Abstract
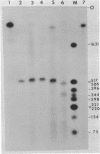
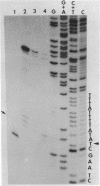
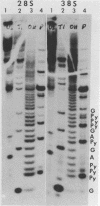
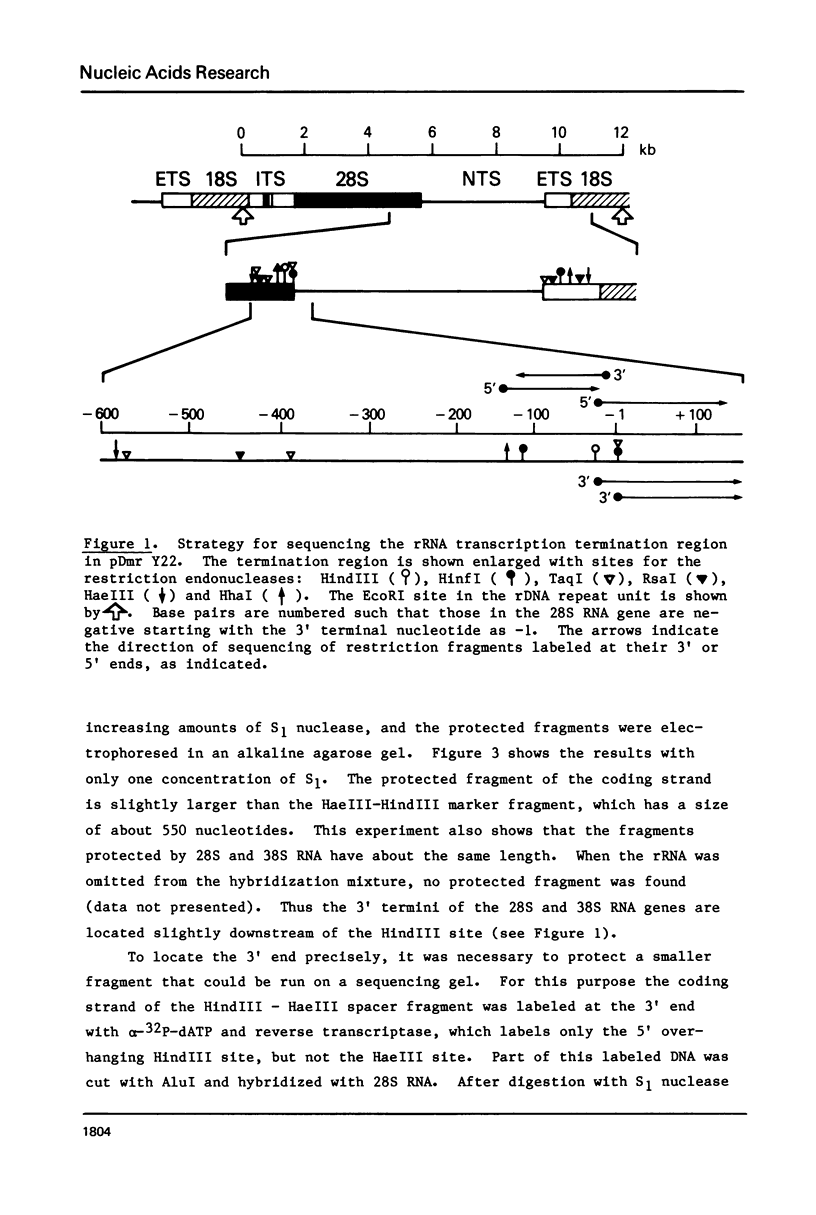
The sequence of 300 nucleotides surrounding the transcription termination site for ribosomal RNA in Drosophila melanogaster has been determined. The precise position of the RNA 3'-end was located by sizing rDNA fragments protected from S1 nuclease by hybridization with cytoplasmic 28S rRNA or with nuclear 38S pre-rRNA. Next, the sequence of the first 10 nucleotides at the 3'-end of 28S and 38S rRNA was determined directly. The results indicate the absence of 3'-terminal processing of the 38S pre-rRNA. The nucleotide sequence from a position 27 nucleotides upstream to 150 nucleotides downstream from the termination site was identical in three uninterrupted rRNA genes and one interrupted gene, except for a single base change in the spacer region of one clone. The terminal 100 nucleotides of Drosophila 28S rRNA have more than 60 per cent homology with the corresponding region of Xenopus 28S rRNA and yeast 26S rRNA.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Biswas B. B., Ganguly A., Das A. Eukaryotic RNA polymerases and the factors that control them. Prog Nucleic Acid Res Mol Biol. 1975;15(0):145–184. doi: 10.1016/s0079-6603(08)60119-1. [DOI] [PubMed] [Google Scholar]
- Brown D. D., Littna E. Synthesis and accumulation of DNA-like RNA during embryogenesis of Xenopus laevis. J Mol Biol. 1966 Sep;20(1):81–94. doi: 10.1016/0022-2836(66)90119-7. [DOI] [PubMed] [Google Scholar]
- Dawid I. B., Wellauer P. K., Long E. O. Ribosomal DNA in Drosophila melanogaster. I. Isolation and characterization of cloned fragments. J Mol Biol. 1978 Dec 25;126(4):749–768. doi: 10.1016/0022-2836(78)90018-9. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England T. E., Bruce A. G., Uhlenbeck O. C. Specific labeling of 3' termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65(1):65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- Gurdon J. B., Woodland H. R. The influence of the cytoplasm on the nucleus during cell differentiation, with special reference to RNA synthesis during amphibian cleavage. Proc R Soc Lond B Biol Sci. 1969 Apr 15;173(1030):99–111. doi: 10.1098/rspb.1969.0042. [DOI] [PubMed] [Google Scholar]
- Hamada H., Kominami R., Muramatsu M. 3'-terminal processing of ribosomal RNA precursors in mammalian cells. Nucleic Acids Res. 1980 Feb 25;8(4):889–903. [PMC free article] [PubMed] [Google Scholar]
- Jolly D. J., Thomas C. A., Jr Nuclear RNA transcripts from Drosophila melanogaster ribosomal RNA genes containing introns. Nucleic Acids Res. 1980 Jan 11;8(1):67–84. doi: 10.1093/nar/8.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn L. J., Brown D. D. Nucleotide sequence of Xenopus borealis oocyte 5S DNA: comparison of sequences that flank several related eucaryotic genes. Cell. 1978 Dec;15(4):1145–1156. doi: 10.1016/0092-8674(78)90042-9. [DOI] [PubMed] [Google Scholar]
- Levis R., Penman S. Processing steps and methylation in the formation of the ribosomal RNA of cultured Drosophila cells. J Mol Biol. 1978 May 15;121(2):219–238. doi: 10.1016/s0022-2836(78)80006-0. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Alternative pathways in the processing of ribosomal RNA precursor in Drosophila melanogaster. J Mol Biol. 1980 Apr 25;138(4):873–878. doi: 10.1016/0022-2836(80)90070-4. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Expression of ribosomal DNA insertions in Drosophila melanogaster. Cell. 1979 Dec;18(4):1185–1196. doi: 10.1016/0092-8674(79)90231-9. [DOI] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Pavlakis G. N., Jordan B. R., Wurst R. M., Vournakis J. N. Sequence and secondary structure of Drosophila melanogaster 5.8S and 2S rRNAs and of the processing site between them. Nucleic Acids Res. 1979 Dec 20;7(8):2213–2238. doi: 10.1093/nar/7.8.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae P. M., Kohorn B. D., Wade R. P. The 10 kb Drosophila virilis 28S rDNA intervening sequence is flanked by a direct repeat of 14 base pairs of coding sequence. Nucleic Acids Res. 1980 Aug 25;8(16):3491–3504. doi: 10.1093/nar/8.16.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair J. H., Brown D. D. Retention of common nucleotide sequences in the ribosomal deoxyribonucleic acid of eukaryotes and some of their physical characteristics. Biochemistry. 1971 Jul 6;10(14):2761–2769. doi: 10.1021/bi00790a017. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Tabak H. F., Flavell R. A. A method for the recovery of DNA from agarose gels. Nucleic Acids Res. 1978 Jul;5(7):2321–2332. doi: 10.1093/nar/5.7.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano Y., Kominami R., Mishima Y., Muramatsu M. The nucleotide sequence of the putative transcription initiation site of a cloned ribosomal RNA gene of the mouse. Nucleic Acids Res. 1980 Dec 20;8(24):6043–6058. doi: 10.1093/nar/8.24.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Jonge P., Leer R. J., Planta R. J. The transcription termination site of the ribosomal RNA operon in yeast. Nucleic Acids Res. 1980 Nov 25;8(22):5179–5192. doi: 10.1093/nar/8.22.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellauer P. K., Dawid I. B., Kelley D. E., Perry R. P. Secondary structure maps of ribosomal RNA. II. Processing of mouse L-cell ribosomal RNA and variations in the processing pathway. J Mol Biol. 1974 Oct 25;89(2):397–407. doi: 10.1016/0022-2836(74)90527-0. [DOI] [PubMed] [Google Scholar]
- Wieslander L. A simple method to recover intact high molecular weight RNA and DNA after electrophoretic separation in low gelling temperature agarose gels. Anal Biochem. 1979 Oct 1;98(2):305–309. doi: 10.1016/0003-2697(79)90145-3. [DOI] [PubMed] [Google Scholar]