Abstract
Protein kinase R (PKR) is an interferon-induced kinase that plays a pivotal role in the innate immunity pathway. PKR is activated to undergo autophosphorylation upon binding to double-stranded RNAs or RNAs that contain duplex regions. Activated PKR phosphorylates the α subunit of eukaryotic initiation factor 2, thereby inhibiting protein synthesis. PKR is also activated by heparin, a highly sulfated glycosaminoglycan. We have used biophysical methods to define the mechanism of PKR activation by heparin. Heparins as short as hexasaccharide bind strongly to PKR and activate autophosphorylation. In contrast to double-stranded RNA, heparin activates PKR by binding to the kinase domain. Analytical ultracentrifugation measurements support a thermodynamic linkage model where heparin binding allosterically enhances PKR dimerization, thereby activating the kinase. These results indicate that PKR can be activated by small molecules and represents a viable target for the development of novel antiviral agents.
Keywords: analytical ultracentrifugation, drug discovery, innate immunity, protein kinase R
Introduction
The RNA-activated kinase, protein kinase R (PKR), plays a key role in the innate immunity response to viral infection.1,2 In addition, PKR mediates apoptosis induced by a variety of stimuli3,4 and functions in the control of cell growth and proliferation and as a tumor-suppressor protein.5,6 PKR is induced by interferon in a latent form that is activated by binding double-stranded RNA (dsRNA) to undergo autophosphorylation. An additional class of RNA activators, which contain a 5′-triphosphate and a 16-bp stem flanked by short single-stranded regions, has been described.7 The protein activator of PKR, PACT, activates PKR in response to a variety of cellular stresses.8,9 The most well-characterized cellular substrate is the α subunit of eukaryotic initiation factor 2 (eIF2α). Phosphorylation of eIF2α at serine 51 blocks the recycling of eIF2 between the GTP-bound state and the GDP-bound state, thereby inhibiting initiation of protein synthesis. Thus, production of dsRNA during viral infection10 results in PKR activation and inhibition of viral and host protein syntheses.
PKR consists of an N-terminal dsRNA binding domain (dsRBD), containing two tandem copies of the dsRNA binding motif11 and a C-terminal kinase, with an ~90-amino-acid linker lying between these domains. Each of the dsRNA binding motifs has the typical αβββα fold, with a short unstructured region lying between them.12 The kinase domain adopts a bilobal structure typical of protein kinases.13
Although an autoinhibition mechanism was originally proposed for the activation of PKR by dsRNA, recent data in support of a dimerization model have accumulated.14,15 PKR dimerizes weakly in solution, and dimerization is sufficient to activate PKR in the absence of RNA.16 The dimer interface has been localized to the N-terminal lobe of the kinase, and a potential allosteric pathway linking the dimer interface to the kinase active site has been identified.13,17 Activation of PKR by dsRNA follows a “bell-shaped” curve where low RNA concentrations activate but higher concentrations inhibit.18,19 These results can be rationalized in a model where low concentrations of dsRNA favor the assembly of multiple PKRs on a single dsRNA, whereas high dsRNA concentrations dilute PKR monomers onto separate molecules of dsRNA.20 Consistent with the dimerization model, a minimum of 30 bp of dsRNA are required to bind two PKRs and to activate autophosphorylation, supporting a model where the role of dsRNA is to bring two or more PKR monomers in close proximity to enhance dimerization via the kinase domain.21–23
There are several classes of compounds that are capable of inducing PKR autophosphorylation in the absence of dsRNA or PACT. PKR is activated by polyanions, including heparin, a highly sulfated glycosaminoglycan.24 In contrast to dsRNA, heparin does not mediate intermolecular autophosphorylation,25 and it has been suggested that heparin activation does not involve PKR dimerization.26 Activation does not require the presence of dsRBD,27 indicating that heparin does not bind at the same site as dsRNA. Deletion mutagenesis studies have located the binding site at a region within the kinase domain.28 The minimally sized heparin reported to be capable of activating PKR is an octasaccharide.25 More recently, two small-molecule PKR activators have been reported.29,30 These results suggest that PKR is a viable target for the development of novel antiviral agents that enhance the innate immune response. Here, we have characterized the nature of the interaction of PKR with a series of heparin oligosaccharides. Our data support a novel activation mechanism where heparin binding is thermodynamically linked to enhanced PKR dimerization.
Results
PKR activation by heparin oligosaccharides
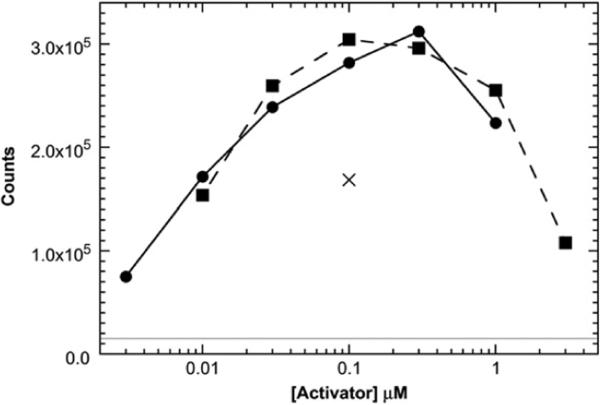
We have confirmed an early report25 indicating that short heparin-derived oligosaccharides can activate PKR. Autophosphorylation of PKR is stimulated by both heparin octasaccharide (dp8) and heparin hexasaccharide (dp6) (Fig. 1; also see Fig. S1 for the gel PhosphorImager scan). In earlier studies,25 dp6 was considerably less active than dp8; the origin of this discrepancy is not clear. Smaller heparins do not elicit significant activation. The maximal activation level is comparable to that elicited by a small 40-bp dsRNA. Like dsRNA, heparin oligosaccharides show a bell-shaped activation curve where higher concentrations inhibit.
Fig. 1.
Activation of PKR by dp8 (squares; broken line) and dp6 (circles; continuous line). Activation of PKR by 100 nM of a 40-bp dsRNA (×) is included for comparison. The continuous horizontal line represents the background autophosphorylation of PKR in the absence of activator. Activation assays were performed at 100 nM PKR in P50 buffer [20 mM Hepes (pH 7.5), 50 mM KCl, 5 mM MgCl2, and 0.1 mM tris(2-carboxyethyl)phosphine] for 20 min at 32 °C. Reactions contained 0.4 mM ATP and 2 μCi of [γ-32P]ATP. Figure S1 shows the PhosphorImager gel scan used to generate the data in this figure.
Analysis of heparin binding to PKR
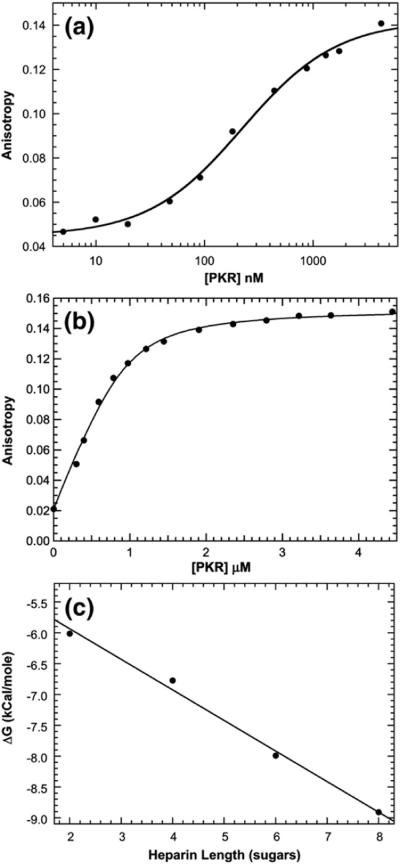
A homogeneous fluorescence anisotropy assay was established to characterize the binding of short heparin oligosaccharides to PKR. Labeled heparin oligosaccharides were prepared by a reaction with BODIPY-FL-hydrazide (4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-propionic acid hydrazide) at the reducing end.31 Binding of these BODIPY–heparin conjugates to PKR results in an increase in steady-state emission anisotropy due to a reduced mobility of the BODIPY fluorophore. Figure 2a shows a titration of BODIPY-labeled dp8 (bdp8) with PKR performed at 15 nM bdp8. The data fitted well to a simple hyperbolic binding function with Kd = 224 ± 27 nM. The relative stoichiometry of heparin binding to PKR was determined by anisotropy titrations performed at higher concentrations of bdp8 well above Kd. Figure 2b shows a data set obtained at 1 μM bdp8. The best-fit stoichiometry of N=1.07±0.07 bdp8/PKR indicates that this heparin oligosaccharide binds to PKR in a 1:1 ratio. In control experiments, potential contributions of the BODIPY fluorophore to the binding affinity were assessed by competing 500 nM bdp8+500 nM PKR with unlabeled ligand (dp8). Addition of unlabeled heparin oligosaccharide reduces the emission anisotropy of bdp8, indicating that these two ligands compete for binding. The competition titration was fitted as previously described32 to obtain Kd=262±21 nM for dp8 binding to PKR. Thus, fluorescence labeling does not appreciably alter binding energetics. However, labeling strongly reduces the ability of dp8 to activate PKR autophosphorylation (data not shown). Note that this observation does not affect the interpretation of the linkage analysis performed below using unlabeled dp8.
Fig. 2.
Binding of heparin oligosaccharide to PKR measured by anisotropy titration. (a) Kd measurement for bdp8. The fluorescence anisotropy of 15 nM bdp8 was measured at increasing concentrations of PKR at an excitation wavelength of 488 nm and at an emission wavelength of 515 nm, with spectral bandwidths of 6 nm for excitation and emission, respectively. Each data point represents the average of three 5-s measurements. The continuous line is a fit of the data to a hyperbolic binding model (Eq. (3)) to obtain Kd=224±27 nM, rf=0.044±0.002, and rb=0.142±0.002. (b) Stoichiometry analysis for bdp8. The fluorescence anisotropies of 1 μM (●) bdp8 were recorded as in (a). The continuous line is a fit of the data to a binding model incorporating the depletion of free ligand. The best-fit parameters are N= 1.07±0.07 and rb=0.153±0.002. (c) Dependence of heparin oligosaccharide binding affinity on length. The Kd values for the binding of BODIPY-labeled dp2–bdp8 were measured, and the binding free energies were calculated using ΔG°=−RTln(1/Kd).
Shorter heparin oligosaccharides were also labeled with BODIPY to determine the effect of length on binding affinity. The Kd values increase dramatically with decreasing oligosaccharide length such that Kd=32.6±7.4 μM for heparin disaccharide (dp2; or almost 150-fold weaker binding affinity than dp8). Interestingly, binding free energies decrease linearly with decreasing length over the range of dp8–dp2 (Fig. 2c). The fact that the energetic contributions of each disaccharide unit are additive suggests that the oligosaccharides interact nonspecifically with an extended positively charged binding pocket on PKR. Consistent with a large electrostatic contribution, the binding affinity is also strongly dependent on salt concentration. For dp8, the Kd increases from 224 nM in 75 mM NaCl (Fig. 2a) to 2.61±0.05 μM in 100 mM NaCl and 19.0±1.9 μM in 125 mM NaCl (data not shown).
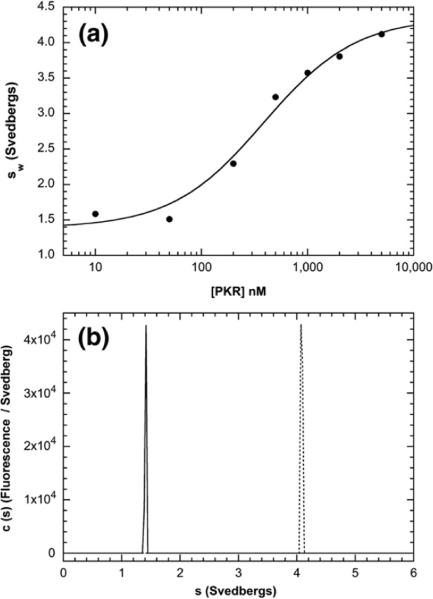
Because of the importance of dimerization in the activation of PKR by dsRNA,15,33 we hypothesized that heparin may activate by altering the PKR monomer–dimer equilibrium. Although PKR is predominantly monomeric at the concentrations employed for the fluorescence anisotropy titrations described above (the Kd for the dimerization of free enzyme is about 500 μM16), heparin binding may alter PKR dimerization. In principle, dimerization should result in a measurable increase in the rotational correlation time of a PKR–heparin complex. However, it is difficult to sort out such complex equilibria by analysis of steady-state fluorescence anisotropy alone. Therefore, fluorescence-detected sedimentation velocity was used to define the hydrodynamic properties of the complex (or complexes) formed upon the binding of bdp8 to PKR under the same conditions employed in the anisotropy assays. The velocity data were analyzed using the time-derivative method34,35 to obtain the weight-average sedimentation coefficients (sw) as a function of [PKR] (Fig. 3a). The sedimentation coefficient of bdp8 alone is about 1.4 S, consistent with a recent study of the hydrodynamic properties of short heparin oligosaccharides.36 Addition of PKR results in an increase in sw due to the formation of a PKR–heparin complex. The isotherm of sw as a function of [PKR] was fitted to obtain a Kd of 387±89 nM and a sedimentation coefficient for the complex of 4.35±0.16 S. The Kd is close to the value derived from the anisotropy titration (Fig. 2a). The sedimentation coefficient is greater than that of the PKR monomer (about 3.5 S16) but is less than that of the dimer (about 5.0 S based on the value obtained with the phosphorylated PKR dimer37). The sedimentation coefficient is consistent with a PKR–dp8 complex (Table S1). Taken together with stoichiometry analysis (Fig. 2b), these results indicate that bdp8 binds to the PKR monomer to form a 1:1 complex. The homogeneity of the complex was assessed in a separate sedimentation velocity experiment in which 250 nM bdp8 was titrated with a 10-fold excess of PKR. Under these conditions, where essentially all of the bdp8 is bound to PKR and [PKR]>>Kd, the system can be treated as essentially nonequilibrating, and the c(s) method can be applied to produce high-resolution sedimentation coefficient distributions.38 Figure 3b shows that addition of PKR cleanly converts bdp8 into a homogeneous complex with an apparent sedimentation coefficient of about 4.1 S, which agrees reasonably well with the value obtained from fitting the weight-average data.
Fig. 3.
bdp8 binding to PKR analyzed by fluorescence-detected sedimentation velocity. (a) PKR titration. Samples contained 15 nM bdp8 and variable concentrations of PKR. Weight-average sedimentation coefficients (sw) were obtained by the integration of g(s*) distributions and were fitted to a hyperbolic binding model to give the following best-fit parameters: Kd=387±89 nM, sw(bdp8)=1.38±0.10 S, and sw(bdp8–PKR complex)=4.35±0.16 S. (b) c(s) distributions of 250 nM bdp8 ( ) and 250 nM bdp8+10 Eq of PKR (----). Conditions: rotor speed, 50,000 rpm; temperature, 20 °C; fluorescence optics.
) and 250 nM bdp8+10 Eq of PKR (----). Conditions: rotor speed, 50,000 rpm; temperature, 20 °C; fluorescence optics.
Analysis of the heparin-induced dimerization of PKR
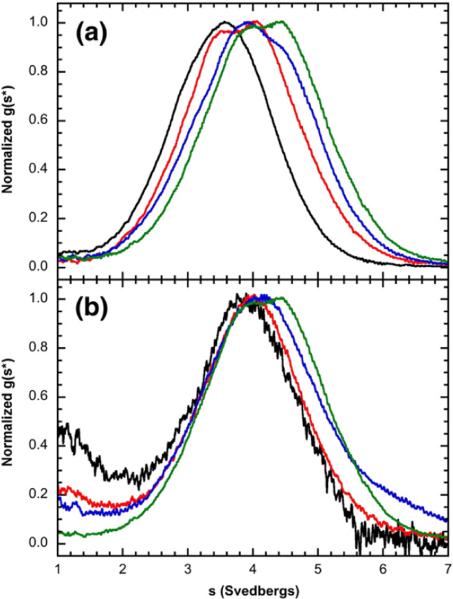
The effects of dp8 binding on the association state of PKR were probed at higher protein concentrations (up to 16 μM, ~1 mg/ml) using sedimentation velocity with interference optics to monitor both the protein and the ligand. Initially, the data were analyzed using the time-derivative approach to define the qualitative effects of heparin binding on the PKR association state.39 The resulting normalized g(s*) distributions provide insight into the nature of the reaction scheme and were over-plotted to define the qualitative effects of heparin binding on the PKR association state. As observed with fluorescently labeled heparin, the 1:1 PKR/dp8 complex has a sedimentation coefficient of about 4 S (Fig. 4a). However, at higher dp8 concentrations, the distributions continue to shift to the right, indicating the formation of a larger species. Because only one dp8 binds to the PKR monomer, the increase in the sedimentation coefficient must reflect the self-association of PKR rather than the binding of multiple heparins to a PKR monomer. Furthermore, the g(s*) distributions shift to the right with increasing PKR concentration in the presence of a saturating, high concentration of dp8 (60 μM) (Fig. 4b). The peak maximum increases from ~4.0 S at 2 μM PKR to about 4.4 S at 16 μM PKR. In contrast, the sedimentation coefficient of unliganded PKR increases only by about 0.1 S as the protein is increased from 1.6 μM to 25 μM,16 indicating that binding of heparin oligosaccharide to PKR enhances dimerization.
Fig. 4.
Sedimentation velocity analysis of dp8 binding to PKR. (a) Normalized g(s*) distributions obtained at a fixed concentration of PKR (16 μM) with the indicated concentrations of dp8: 0 μM (black), 8 μM (red), 16 μM (blue), and 32 μM (green). The distributions are normalized by peak maximum. (b) Normalized g(s*) distributions obtained at a fixed concentration of dp8 (60 μM) with the indicated concentrations of PKR: 2 μM (black), 4 μM (red), 8 μM (blue), and 16 μM (green). The distributions are normalized by peak maximum. Conditions: rotor speed, 50,000 rpm; temperature, 20 °C; interference optics; scan interval, 1 min. Note that the free dp8 is not resolved in these distributions because the scan range was chosen to emphasize the higher-S features and because diffusional broadening reduces the resolution of the low-S region in the time-derivative method.
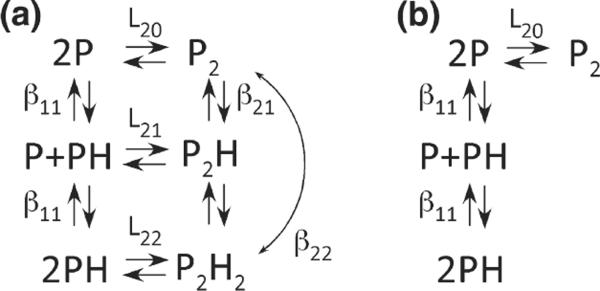
In many allosteric systems, protein self-association is thermodynamically linked to ligand binding,40 and analytical ultracentrifugation is a useful method for defining linkage relationships, as we have recently demonstrated in our analysis of the dimerization of SecA in the presence of signal peptide.41 The thermodynamic linkage of heparin binding and PKR dimerization was quantitatively resolved by a global analysis of sedimentation velocity data obtained at multiple PKR and dp8 concentrations using the program SEDANAL.42 Figure 5a depicts the standard Wyman–Gill linkage model40 that we have employed to analyze the data. In this case, the linkage free-energy coupling dimerization and ligand binding ΔGC is given by:
| (1) |
where R is the gas constant, T is the temperature, and the binding constants are defined in the legend to Fig. 5.
Fig. 5.
Models for thermodynamic linkage of PKR dimerization and heparin binding. (a) Linked model. (b) Unlinked model. P, PKR; H, heparin. The binding constants are defined as follows: L20 is the dimerization constant for unliganded PKR, L22 is the dimerization constant for the PH complex, L21 is the dimerization constant for the interaction of P and PH, β11 is the constant for heparin binding to the PKR monomer, β21 is the constant for heparin binding to the PKR dimer, and β22 is the overall constant for the interaction of two heparin monomers with the PKR dimer to produce P2H2. The nomenclature was adapted from Fig. 6-5 in Wyman and Gill.40
When fitting data using such a complex model containing multiple binding constants and sedimentation coefficients as adjustable parameters, it is useful to constrain as many of them as possible to obtain narrow confidence intervals on the parameters of interest. The binding constant for the interaction of unlabeled dp8 with PKR monomer (β11) was fixed based on the anisotropy binding measurements. The dimerization of unliganded PKR had been previously characterized (Kd ~500 μM).16 However, these measurements were performed under slightly different conditions (200 mM NaCl versus 75 mM NaCl in the present study). Dimerization of PKR was determined in 75 mM NaCl by sedimentation velocity experiments over a protein concentration range of 0.28–2.0 mg/ml. The best-fit value of the association constant L20=2.48 (2.24, 2.74)×103 M−1 corresponds to a slightly enhanced dimerization (Kd=404 μM) relative to the value obtained in 200 mM NaCl. Several sedimentation coefficients were constrained (see the legend to Fig. 6). We also made the reasonable assumption that the dimer association constant for P binding to PH (L21) is equal to the geometric mean of the association constants for P+P(L20) and PH+PH(L22). However, the quality of the fit and the magnitude of ΔGC are not strongly dependent on this assumption (see the text below).
Fig. 6.
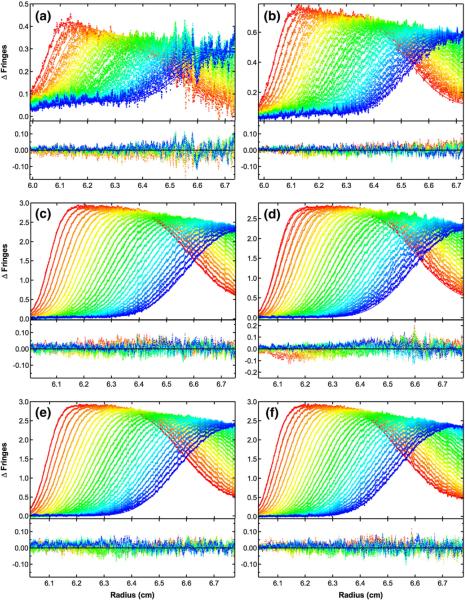
Global analysis of sedimentation velocity difference curves for dp8 binding to PKR. The data were subtracted in pairs for six data channels collected at the following concentrations: (a) 2 μM PKR+60 μM dp8; (b) 4 μM PKR+60 μM dp8; (c) 16 μM PKR+60 μM dp8; (d) 16 μM PKR+30 μM dp8; (e) 16 μM PKR+16 μM dp8; (f) 16 μM PKR+8 μM dp8. The data (58 difference scans for each channel) were globally fitted to the binding model depicted in Fig. 4a with an RMSD of 0.0196 fringes. The value of β11 was fixed at 3.82×106 M−1 based on the fluorescence anisotropy competition of bdp8 with dp8; L20 was fixed at 2.48×103 M−1 from the sedimentation velocity analysis of PKR dimerization; L21 was constrained as the geometric mean of L20 and L22; sdp8 was fixed at 1.17 S from published data36; sPKR was fixed at 3.52 S16; s(PKR)2 was estimated to be 5.0 S based on the sedimentation analysis of the phosphorylated PKR dimer37; s(PKR)2–dp8 was estimated as 5.5 S; and the ratio of s(PKR–dp8)2 to sPKR–dp8 was constrained to be 1.5. The top panels show the data (points) and fit (continuous lines), and the bottom panels show the residuals (points). The best-fit parameters are shown in Table 1. Conditions: rotor speed, 50,000 rpm; temperature, 20 °C; interference optics; scan interval, 1 min.
Figure 6 shows the global fit of six data sets containing multiple heparin and PKR concentrations to the linkage model (Fig. 5a) using the constraints described above, and Table 1 contains the best-fit parameters. The data fitted well to this model, as indicated by the absence of large systematic deviations in the residuals and the low RMSD of 0.0196 fringes. The confidence limits on the parameters are narrow, indicating that the parameters are well defined by the data. Most importantly, the best-fit value of L22=7.19 (6.37, 8.06)×103 M−1 indicates that dp8 binding enhances PKR dimerization, corresponding to a coupling energy of ΔGC=− 0.62 kcal/mol. When we attempted to fit the data to a model where heparin binding and PKR dimerization are mutually exclusive (Fig. 5b), the RMSD increased substantially, indicating that the PKR–heparin complexes are indeed able to dimerize. Furthermore, the data also do not fit as well to a model in which L22 is constrained to be equal to L20. Although the increase in the RMSD is smaller than the increase for model B, it is well outside the 95% confidence range defined by the F test.
Table 1.
Equilibrium binding constants for the interaction of dp8 and bdp8 with PKR
| Model | Method | β11 (M−1a | L22 (M−1)a | s(PH)b | s(P2H2)b | RMSDc |
|---|---|---|---|---|---|---|
| Anisotropyd | 4.47±0.54 × 106 | — | — | — | — | |
| Anisotropy competition | 3.82±0.54 × 106 | — | — | — | — | |
| FDSVd,e | 2.58±0.60 × 106 | — | 4.35 | — | — | |
| Model Af | SVg | 3.82 × 106 (fixed) | 7.19 (6.37, 8.10) × 103 | 3.98 | 5.98 | 0.0196 |
| Model Bf | SVg | 3.82 × 106 (fixed) | — | 4.26 | — | 0.0256 |
| Model Af | SVg | 3.82 × 106 (fixed) | 2.48 × 103 (fixed) | 4.14 | 6.21 | 0.0218 |
Parameters were obtained by nonlinear least-squares analysis using the indicated model and the experimental method. The values in parentheses represent the 95% joint confidence intervals.
See Fig. 5 for the definitions of the equilibrium constants.
Uncorrected sedimentation coefficients (Svedbergs) of PKR-dp8 (PH) and PKR2-dp82 (P2H2).
Root mean square deviation of the fit in fringe units.
Data were obtained using bdp8.
Fluorescence-detected sedimentation velocity.
See Fig. 5 for a definition of the model and the legend to Fig. 6 for a description of the fixed parameters.
Sedimentation velocity using interference optics.
It is not possible to determine the equilibrium contribution of the intermediate species P2H from the present data. In fact, constraining L21=0 to set the population of P2H to zero results in a fit with exactly the same RMSD as the original model, where L21 was fixed as the geometric mean of L20 and L22. However, the best-fit value of L22 increases slightly to L22=7.46×103 M−1. Because the mass and optical properties of P2H and P2H2 are virtually the same, the sedimentation velocity analysis is only sensitive to the sum of their populations. Thus, if the equilibrium population of P2H is set to zero, then L22 rises to increase the population of P2H2.
The heparin binding site on PKR
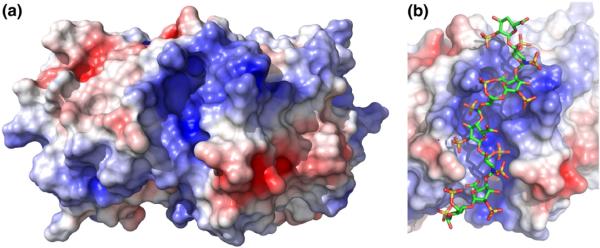
Previous studies have shown that PKR activation does not require the presence of dsRBD27 and have assigned two heparin binding motifs lying within the kinase domain.28 Fluorescence anisotropy measurements reveal that bdp8 binds to a PKR kinase domain construct with Kd= 727±47 nM (Fig. S2). Thus, heparin oligosaccharides do bind to the PKR kinase domain, albeit with slightly lower affinity than observed in the full-length kinase. Examination of the crystal structure of the PKR kinase domain13 reveals a prominent cleft on the kinase domain that represents a potential binding site for negatively charged heparin oligosaccharides (Fig. 7a). The rim of the cleft is lined with several cationic side chains (K304, R307, K310, K314, K408, K409, R413, and K440). The resulting positive charge of the cleft would give rise to a favorable electrostatic contribution to heparin binding. The bottom of the pocket is formed by the aliphatic side chains of L410 and V436. We have tested the contribution of the pocket to heparin binding by mutating these aliphatic residues in the kinase domain. The L410E mutation introduces a negative charge at the base of the pocket. The Kd for L410E is 10,530±1530 nM, corresponding to about a 15-fold reduction in binding affinity (Fig. S2). The V436F mutation blocks the base of the cavity with a bulky phenyl side chain. For this construct, Kd=9500±760 nM, indicating a 12-fold reduction in binding affinity (Fig. S2). Together, these data support a model where heparin binds at the positively charged cleft on the kinase domain.
Fig. 7.
The positively charged cleft on the PKR kinase domain. (a) Electrostatic surface of the kinase domain. A solvent-accessible surface of the PKR kinase domain alone was generated from the crystal structure of a complex of the kinase domain and eIF2α (Protein Data Bank ID: 2A1A) using PyMOL. The dimer interface is located on the top surface of the kinase domain monomer. The electrostatic surface was produced using APBS and was contoured at ±5 kT. (b) Model of dp8 bound to the kinase domain in the positively charged cleft. The dp8 ligand was docked using AutoDock Vina. See Materials and Methods for details.
We have constructed a structural model for dp8 bound to the cleft present in the kinase domain. Short heparin oligosaccharides adopt an extended helical conformation in solution,36,43,44 and this structure is typically retained in protein complexes. A model for dp8 was generated using NMR solution data44 and was docked into the positively charged cleft using AutoDock Vina45 (Fig. 7b). The protein–ligand complex shows good shape complementarity, and the negatively charged sulfates interact favorably with the cationic surface. The dimensions of the pocket are clearly suitable for accommodating heparin oligosaccharides as large as dp8.
Discussion
Our biophysical analysis of the interaction of short heparin oligosaccharides with PKR supports a model where these molecules activate PKR by enhancing dimerization. Heparins as short as hexasaccharides activate the kinase to undergo autophosphorylation. Fluorescence anisotropy and fluorescence-detected sedimentation velocity measurements indicate that a single heparin binds to a PKR monomer with an affinity that is strongly dependent on heparin length and salt concentration. Binding of heparin enhances the dimerization of PKR, and sedimentation velocity data obtained over a broad range of PKR and dp8 concentrations fit a classical linkage thermodynamic model with a coupling free energy of ΔGC=− 0.62 kcal/mol. We propose that this enhancement in PKR dimerization is responsible for activating the kinase.
It is noteworthy that the enhancement of PKR dimerization affinity induced by dp8 is only about 4-fold and would thus appear insufficient to induce autophosphorylation. However, it is important to recognize that the extent of PKR autophosphorylation does not reflect an equilibrium process but represents the end point of a series of autocatalytic reactions. We have previously proposed a chain reaction model to explain how the dimerization of PKR at concentrations ≥0.5 μM induces activation despite a much higher dimerization Kd~500 μM.16 An analogous mechanism may mediate activation by heparin. In the crystal structure of the PKR kinase domain, a plausible allosteric pathway linking the dimer interface to the kinase active site has been identified.13,17 A large body of biophysical and biochemical data implicates dimerization as a key step in PKR activation by RNA.15,21–23
Although PKR dimerization is common to the models for dsRNA-induced and heparin-induced PKR activation, it is likely that the detailed mechanisms are quite different. First, these ligands bind at different locations. Previous studies have determined the following: (1) heparin does not compete with the binding of adenovirus VAI RNA25; (2) dsRBD is not required for the activation of PKR by heparins27; and (3) deletion mutagenesis defines two short heparin binding motifs located at residues 295–300 and 443–448 lying within the kinase domain.28 Our result supports a model where heparins bind to an extended positively charged cleft lying within the kinase domain. This cleft is able to accommodate dp8 with good shape and charge complementarity. This model is supported by the linear dependence of binding free energy on the length of the heparin, which implies an extended binding interface. The model is also consistent with the strong electrostatic contribution to binding affinity. The positively charged cleft partially overlaps with the previously reported C-terminal motif.28
The mechanisms by which dsRNA and heparin enhance PKR dimerization also differ. About 30 bp of dsRNA are required to bind two PKRs and to activate autophosphorylation, supporting a model where dsRNA serves as a binding scaffold that brings two or more PKR monomers into close proximity.21–23 In our model for heparin-induced PKR dimerization, one heparin binds to each PKR monomer rather than serving as a template for the assembly of multiple PKR monomers into a single activator. The length of the minimal activating heparin (dp6) is about 25 Å, which appears too short to simultaneously occupy the binding clefts on two PKR monomers. Furthermore, the clefts face away from the kinase dimerization surface, so it would not be feasible for a single heparin to interact with both monomers. Instead, the binding of heparin likely indirectly modulates dimerization affinity. In fact, there is evidence from sedimentation velocity that heparin binding induces a conformational change in the PKR monomer. The increase in the sedimentation coefficient of the PKR monomer from 3.5 S to 4.0 S upon binding of dp8 is greater than can be explained based on the mass increase of ~2600 Da and corresponds to a decrease in the frictional ratio f/f0 from 1.56 to 1.47 (Table S1). Thus, heparin binding causes PKR to adopt a more compact conformation that may be associated with the increase in dimerization affinity.
Curiously, we found that both dsRNA-induced activation and heparin-induced activation of PKR follow a bell-shaped curve. Previously, a bell-shaped curve for PKR activation by dsRNA18,19 was interpreted as evidence for a dimerization model in which the inhibition at high dsRNA concentrations results from the dilution of PKR dimers lying on a single dsRNA into PKR monomers bound to separate molecules of dsRNA.20 This explanation is not compatible with the linkage model for heparin-induced PKR dimerization (Fig. 5a), where one heparin binds to each PKR monomer and addition of excess heparin would not induce dimer dissociation. A bell-shaped curve would result if P2H2 were inactive and P2H represented the active species; however, there is no direct evidence for such an effect. Alternatively, the inhibition at high heparin concentrations may be associated with a lower-affinity binding of additional heparin (or heparins) at other binding sites that block PKR. The fluorescence anisotropy stoichiometry measurements indicate the formation of a 1:1 PH complex, but we cannot rule out the binding of additional heparin (or heparins) to the dimeric P2H or P2H2 species.
An earlier study proposed that heparin activation of PKR does not proceed via a dimerization mechanism.26 Point mutations that reportedly block PKR dimerization do not affect heparin binding and activation.26 However, these mutations are within dsRBD. We have demonstrated that the isolated dsRBD does not self-associate or interact with full-length PKR,46 and crystallographic and mutagenesis studies have localized the PKR dimer interface within the kinase domain.13,17
Interestingly, a catalytically inactive His-tagged K296R mutant of PKR can be phosphorylated by wild-type PKR upon activation by dsRNA, but not by heparin.25 This result suggests that PKR activated by heparin is incapable of mediating intermolecular autophosphorylation and implies that there is a fundamental difference in the catalytic activities of dsRNA-activated and heparin-activated enzymes. However, heparin-activated PKR can phosphorylate the substrate eIF2α, indicating that this enzyme form is competent for transphosphorylation. Similarly, PKR activated in the absence of either dsRNA or heparin upon incubation at high concentrations is competent to phosphorylate the K296R substrate.16 Thus, the fundamental catalytic activities of PKR activated by the three different routes may be similar. Possibly, K296R PKR becomes incompetent as a substrate upon binding heparin.
Because heparin is found in secretory granules in mast cells and is not present in the cytosol, PKR is not likely to be activated by heparin in vivo. Given that PKR can be activated by multiple polyanionic species,24 the interaction of PKR with heparin is likely a nonspecific reaction that is not relevant to its antiviral role. However, the fact that molecules as small as ~2000 Da (hexasaccharide) are capable of activating PKR indicates that small-molecule activation of PKR is feasible and provides the impetus to search for novel and specific activators of PKR. Potentially, small molecules that bind within the PKR cationic cleft and enhance dimerization similarly to heparin can be identified. There is a precedent for the discovery of synthetic small-molecule enzyme activators.47 Another mediator of the interferon antiviral response, RNase L, is activated by dimerization induced by the binding of 2′,5′-linked oligoadenylates.48,49 High-throughput screening has identified several small-molecule RNase L activators with antiviral properties that act by inducing dimerization.50
Materials and Methods
All reagents used were reagent grade and purchased from Fisher Scientific, except as noted. Heparin oligosaccharides were obtained from V-labs, Inc. (Covington, LA). BODIPY-FL-hydrazide was purchased from Invitrogen, Inc.
BODIPY-FL-hydrazide was coupled to heparin oligosaccharides by a reductive amination of the reducing end carbonyl with the hydrazide amine group present within the fluorophore.31 Heparin oligosaccharide (5 mM) dissolved in water (5 mM) was combined with 5 mM BODIPY-FL-hydrazide dissolved in methanol at a 1:5 ratio and incubated for 1 h at room temperature. The reaction mixture was dried in a SpeedVac and resuspended in 10 μl of dimethyl sulfoxide/glacial acetic acid 17:3 (vol/vol). The reaction mixture was then incubated for 4 h at room temperature, followed by a reduction of the imine group with 10 μl of 1 M sodium borohydride (reducing agent). The mixture was frozen in dry ice, dried in a SpeedVac, and resuspended in a small volume of methanol/100 mM ammonium acetate 1:1 (vol/vol). The mixture was incubated at room temperature for 1 h and diluted 10-fold with 100 mM ammonium acetate. The conjugate was purified on an Oasis HLB column (Waters). The column was rinsed with methanol, followed by 100 mM ammonium acetate. The reaction mixture was applied to the column and incubated for 10 min. The column was washed with 100 mM ammonium acetate, and then a step gradient with increasing concentrations of methanol was used for elution. Unlabeled heparin oligosaccharides do not bind to the column, the conjugate elutes between 20% and 30% methanol, and free dye elutes at 40% methanol and above.
PKR was purified as previously described,16 with the following modifications. The poly(rI)/poly(rC) dsRNA affinity column was replaced with hydroxyapatite (CHT ceramic hydroxyapatite; Bio-Rad). Following the heparin–Sepharose affinity column, PKR-containing fractions were pooled, diluted 1:1 with buffer HA [20 mM potassium phosphate and 10 mM β-mercaptoethanol (pH 7.0)], and applied to a 70-ml CHT column at a flow rate of 1 ml/min. The column was washed with buffer HA until the absorbance and conductivity had returned to baseline, followed by one column volume of 40% HB [HB contains 400 mM potassium phosphate and 10 mM β-mercaptoethanol (pH 7.0)]. PKR was eluted with a gradient of 40–100% HB. The peak fractions elute at ~55% HB. Unless otherwise indicated, measurements were performed in AU75 buffer [75 mM NaCl, 20 mM Hepes, 0.1 mM ethylenediaminetetraacetic acid, and 0.1 mM tris(2-carboxyethyl)phosphine (pH 7.5)]. Equilibration was performed using spin columns packed with Biogel P6 (Bio-Rad). The PKR kinase domain comprising residues 242–551 containing the inactivating mutation K296R was expressed as previously described.51 Site-directed mutants were produced using the Quik-Change method and were verified by DNA sequencing.
Fluorescence anisotropy titrations were conducted using a Jobin-Yvon Horiba Fluoromax-3 spectrofluorometer equipped with Glan–Thompson polarizers. Fluorescence anisotropies of heparin oligosaccharides were measured in a 1 cm×0.3 cm cuvette at 20 °C at an excitation wavelength of 488 nm and at an emission wavelength of 515 nm. Kd measurements were performed using 15 nM labeled heparin oligosaccharide with spectral bandwidths of 6 nm for excitation and emission. Stoichiometries were measured using 1 μM bdp8 with spectral bandwidths of 4 nm for excitation and emission. Fluorescence intensities at each polarizer orientation were integrated for 5 s and corrected for background fluorescence. Each data point represents the average of three measurements. PKR binding kinetics were examined to determine the minimum time needed to reach equilibrium. In all cases, the equilibration time was less than 30 s. Thus, titrations were performed with a 5-min equilibration time between measurements. The binding isotherms were fitted, using the program KaleidaGraph (Synergy Software), to the hyperbolic binding model:
| (2) |
where rave is the average anisotropy, rf and rb are the anisotropies of the free and bound forms of the BODIPY-labeled heparins, and [P] is the PKR concentration. The fluorescence intensity or wavelength maxima of the labeled heparins did not change upon binding to PKR (E.A. and J.L.C., data not shown), so it was not necessary to correct the anisotropies for intensity effects. Fluorescence competition titrations were fitted as previously described32,52 using the program IGOR PRO (Wave-metrics, Inc.). Stoichiometry titrations where [H]>Kd were fitted to a quadratic binding model:
| (3) |
where [C] is the concentration of the PKR–heparin complex, N is the number of heparin oligosaccharide binding sites/PKR, [P]0 is the initial concentration of PKR, and [H]0 is the initial concentration of heparin oligosaccharide. The average anisotropy is then given by:
| (4) |
PKR activation assays were performed as previously described.53
Sedimentation velocity analysis was conducted at 20 °C and 50,000 rpm using either interference optics in a Beckman-Coulter XL-I analytical ultracentrifuge or fluorescence optics in an XL-I analytical ultracentrifuge equipped with an AU-FDS detector (AVIV). Double-sector synthetic boundary centerpieces (Beckman-Coulter) and sapphire windows were used for interference measurements. Menisci were matched as previously described.54 Double-sector SedVel60K centerpieces (Spin Analytical, Inc.) and quartz windows were used for fluorescence measurements. Protein partial specific volumes, extinction coefficients, and solvent densities were calculated using SEDNTERP.55 Sedimentation velocity data were analyzed with the time-derivative method using the program DCDT+ to obtain weight-average sedimentation coefficients and normalized g(s*) distributions. c(s) distributions were calculated using SEDFIT.56 Multiple velocity data sets were globally fitted to alternative heteroassociation models using SEDANAL.42
Structures were presented using PyMOL (Schrödinger LLC). Electrostatic surfaces were generated using APBS,57 with the PyMOL plug-in developed by M. G. Lerner and H. A. Carlson (University of Michigan, Ann Arbor, MI). dp8 was docked onto the kinase domain using AutoDock Vina.45 The coordinates for dp8 were obtained from Protein Data Bank ID 1HPN,44 and the kinase domain coordinates were obtained from Protein Data Bank ID 2A1A.13
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grant AI-53615 to J.L.C.
Abbreviations used
- PKR
protein kinase R
- dsRNA
double-stranded RNA
- eIF2α
α subunit of eukaryotic initiation factor 2
- dsRBD
dsRNA binding domain
- dp8
heparin octasaccharide
- dp6
heparin hexasaccharide
- bdp8
BODIPY-labeled dp8
- dp2
heparin disaccharide
Footnotes
Supplementary Data Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2011.09.025
References
- 1.Toth AM, Zhang P, Das S, George CX, Samuel CE. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:369–434. doi: 10.1016/S0079-6603(06)81010-X. [DOI] [PubMed] [Google Scholar]
- 2.Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barber GN. Host defense, viruses and apoptosis. Cell Death Differ. 2001;8:113–126. doi: 10.1038/sj.cdd.4400823. [DOI] [PubMed] [Google Scholar]
- 4.Garcia MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Koromilas AE, Roy S, Barber GN, Katze MG, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- 6.Meurs E, Galabru J, Barber GN, Katze MG, Hovanessian AG. Tumor supressor function of interferon-induced double-stranded RNA activated protein kinase. Proc. Natl Acad. Sci. USA. 1993;90:232–236. doi: 10.1073/pnas.90.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 5′-Triphosphate-dependent activation of PKR by RNAs with short stem–loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 8.Patel CV, Handy I, Goldsmith T, Patel RC. PACT, a stress-modulated cellular activator of interferon-induced double-stranded RNA-activated protein kinase, PKR. J. Biol. Chem. 2000;275:37993–37998. doi: 10.1074/jbc.M004762200. [DOI] [PubMed] [Google Scholar]
- 9.Patel RC, Sen GC. PACT, a protein activator of the interferon-induced protein kinase PKR. EMBO J. 1998;17:4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded RNA binding motif: interference and much more. Nat. Rev. Mol. Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 12.Nanduri S, Carpick BW, Yang Y, Williams BR, Qin J. Structure of the double-stranded RNA binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 14.Robertson HD, Manche L, Mathews MB. Paradoxical interactions between human delta hepatitis agent RNA and the cellular protein kinase PKR. J. Virol. 1996;70:5611–5617. doi: 10.1128/jvi.70.8.5611-5617.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole JL. Activation of PKR: an open and shut case? Trends Biochem. Sci. 2007;32:57–62. doi: 10.1016/j.tibs.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemaire PA, Lary J, Cole JL. Mechanism of PKR activation: dimerization and kinase activation in the absence of double-stranded RNA. J. Mol. Biol. 2005;345:81–90. doi: 10.1016/j.jmb.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 17.Dey M, Cao C, Dar AC, Tamura T, Ozato K, Sicheri F, Dever TE. Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition. Cell. 2005;122:901–913. doi: 10.1016/j.cell.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 18.Hunter T, Hunt T, Jackson RJ, Robertson HD. The characteristics of inhibition of protein synthesis by double-stranded ribonucleic acid in reticulocyte lysates. J. Biol. Chem. 1975;250:409–417. [PubMed] [Google Scholar]
- 19.Manche L, Green SR, Schmedt C, Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kostura M, Mathews MB. Purification and activation of the double-stranded RNA-dependent eIF-2 kinase DAI. Mol. Cell. Biol. 1989;9:1576–1586. doi: 10.1128/mcb.9.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinicke LA, Wong CJ, Lary J, Nallagatla SR, Diegelman-Parente A, Zheng X, et al. RNA dimerization promotes PKR dimerization and activation. J. Mol. Biol. 2009;390:319–338. doi: 10.1016/j.jmb.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemaire PA, Anderson E, Lary J, Cole JL. Mechanism of PKR activation by dsRNA. J. Mol. Biol. 2008;381:351–360. doi: 10.1016/j.jmb.2008.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nallagatla SR, Toroney R, Bevilacqua PC. Regulation of innate immunity through RNA structure and the protein kinase PKR. Curr. Opin. Struct. Biol. 2011;21:119–127. doi: 10.1016/j.sbi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hovanessian AG, Galabru J. The double-stranded RNA-dependent protein kinase is also activated by heparin. Eur. J. Biochem. 1987;167:467–473. doi: 10.1111/j.1432-1033.1987.tb13360.x. [DOI] [PubMed] [Google Scholar]
- 25.George CX, Thomis DC, McCormack SJ, Svahn CM, Samuel CE. Characterization of the heparin-mediated activation of PKR, the interferoninducible RNA-dependent protein kinase. Virology. 1996;221:180–188. doi: 10.1006/viro.1996.0364. [DOI] [PubMed] [Google Scholar]
- 26.Patel RC, Sen GC. Requirement of PKR dimerization mediated by specific hydrophobic residues for its activation by double-stranded RNA and its antigrowth effects in yeast. Mol. Cell. Biol. 1998;18:7009–7019. doi: 10.1128/mcb.18.12.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel RC, Stanton P, Sen GC. Role of the amino-terminal residues of the interferon-induced protein kinase in its activation by double-stranded RNA and heparin. J. Biol. Chem. 1994;269:18593–18598. [PubMed] [Google Scholar]
- 28.Fasciano S, Hutchins B, Handy I, Patel RC. Identification of the heparin-binding domains of the interferon-induced protein kinase, PKR. FEBS J. 2005;272:1425–1439. doi: 10.1111/j.1742-4658.2005.04575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu W, Hofstetter W, Wei X, Guo W, Zhou Y, Pataer A, et al. Double-stranded RNA-dependent protein kinase-dependent apoptosis induction by a novel small compound. J. Pharmacol. Exp. Ther. 2009;328:866–872. doi: 10.1124/jpet.108.141754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elazar M, Liu M, McKenna SA, Liu P, Gehrig EA, Puglisi JD, et al. The anti-hepatitis C agent nitazoxanide induces phosphorylation of eukaryotic initiation factor 2alpha via protein kinase activated by double-stranded RNA activation. Gastroenterology. 2009;137:1827–1835. doi: 10.1053/j.gastro.2009.07.056. [DOI] [PubMed] [Google Scholar]
- 31.Skidmore M, Atrih A, Yates E, Turnbull JE. Labelling heparan sulphate saccharides with chromophore, fluorescence and mass tags for HPLC and MS separations. Methods Mol. Biol. 2009;534:157–169. doi: 10.1007/978-1-59745-022-5_12. [DOI] [PubMed] [Google Scholar]
- 32.Lemaire PA, Tessmer I, Craig R, Erie DA, Cole JL. Unactivated PKR exists in an open conformation capable of binding nucleotides. Biochemistry. 2006;45:9074–9084. doi: 10.1021/bi060567d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole JL. Analysis of PKR activation using analytical ultracentrifugation. Macromol. Biosci. 2010;10:703–713. doi: 10.1002/mabi.201000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal. Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 35.Stafford WF. Boundary analysis in sedimentation transport experiments: a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal. Biochem. 1992;203:295–301. doi: 10.1016/0003-2697(92)90316-y. [DOI] [PubMed] [Google Scholar]
- 36.Khan S, Gor J, Mulloy B, Perkins SJ. Semi-rigid solution structures of heparin by constrained X-ray scattering modelling: new insight into heparin–protein complexes. J. Mol. Biol. 2010;395:504–521. doi: 10.1016/j.jmb.2009.10.064. [DOI] [PubMed] [Google Scholar]
- 37.Lemaire PA. Biochemical and Biophysical Analysis of the Activation of Protein Kinase R. PhD Thesis. University of Connecticut; 2006. [Google Scholar]
- 38.Dam J, Velikovsky CA, Mariuzza RA, Urbanke C, Schuck P. Sedimentation velocity analysis of heterogeneous protein–protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s) Biophys. J. 2005;89:619–634. doi: 10.1529/biophysj.105.059568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong CJ, Launer-Felty K, Cole JL. Analysis of PKR–RNA interactions by sedimentation velocity. Methods Enzymol. 2011;488:59–79. doi: 10.1016/B978-0-12-381268-1.00003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wyman J, Gill SJ. Binding and Linkage. University Science Books; Mill Valley, CA: 1990. [Google Scholar]
- 41.Wowor AJ, Yu D, Kendall DA, Cole JL. Energetics of SecA dimerization. J. Mol. Biol. 2011;408:87–98. doi: 10.1016/j.jmb.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stafford WF, Sherwood PJ. Analysis of heterologous interacting systems by sedimentation velocity: curve fitting algorithms for estimation of sedimentation coefficients, equilibrium and kinetic constants. Biophys. Chem. 2004;108:231–243. doi: 10.1016/j.bpc.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 43.Pavlov G, Finet S, Tatarenko K, Korneeva E, Ebel C. Conformation of heparin studied with macromolecular hydrodynamic methods and X-ray scattering. Eur. Biophys. J. 2003;32:437–449. doi: 10.1007/s00249-003-0316-9. [DOI] [PubMed] [Google Scholar]
- 44.Mulloy B, Forster MJ, Jones C, Davies DB. NMR and molecular-modelling studies of the solution conformation of heparin. Biochem. J. 1993;293:849–858. doi: 10.1042/bj2930849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ucci JW, Kobayashi Y, Choi G, Alexandrescu AT, Cole JL. Mechanism of interaction of the double-stranded RNA (dsRNA) binding domain of protein kinase R with short dsRNA sequences. Biochemistry. 2007;46:55–65. doi: 10.1021/bi061531o. [DOI] [PubMed] [Google Scholar]
- 47.Zorn JA, Wells JA. Turning enzymes ON with small molecules. Nat. Chem. Biol. 2010;6:179–188. doi: 10.1038/nchembio.318. [DOI] [PubMed] [Google Scholar]
- 48.Cole JL, Carroll SS, Blue ES, Viscount T, Kuo LC. Activation of RNase L by 2′,5′-oligoadenylates. Biophysical characterization. J. Biol. Chem. 1997;272:19187–19192. doi: 10.1074/jbc.272.31.19187. [DOI] [PubMed] [Google Scholar]
- 49.Dong B, Silverman RH. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J. Biol. Chem. 1995;270:4133–4137. doi: 10.1074/jbc.270.8.4133. [DOI] [PubMed] [Google Scholar]
- 50.Thakur CS, Jha BK, Dong B, Das Gupta J, Silverman KM, Mao H, et al. Small-molecule activators of RNase L with broad-spectrum antiviral activity. Proc. Natl Acad. Sci. USA. 2007;104:9585–9590. doi: 10.1073/pnas.0700590104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson E, Cole JL. Domain stabilities in protein kinase R (PKR): evidence for weak interdo-main interactions. Biochemistry. 2008;47:4887–4897. doi: 10.1021/bi702211j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Talavera MA, De La Cruz EM. Equilibrium and kinetic analysis of nucleotide binding to the DEAD-box RNA helicase DbpA. Biochemistry. 2005;44:959–970. doi: 10.1021/bi048253i. [DOI] [PubMed] [Google Scholar]
- 53.Anderson E, Quartararo C, Brown RS, Shi Y, Yao X, Cole JL. Analysis of monomeric and dimeric phosphorylated forms of protein kinase R. Biochemistry. 2010;49:1217–1225. doi: 10.1021/bi901873p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cole JL, Lary JW, Moody PT, Laue TM. Analytical ultracentrifugation: sedimentation velocity and sedimentation equilibrium. Methods Cell Biol. 2008;84:143–179. doi: 10.1016/S0091-679X(07)84006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding S, Rowe A, Horton J, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge, UK: 1992. pp. 90–125. [Google Scholar]
- 56.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.