biophysica Acta. 2000;1458(2-3):428–442. doi: 10.1016/s0005-2728(00)00092-x. [DOI] [PubMed] [Google Scholar]
- 55.Karrasch S, Walker JE. Novel features in the structure of bovine ATP synthase. Journal of Molecular Biology. 1999;290(2):379–384. doi: 10.1006/jmbi.1999.2897. [DOI] [PubMed] [Google Scholar]
- 56.Senior AE. ATP synthesis by oxidative phosphorylation. Physiological Reviews. 1988;68(1):177–231. doi: 10.1152/physrev.1988.68.1.177. [DOI] [PubMed] [Google Scholar]
- 57.Diez M, Zimmermann B, Borsch M, et al. Proton-powered subunit rotation in single membrane-bound F F-ATP synthase. Nature Structural and Molecular Biology. 2004;11(2):135–141. doi: 10.1038/nsmb718. [DOI] [PubMed] [Google Scholar]
- 58.Itoh H, Takahashi A, Adachi K, et al. Mechanically driven ATP synthesis by F-ATPase. Nature. 2004;427(6973):465–468. doi: 10.1038/nature02212. [DOI] [PubMed] [Google Scholar]
- 59.Pedersen SS, Smith OR, De Vries J, Appels A, Denollet J. Course of anxiety symptoms over an 18-month period in exhausted patients post percutaneous coronary intervention. Psychosomatic Medicine. 2008;70(3):349–355. doi: 10.1097/PSY.0b013e3181656540. [DOI] [PubMed] [Google Scholar]
- 60.Leslie AG, Walker JE. Structural model of F-ATPase and the implications for rotary catalysis. Philosophical Transactions of the Royal Society B. 2000;355(1396):465–471. doi: 10.1098/rstb.2000.0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Menz RI, Walker JE, Leslie AG. Structure of bovine mitochondrial F-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell. 2001;106(3):331–341. doi: 10.1016/s0092-8674(01)00452-4. [DOI] [PubMed] [Google Scholar]
- 62.Boyer PD. A research journey with ATP synthase. Journal of Biological Chemistry. 2002;277(42):39045–39061. doi: 10.1074/jbc.X200001200. [DOI] [PubMed] [Google Scholar]
- 63.Kinosita K, Jr., Yasuda R, Noji H, Ishiwata S, Yoshida M. F-ATPase: a rotary motor made of a single molecule. Cell. 1998;93(1):21–24. doi: 10.1016/s0092-8674(00)81142-3. [DOI] [PubMed] [Google Scholar]
- 64.Nishizaka T, Oiwa K, Noji H, et al. Chemomechanical coupling in F-ATPase revealed by simultaneous observation of nucleotide kinetics and rotation. Nature Structural and Molecular Biology. 2004;11(2):142–148. doi: 10.1038/nsmb721. [DOI] [PubMed] [Google Scholar]
- 65.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr. Direct observation of the rotation of F-ATPase. Nature. 1997;386(6622):299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 66.Senior AE, Weber J. Happy motoring with ATP synthase. Nature Structural and Molecular Biology. 2004;11(2):110–112. doi: 10.1038/nsmb0204-110. [DOI] [PubMed] [Google Scholar]
- 67.Yasuda R, Noji H, Kinosita K, Jr., Yoshida M. F-ATPase is a highly efficient molecular motor that rotates with discrete 120°steps. Cell. 1998;93(7):1117–1124. doi: 10.1016/s0092-8674(00)81456-7. [DOI] [PubMed] [Google Scholar]
- 68.Yoshida M, Muneyuki E, Hisabori T. ATP synthase—a marvellous rotary engine of the cell. Nature Reviews Molecular Cell Biology. 2001;2(9):669–677. doi: 10.1038/35089509. [DOI] [PubMed] [Google Scholar]
- 69.Whitesides GM. The ‘right’ size in nanobiotechnology. Nature Biotechnology. 2003;21(10):1161–1165. doi: 10.1038/nbt872. [DOI] [PubMed] [Google Scholar]
- 70.Sayle RA, Milner-White EJ. RASMOL: biomolecular graphics for all. Trends in Biochemical Sciences. 1995;20(9):374–376. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
- 71.Dou C, Fortes PA, Allison WS. The α(βYW)γ subcomplex of the F-ATpase from the thermophilic Bacillus PS3 fails to dissociate ADP when MgATP is hydrolyzed at a single catalytic site and attains maximal velocity when three catalytic sites are saturated with MgATP. Biochemistry. 1998;37(47):16757–16764. doi: 10.1021/bi981717q. [DOI] [PubMed] [Google Scholar]
- 72.Lobau S, Weber J, Senior AE. Catalytic Site Nucleotide Binding and Hydrolysis in F1Fo-ATP Synthase. Biochemistry. 1998;37(30):10846–10853. doi: 10.1021/bi9807153. [DOI] [PubMed] [Google Scholar]
- 73.Weber J, Hammond ST, Wilke-Mounts S, Senior AE. Mg coordination in catalytic sites of F-ATPase. Biochemistry. 1998;37(2):608–614. doi: 10.1021/bi972370e. [DOI] [PubMed] [Google Scholar]
- 74.Weber J, Wilke-Mounts S, Lee RS, Grell E, Senior AE. Specific placement of tryptophan in the catalytic sites of Escherichia coli F-ATPase provides a direct probe of nucleotide binding: maximal ATP hydrolysis occurs with three sites occupied. Journal of Biological Chemistry. 1993;268(27):20126–20133. [PubMed] [Google Scholar]
- 75.Gibbons C, Montgomery MG, Leslie AG, Walker JE. The structure of the central stalk in bovine F-ATPase at 2.4 Å resolution. Nature Structural Biology. 2000;7(11):1055–1061. doi: 10.1038/80981. [DOI] [PubMed] [Google Scholar]
- 76.Rastogi VK, Girvin ME. Structural changes linked to proton translocation by subunit c of the ATP synthase. Nature. 1999;402(6759):263–268. doi: 10.1038/46224. [DOI] [PubMed] [Google Scholar]
- 77.Lobau S, Weber J, Wilke-Mounts S, Senior AE. F-ATPase, roles of three catalytic site residues. Journal of Biological Chemistry. 1997;272(6):3648–3656. doi: 10.1074/jbc.272.6.3648. [DOI] [PubMed] [Google Scholar]
- 78.Nadanaciva S, Weber J, Senior AE. The role of β-Arg-182, an essential catalytic site residue in Escherichia coli F-ATPase. Biochemistry. 1999;38(24):7670–7677. doi: 10.1021/bi990663x. [DOI] [PubMed] [Google Scholar]
- 79.Nadanaciva S, Weber J, Senior AE. Binding of the transition state analog MgADP-fluoroaluminate to F- ATPase. Journal of Biological Chemistry. 1999;274(11):7052–7058. doi: 10.1074/jbc.274.11.7052. [DOI] [PubMed] [Google Scholar]
- 80.Nadanaciva S, Weber J, Senior AE. New probes of the F-ATPase catalytic transition state reveal that two of the three catalytic sites can assume a transition state conformation simultaneously. Biochemistry. 2000;39(31):9583–9590. doi: 10.1021/bi000941o. [DOI] [PubMed] [Google Scholar]
- 81.Nadanaciva S, Weber J, Wilke-Mounts S, Senior AE. Importance of F-ATPase residue α-Arg-376 for catalytic transition state stabilization. Biochemistry. 1999;38(47):15493–15499. doi: 10.1021/bi9917683. [DOI] [PubMed] [Google Scholar]
- 82.Ahmad Z, Senior AE. Role of βAsn-243 in the phosphate-binding subdomain of catalytic sites of Escherichia coli F-ATPase. Journal of Biological Chemistry. 2004;279(44):46057–46064. doi: 10.1074/jbc.M407608200. [DOI] [PubMed] [Google Scholar]
- 83.Ahmad Z, Senior AE. Modulation of charge in the phosphate binding site of Escherichia coli ATP synthase. Journal of Biological Chemistry. 2005;280(30):27981–27989. doi: 10.1074/jbc.M503955200. [DOI] [PubMed] [Google Scholar]
- 84.Ahmad Z, Senior AE. Involvement of ATP synthase residues αArg-376, βArg-182, and βLys-155 in Pi binding. The FEBS Letters. 2005;579(2):523–528. doi: 10.1016/j.febslet.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 85.Ahmad Z, Senior AE. Inhibition of the ATPase activity of Escherichia coli ATP synthase by magnesium fluoride. The FEBS Letters. 2006;580(2):517–520. doi: 10.1016/j.febslet.2005.12.057. [DOI] [PubMed] [Google Scholar]
- 86.Brudecki LE, Grindstaff JJ, Ahmad Z. Role of αPhe-291 residue in the phosphate-binding subdomain of catalytic sites of Escherichia coli ATP synthase. Archives of Biochemistry and Biophysics. 2008;471(2):168–175. doi: 10.1016/j.abb.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 87.Kagawa R, Montgomery MG, Braig K, Leslie AG, Walker JE. The structure of bovine F-ATPase inhibited by ADP and beryllium fluoride. EMBO Journal. 2004;23(14):2734–2744. doi: 10.1038/sj.emboj.7600293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Braig K, Menz RI, Montgomery MG, Leslie AG, Walker JE. Structure of bovine mitochondrial F-ATPase inhibited by MgADP and aluminium fluoride. Structure. 2000;8(6):567–573. doi: 10.1016/s0969-2126(00)00145-3. [DOI] [PubMed] [Google Scholar]
- 89.Noumi T, Taniai M, Kanazawa H, Futai M. Replacement of arginine 246 by histidine in the β subunit of Escherichia coli H-ATPase resulted in loss of multi-site ATPase activity. Journal of Biological Chemistry. 1986;261(20):9196–9201. [PubMed] [Google Scholar]
- 90.Parsonage D, Duncan TM, Wilke-Mounts S, Kironde FA, Hatch L, Senior AE. The defective proton-ATPase of uncD mutants of Escherichia coli. Identification by DNA sequencing of residues in the beta-subunit which are essential for catalysis or normal assembly. Journal of Biological Chemistry. 1987;262(13):6301–6307. [PubMed] [Google Scholar]
- 91.al-Shawi MK, Parsonage D, Senior AE. Kinetic characterization of the unisite catalytic pathway of seven β-subunit mutant F-ATPases from Escherichia coli . Journal of Biological Chemistry. 1989;264(26):15376–15383. [PubMed] [Google Scholar]
- 92.Bockmann RA, Grubmuler H. Nanoseconds molecular dynamics simulation of primary mechanical energy transfer steps in F-ATP synthase. Nature Structural Biology. 2002;9(3):198–202. doi: 10.1038/nsb760. [DOI] [PubMed] [Google Scholar]
- 93.Dittrich M, Hayashi S, Schulten K. On the mechanism of ATP hydrolysis in F-ATPase. Biophysical Journal. 2003;85(4):2253–2266. doi: 10.1016/s0006-3495(03)74650-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang W, Gao YQ, Cui Q, Ma J, Karplus M. The missing link between thermodynamics and structure in F-ATPase. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(3):874–879. doi: 10.1073/pnas.0337432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weber J. ATP synthase-the structure of the stator stalk. Trends in Biochemical Sciences. 2007;32(2):53–56. doi: 10.1016/j.tibs.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weber J, Senior AE. Location and properties of pyrophosphate-binding sites in Escherichia coli F-ATPase. Journal of Biological Chemistry. 1995;270(21):12653–12658. doi: 10.1074/jbc.270.21.12653. [DOI] [PubMed] [Google Scholar]
- 97.Perez JA, Greenfield AJ, Sutton R, Ferguson SJ. Characterisation of phosphate binding to mitochondrial and bacterial membrane-bound ATP synthase by studies of inhibition with 4-chloro-7-nitrobenzofurazan. The FEBS Letters. 1986;198(1):113–118. doi: 10.1016/0014-5793(86)81195-4. [DOI] [PubMed] [Google Scholar]
- 98.Orriss GL, Leslie AGW, Braig K, Walker JE. Bovine F-ATPase covalently inhibited with 4-chloro-7-nitrobenzofurazan: the structure provides further support for a rotary catalytic mechanism. Structure. 1998;6(7):831–837. doi: 10.1016/s0969-2126(98)00085-9. [DOI] [PubMed] [Google Scholar]
- 99.Penefsky HS. Pi binding by the F-ATPase of beef heart mitochondria and of the Escherichia coli plasma membrane. The FEBS Letters. 2005;579(10):2250–2252. doi: 10.1016/j.febslet.2005.02.072. [DOI] [PubMed] [Google Scholar]
- 100.Copley RR, Barton GJ. A structural analysis of phosphate and sulphate binding sites in proteins estimation of propensities for binding and conservation of phosphate binding sites. Journal of Molecular Biology. 1994;242(4):321–329. doi: 10.1006/jmbi.1994.1583. [DOI] [PubMed] [Google Scholar]
- 101.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]


