Abstract
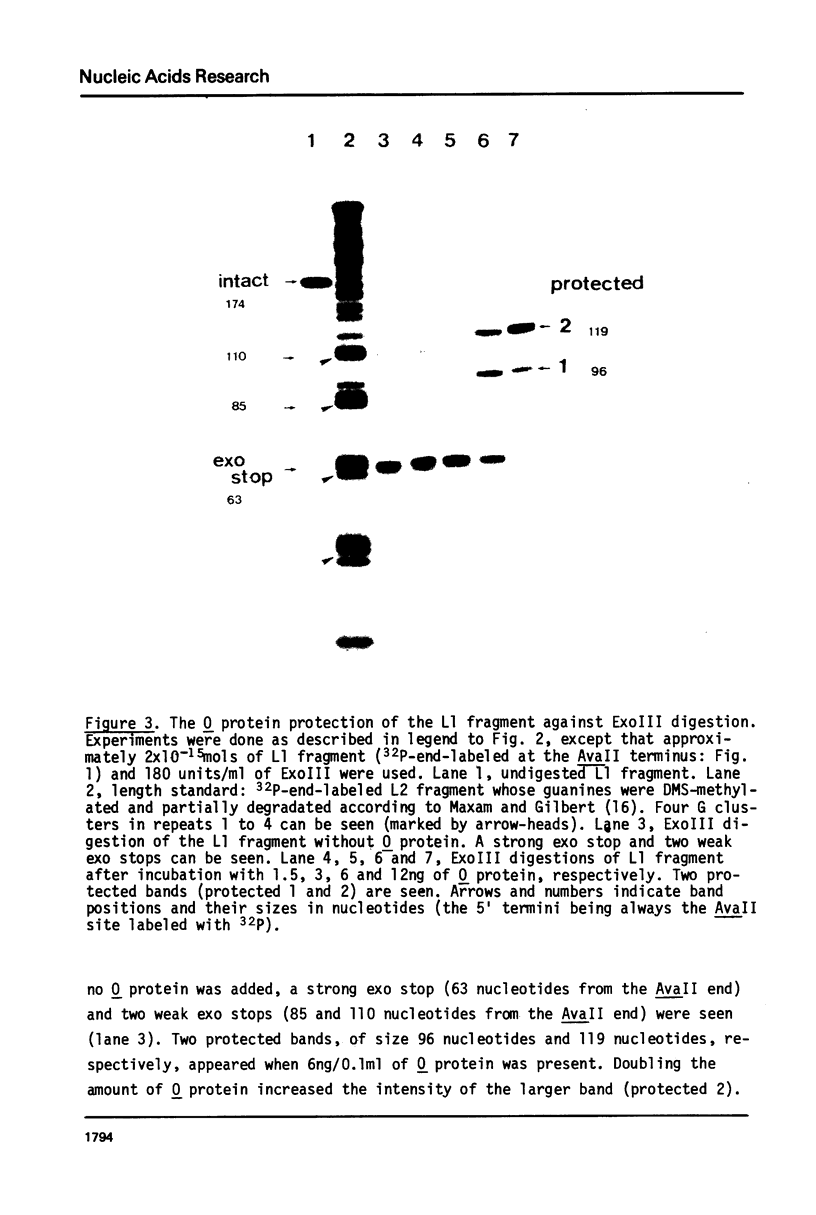
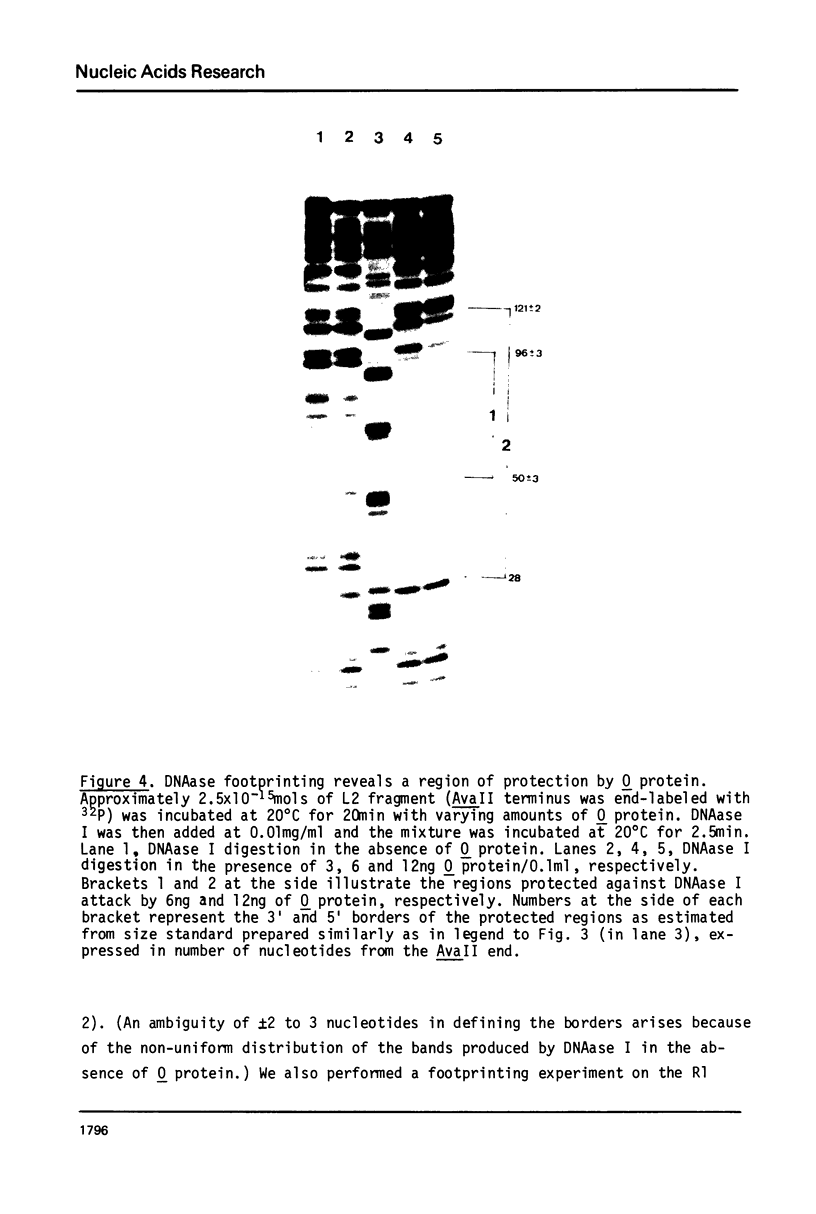
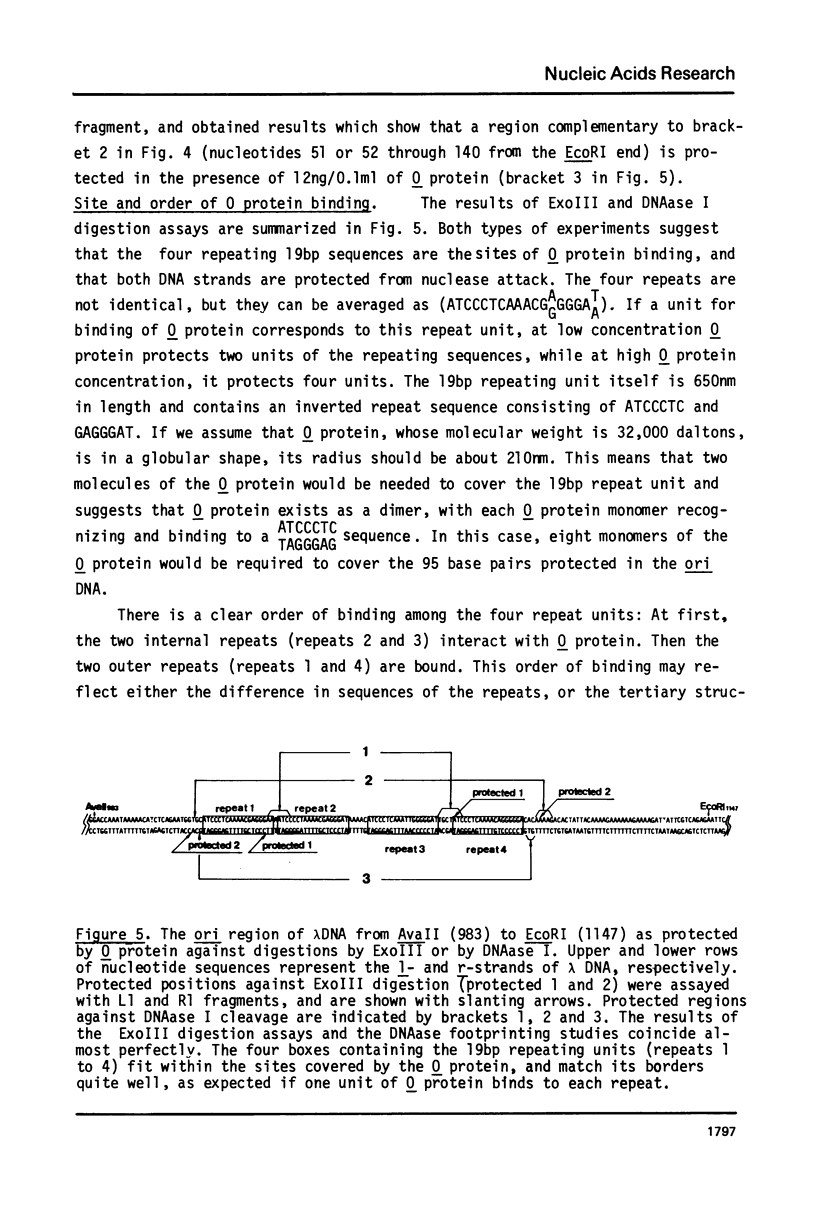
The bacteriophage lambda O protein is needed for initiation of lambda DNA replication. Several lines of evidence suggest that initiation requires that this protein interacts with a specific sequence called ori (for origin) in lambda DNA. We have purified this protein to near homogeneity and studied the protection against nuclease cleavage of the origin DNA sequences. Our data demonstrate that the O protein binds within an interval of about 95 base pairs (bp), which contains four tandemly arranged 19bp repeating sequences, ATCCCTCAAAACGA (G)GG GAT(A). At a low concentration of O protein, the inner two repeats are primarily covered, while binding to the outer two repeats requires a high concentration of O protein. From the molecular size of O protein (32,000 daltons), and the internal symmetry in each 19bp repeat, we inferred that the O protein may bind in dimeric form, and that the 95bp region may be filled only when four such dimers have bound. This interaction is discussed in connection with the "activation" of the ori by O protein leading to initiation of DNA synthesis.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denniston-Thompson K., Moore D. D., Kruger K. E., Furth M. E., Blattner F. R. Physical structure of the replication origin of bacteriophage lambda. Science. 1977 Dec 9;198(4321):1051–1056. doi: 10.1126/science.929187. [DOI] [PubMed] [Google Scholar]
- Furth M. E., McLeester C., Dove W. F. Specificity determinants for bacteriophage lambda DNA replication. I. A chain of interactions that controls the initiation of replication. J Mol Biol. 1978 Dec 5;126(2):195–225. doi: 10.1016/0022-2836(78)90359-5. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R., Hobom G. DNA sequences and structural homologies of the replication origins of lambdoid bacteriophages. Nature. 1979 Feb 22;277(5698):621–627. doi: 10.1038/277621a0. [DOI] [PubMed] [Google Scholar]
- Hirose S., Okazaki R., Tamanoi F. Mechanism of DNA chain growth. XI. Structure of RNA-linked DNA fragments of Escherichia coli. J Mol Biol. 1973 Jul 15;77(4):501–517. doi: 10.1016/0022-2836(73)90219-2. [DOI] [PubMed] [Google Scholar]
- Hobom G., Grosschedl R., Lusky M., Scherer G., Schwarz E., Kössel H. Functional analysis of the replicator structure of lambdoid bacteriophage DNAs. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):165–178. doi: 10.1101/sqb.1979.043.01.023. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore D. D., Denniston-Thompson K., Kruger K. E., Furth M. E., Williams B. G., Daniels D. L., Blattner F. R. Dissection and comparative anatomy of the origins of replication of lambdoid phages. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):155–163. doi: 10.1101/sqb.1979.043.01.022. [DOI] [PubMed] [Google Scholar]
- Murotsu T., Tanaka H., Imaji M., Koga H., Matsubara K., Horiuchi T. Purification and properties of a lambda operator-binding protein which is expected to be autorepressor (tof protein) from E. coli carrying lambdadv plasmid. Mol Gen Genet. 1977 Nov 29;157(2):139–147. doi: 10.1007/BF00267391. [DOI] [PubMed] [Google Scholar]
- Ptashne M., Jeffrey A., Johnson A. D., Maurer R., Meyer B. J., Pabo C. O., Roberts T. M., Sauer R. T. How the lambda repressor and cro work. Cell. 1980 Jan;19(1):1–11. doi: 10.1016/0092-8674(80)90383-9. [DOI] [PubMed] [Google Scholar]
- RICHARDSON C. C., KORNBERG A. A DEOXYRIBONUCLEIC ACID PHOSPHATASE-EXONUCLEASE FROM ESCHERICHIA COLI. I. PURIFICATION OF THE ENZYME AND CHARACTERIZATION OF THE PHOSPHATASE ACTIVITY. J Biol Chem. 1964 Jan;239:242–250. [PubMed] [Google Scholar]
- RICHARDSON C. C., LEHMAN I. R., KORNBERG A. A DEOXYRIBONUCLEIC ACID PHOSPHATASE-EXONUCLEASE FROM ESCHERICHIA COLI. II. CHARACTERIZATION OF THE EXONUCLEASE ACTIVITY. J Biol Chem. 1964 Jan;239:251–258. [PubMed] [Google Scholar]
- Rambach A. Replicator mutants of bacteriophage lambda: characterization of two subclasses. Virology. 1973 Jul;54(1):270–277. doi: 10.1016/0042-6822(73)90136-0. [DOI] [PubMed] [Google Scholar]
- Scherer G. Nucleotide sequence of the O gene and of the origin of replication in bacteriophage lambda DNA. Nucleic Acids Res. 1978 Sep;5(9):3141–3156. doi: 10.1093/nar/5.9.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnös M., Inman R. B. Position of branch points in replicating lambda DNA. J Mol Biol. 1970 Jul 14;51(1):61–73. doi: 10.1016/0022-2836(70)90270-6. [DOI] [PubMed] [Google Scholar]
- Shalloway D., Kleinberger T., Livingston D. M. Mapping of SV40 DNA replication origin region binding sites for the SV40 T antigen by protection against exonuclease III digestion. Cell. 1980 Jun;20(2):411–422. doi: 10.1016/0092-8674(80)90627-3. [DOI] [PubMed] [Google Scholar]
- Skalka A. M. DNA replication--bacteriophage lambda. Curr Top Microbiol Immunol. 1977;78:201–237. [PubMed] [Google Scholar]
- Szybalski E. H., Szybalski W. A comprehensive molecular map of bacteriophage lambda. Gene. 1979 Nov;7(3-4):217–270. doi: 10.1016/0378-1119(79)90047-7. [DOI] [PubMed] [Google Scholar]