Abstract
We recently reported a chemical genetic method for generating bivalent inhibitors of protein kinases. This method relies on the use of the DNA repair enzyme O6-alkylguanine-DNA alkyltransferase (AGT) to display an ATP-competitive inhibitor and a ligand that targets a secondary binding domain. With this method potent and selective inhibitors of the tyrosine kinases SRC and ABL were identified. Here, we dissect the molecular determinants of the potency and selectivity of these bivalent ligands. Systematic analysis of ATP-competitive inhibitors with varying linker lengths revealed that SRC and ABL have differential sensitivities to ligand presentation. Generation of bivalent constructs that contain ligands with differential affinities for the ATP-binding sites and SH3 domains of SRC and ABL demonstrated the modular nature of inhibitors based on the AGT scaffold. Furthermore, these studies revealed that the interaction between the SH3 domain ligand and the kinase SH3 domain is the major selectivity determinant amongst closely-related tyrosine kinases. Finally, the potency of bivalent inhibitors against distinct phospho-isoforms of SRC was determined. Overall, these results provide insight into how individual ligands can be modified to provide more potent and selective bivalent inhibitors of protein kinases.
Introduction
Protein kinases are an important family of signaling enzymes that use the cofactor adenosine-5′-triphosphate (ATP) to phosphorylate intra-cellular protein substrates.1 These phosphorylation events are important regulators of signal transduction pathways in cells and the activities of protein kinases are tightly regulated. Aberrant protein kinase activity has been correlated with a number of disease states.2–4 For this reason, there has been a great deal of interest in the development of tools that are able to control protein kinase function. Ligands that are able to selectively block the catalytic activity of protein kinases are valuable tools for studying signal transduction and can provide insight into kinase regulation. However, it is extremely challenging to generate selective ligands for specific kinases due the large size of this enzyme family (> 500 kinases in humans). Therefore, new strategies that facilitate the discovery of selective kinase ligands are of general interest. In addition to the important role of selective inhibitors as functional tools to study kinase function, selective ligands can also provide insight into the regulation and dynamics of kinase activity.
Bivalent ligands that target two distinct binding sites have proven to be potent and selective kinase inhibitors. All protein kinases are bisubstrate enzymes that contain separate ATP- and protein substrate-binding sites. In addition, many protein kinases contain other binding sites that are either located in the catalytic domain or in separate functional domains. These binding sites regulate kinase function and are responsible for proper cellular localization. The interplay between the regulatory and binding sites of protein kinases is believed to be a major contributor to intra-cellular signaling specificity. Therefore, kinases contain diverse binding sites that can potentially be targeted with bivalent inhibitors. A number of strategies have been developed for the generation of bivalent inhibitors of protein kinases.5–13 One successful approach is the use of bisubstrate inhibitors that simultaneously target both the ATP- and protein substrate-binding sites of protein kinases. For example, bisubstrate inhibitors of the serine/threonine kinase cAMP-dependent protein kinase (PKA) have been generated by linking an ATP-competitive small molecule inhibitor to a short pseudo-substrate peptide through a flexible tether.6 Cole and co-workers have successfully identified bisubstrate inhibitors of PKA and the tyrosine kinase Insulin Receptor Kinase (IRK) by linking ATPγS to peptide ligands that occupy the substrate binding sites of these kinases.7, 8 Schepartz and co-workers have demonstrated that the promiscuous kinase inhibitor K252a can be converted into a selective bisubstrate inhibitor of PKA by tethering it to a miniature protein that contains a specific binding epitope for this kinase.9 Furthermore, Lawrence and co-workers were able to use directed molecular evolution to generate a bisubstrate inhibitor of the serine/threonine kinase AKT from a protein substrate-competitive peptide ligand.10 Bivalent inhibitors possessing ligands that target sites that are not involved in substrate binding have also been developed. Ghosh and co-workers used a non-covalent fragment assembly technique to discover a cyclic peptide/staurosporine conjugate that is an extremely potent inhibitor of PKA. While staurosporine targets the ATP-binding cleft of this kinase, kinetic analysis demonstrated that the cyclic peptide is non-competitive with a peptide substrate.11, 12 Finally, bivalent inhibitors that target the protein substrate-binding sites and the SRC homology 2 (SH2) domains of SRC-family kinases have been described. These inhibitors were found to potently block the catalytic activity of several SRC-family kinase members and demonstrated impressive selectivity within this tyrosine kinase subfamily.13, 14 An important attribute of previously described bivalent inhibitors is their increased potency compared to their monovalent ligand components. In addition, many bivalent inhibitors exhibit increased selectivity for their desired targets.
Recently, we reported a chemical genetic method for generating bivalent inhibitors of the tyrosine kinases SRC and ABL.15 This system relies on the use of the DNA repair enzyme O6-alkylguanine-DNA alkyltransferase (AGT) to display an SH3 domain ligand and an ATP-competitive inhibitor.16–18 By linking an ATP-competitive inhibitor to an AGT fusion protein containing a polyproline (PP) motif peptide that is selective for the SRC homology 3 (SH3) domain of ABL, a bivalent inhibitor that is highly selective for this kinase was generated. A potent and selective inhibitor of SRC was obtained by linking the same ATP-competitive inhibitor to an AGT fusion protein that contains a SRC-family selective SH3 domain ligand. Thus, bivalent inhibitor selectivity is conferred by an interaction outside of the catalytic domain. As most secondary binding domain interactions are less conserved than binding sites in the catalytic domain, this method should allow for the identification of highly selective bivalent ligands for a number of kinases. A unique aspect of our method is that both ligands are displayed from a protein scaffold. Therefore, if both ligands are oriented properly a minimal entropic penalty will be paid due to the rigidity of the AGT protein. However, the overall size and lack of flexibility of the AGT protein presents a number of challenges for obtaining optimal ligand display. Here we dissect the interactions of bivalent inhibitors based on the AGT scaffold with SRC and ABL. First, the effect of the tether length between AGT and the ATP-competitive inhibitor is explored. Next, we analyze how the affinity of each individual ligand (ATP-competitive inhibitor and SH3 domain ligand) contributes to overall bivalent inhibitor potency and selectivity. Finally, we compare the potencies of our bivalent inhibitors against different phospho-isoforms of SRC. The results from these studies provide insight into how more potent and selective bivalent inhibitors can be obtained.
2. Results and Discussion
Architecture of AGT-Based Bivalent Inhibitors and Tyrosine Kinase Constructs
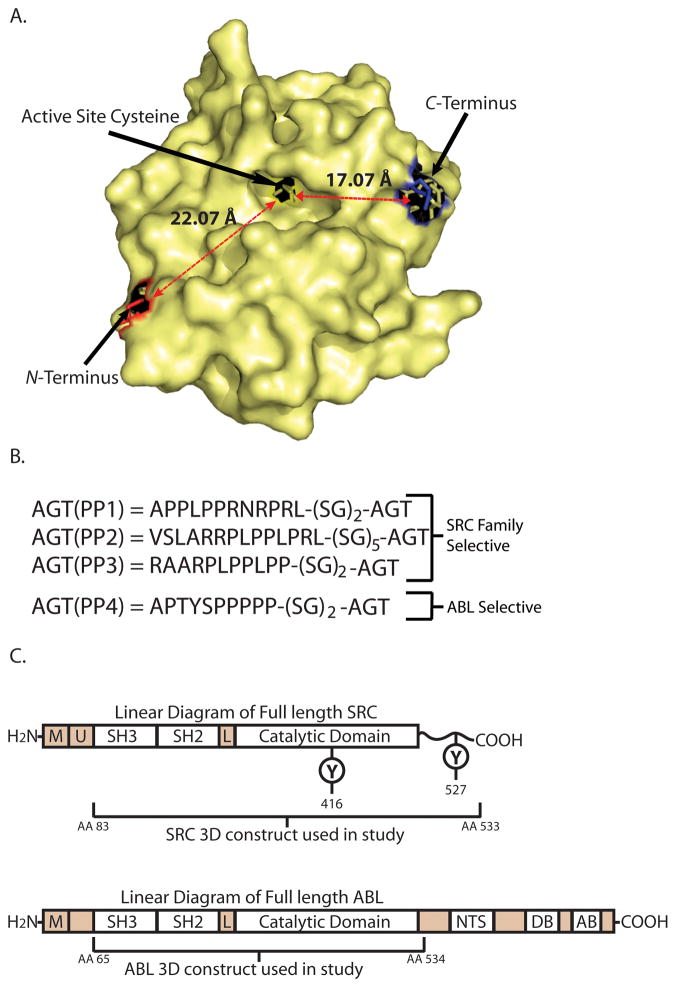
A key component of any bivalent inhibitor strategy is the linker connecting the two ligands. We have used the 20 kDa protein O6-alkylguanine-DNA alkyltransferase (AGT) as the linker in our strategy due to the unique ability of this enzyme to self-label an active site cysteine with a diverse range of O6-benzylguanine derivatives.16–18 This property allows for the rapid, selective, and covalent linkage of diverse ATP-competitive kinase inhibitors to the AGT protein scaffold. Ligands displayed from the active site of AGT are relatively accessible to potential binding partners because the modified cysteine is located in a relatively shallow hydrophobic pocket (Figure 1A). Additional binding elements can be presented from the AGT scaffold by generating AGT fusion proteins that contain N- or C-terminal peptide ligands (Figure 1A). Both the N- and C-terminus of AGT are located on the same face as the active site, which should allow for favorable bivalent interactions with target proteins (Figure 1A). In addition, the N- and C-termini of AGT are relatively close to the active site (the N-terminus is ≈22 Å and the C-terminus is ≈17 Å from the modified cysteine).
Figure 1.
AGT and Kinase Constructs. (A). A crystal structure of AGT denoting the relative location and distances of the N- and C-terminus from the active site cysteine (PDP entry 1EH8). (B). The four PP motif AGT fusions that were used in this study. (C). Structures of full length SRC and ABL. The domains contained in the SRC-3D and ABL-3D constructs used in this study are indicated. The location of two regulatory phosphorylation sites (Y461, and Y527) are shown on the linear diagram of SRC: [M] = membrane binding region, [U] = unique region, [L] = linker, [NTS] = nuclear-transport signal, [DB] = DNA binding domain, [AB] = actin-binding domain.
In previous studies, we observed little difference between SH3 domain ligands that were displayed from N- or C-terminus of AGT.15 For this reason, only N-terminal peptide fusion constructs were used in this study. When determining which AGT-peptide fusions to generate, we selected AGT constructs that contain the peptides APPLPPRNRPRL, VSLARRPLPPLPRL, RAARPLPPLPP, APTYSPPPPP (AGT(PP1)-AGT(PP4), Figure 1B). These PP motif peptides have previously been characterized to bind with micromolar affinity to the SH3 domains of either SRC or ABL kinase.19–22 Based on prior studies, the PP motifs in AGT(PP1), AGT(PP2), and AGT(PP3) should show selectivity towards the SH3 domain of SRC and the PP motif in AGT(PP4) should be selective for the SH3 domain ABL. His6-tagged versions of AGT(PP1), AGT(PP2), AGT(PP3), and AGT(PP4) were generated by overlap extension PCR, expressed in bacteria, and purified by immobilized metal ion affinity chromatography.
SRC and ABL are both multi-domain non-receptor tyrosine kinases. Although SRC and ABL have very different C-terminal regions, they share highly homologous SH3, SH2, and catalytic domains (Figure 1C).23 In order to simplify the analysis of our bivalent inhibitors with SRC and ABL, we have utilized protein constructs that contain only the SH3, SH2, and catalytic domains of these kinases. The SRC construct used in this study (SRC-3D) contains residues 83–533 of this kinase and the ABL construct (ABL-3D) contains residues 65–534. For selectivity studies, constructs that contain only the SH3, SH2, and catalytic domains of the SRC-family kinases HCK and LCK were also generated. All kinase constructs were co-expressed in E. coli with the tyrosine phosphatase YopH, which ensures that they are quantitatively dephosphorylated at all regulatory positions.24 Lack of tyrosine phosphorylation was confirmed by Western Blot analysis (data not shown).
ATP-Competitive Inhibitor Tether Length
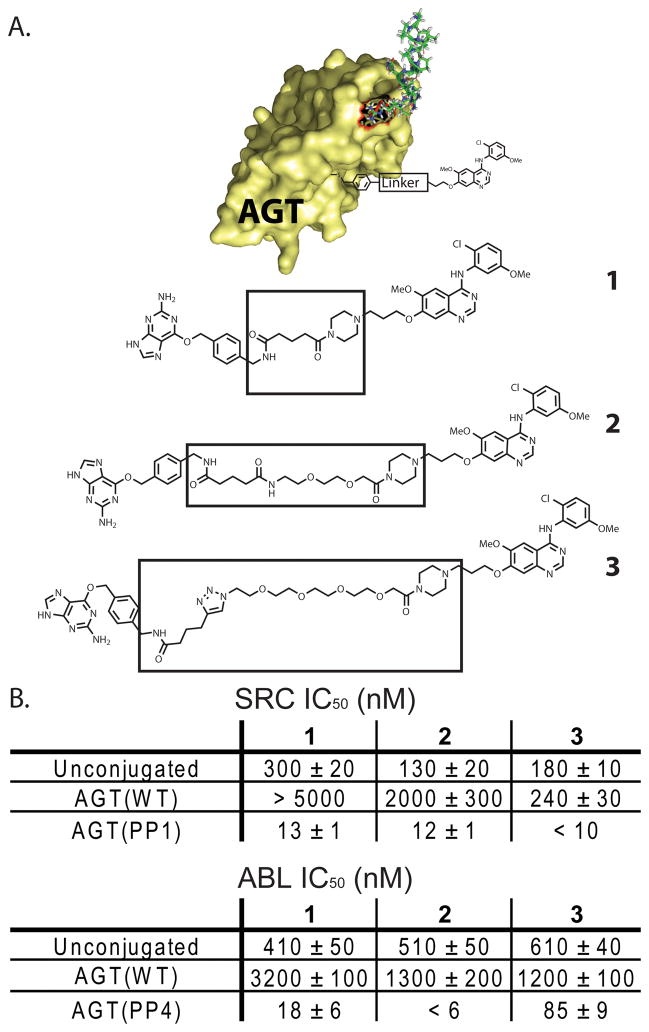
Obtaining the proper display of each ligand from the AGT scaffold is an important factor in the design of potent bivalent kinase inhibitors. If the tether connecting a ligand to the AGT scaffold is too short, potency may be compromised due to a sterically obstructed interaction. However, if the tether is longer than the optimal distance, an energetic penalty in conformational entropy will be observed. We have previously demonstrated that bivalent inhibitors based on the AGT scaffold are not greatly affected by the distance between the SH3 domain ligand and the AGT scaffold.15 This is true for SH3 domain ligands displayed from the N- or C-terminus of AGT.15 To further investigate the role of ligand display, we systematically analyzed how the length of the linker between the active site cysteine of AGT and the ATP-competitive ligand affected the overall potency of bivalent kinase inhibitors. A series of constructs that contain the same ATP-competitive inhibitor but variable linker lengths were generated and tested for their ability to inhibit SRC and ABL (1–3) (Figure 2A). These inhibitors are all based on a 4-anilinoquinazoline scaffold that is a moderately potent inhibitor of SRC and ABL kinases.25, 26 Unconjugated derivatives 1–3 are nearly equipotent inhibitors of SRC, with 1 demonstrating a slightly reduced potency against this enzyme (Figure 2B). For ABL, all three unconjugated inhibitors (1–3) are identical in potency. When 1–3 are conjugated to AGT(WT), a decrease in potency is observed compared to the unconjugated inhibitors. It would be expected that AGT(WT) conjugates that contain a longer tether length between the protein and inhibitor would have lower IC50s against SRC and ABL due to increased accessibility of the ATP-competitive inhibitor. Indeed, this is observed for SRC, as AGT(WT)-3 is at least 20 times more potent than AGT(WT)-1. A similar, but less striking, trend is observed for these conjugates against ABL. 1–3 are all significantly more potent inhibitors of SRC when conjugated to a PP motif-containing AGT fusion protein (AGT(PP1)). While AGT(PP1)-3, which contains the longest linker length, is the most potent inhibitor of SRC, there is not a strong linker length dependence for this kinase. In contrast, the length of the tether between the ATP-competitive inhibitor and AGT scaffold has a much larger effect on the ability of bivalent inhibitors to block the catalytic activity of ABL (Figure 2B). AGT(PP4)-2, which contains an intermediate linker length, is at least three-fold more potent than AGT(PP4)-1 and at least fourteen-fold more potent than AGT(PP4)-3. The more distinct linker length dependence observed for ABL may reflect a difference in the orientation between its SH3 domain and catalytic domain relative to SRC. However, further structural studies are needed to confirm this.
Figure 2.
IC50 values of bivalent inhibitors with variable tether lengths between the ATP-competitive inhibitor and AGT. (A). Chemical structures of BG-linked quinazoline inhibitors with variable linker lengths. Each inhibitor contains a benzyl guanine (BG) moiety linked to a chloromethoxyaniline quinazoline inhibitor through a flexible tether. (B). In vitro activities of unconjugated inhibitors 1, 2, and 3 and bivalent conjugates AGT(WT)-1, AGT(WT)-2, AGT(WT)-3, AGT(PP1)-1, AGT(PP1)-2, AGT(PP1)-3 against SRC-3D. IC50 values of unconjugated 1, 2, and 3 and bivalent conjugates AGT(WT)-1, AGT(WT)-2, AGT(WT)-3, AGT(PP4)-1, AGT(PP4)-2, AGT(PP4)-3 against ABL-3D. All protein-small molecule conjugates were prepared in two independent labeling reactions, and values shown are the average of four assays ± SEM.
Contribution of the ATP-Competitive Inhibitor
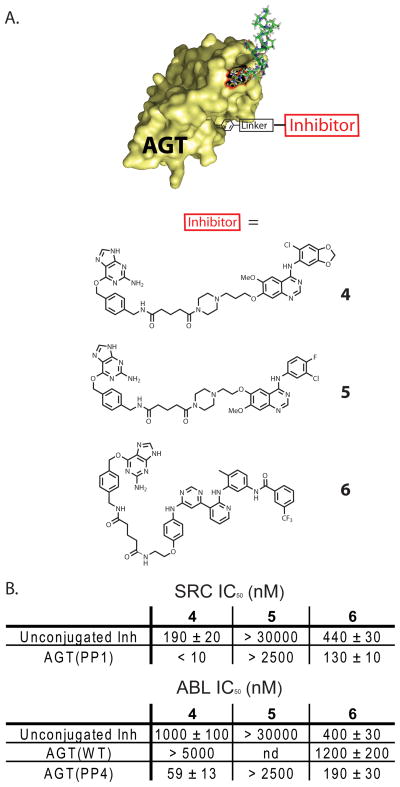
Next, we explored how the affinity of the ATP-competitive ligand that is displayed from the AGT scaffold affects bivalent inhibitor potency. To test this, a small panel of BG-linked inhibitors that contain ATP-competitive ligands with variable affinities for the ATP-binding sites of SRC and ABL were generated (4, 5, and 6, Figure 3A). All three BG-linked conjugates have a tether length roughly equivalent to parent compound 1. Analogue 4 is based on the same 4-anilinoquinazoline scaffold as parent compound 1 but contains 5-chlorobenzo[1,3]dioxol-4-ylamine at the 4-position rather than 2-chloro-5-methoxyaniline.26 This substitution results in unconjugated analogue 4 being a 1.5-fold more potent inhibitor of SRC (IC50 = 190 ± 20 nM) and a 2.5-fold weaker inhibitor of ABL (IC50 = 1000 ± 90 nM) (Figure 3B) than parent derivative 1. Analogue 5 is a BG-derivatized version of the highly selective epidermal growth factor receptor kinase (EGFR) inhibitor, gefitinib.27 Despite being structurally similar to 1, compound 5 shows minimal inhibition of SRC and ABL at the highest concentration tested (30 μM) (Figure 3B). Therefore, the selectivity profile of the BG-derivatized version of this inhibitor is similar to its parent compound gefitinib.28, 29 Pyrimidinepyridine 6 is a BG-linked version of a previously-described equipotent inhibitor of SRC and ABL.30 Despite being structurally distinct from 1, 4, and 5, inhibitors based on the pyrimidinepyridine scaffold make similar hydrogen bonds to the hinge region of the ATP-binding site and can be modified with a flexible linker without loss of activity. In contrast to 1, 4, and 5, pyrimidinepyridine inhibitors do not bind the active conformation of their kinase targets but rather to an inactive form called the DFG-out conformation. Analogue 6 is an equipotent inhibitor of SRC (IC50 = 440 ± 30 nM) and ABL (IC50 = 400 ± 30 nM).
Figure 3.
IC50 values of various ATP-competitive inhibitors conjugated to AGT(PP1). (A). Chemical structures of BG-linked, ATP-competitive kinase inhibitors 4–6. (B). In vitro activities of unconjugated inhibitors 4, 5, and 6 and bivalent conjugates AGT(PP1)-4, AGT(PP1)-5, AGT(PP1)-6 against SRC-3D. In vitro activities of unconjugated 4, 5, 6 and bivalent conjugates AGT(WT)-4, AGT(WT)-6, AGT(PP4)-4, AGT(PP4)-5, AGT(PP4)-6 against ABL-3D. All protein-small molecule conjugates were prepared in two independent labeling reactions, and values shown are the average of four assays ± SEM.
4–6 were conjugated to either AGT(PP1) or AGT(PP4) and tested for their ability to inhibit SRC or ABL. The AGT(PP1)-4 conjugate is a more potent inhibitor of SRC than AGT(PP1)-1 (Figure 3A), which reflects the increased affinity of inhibitor 4 for the ATP-binding site of SRC. Both AGT(PP1)-1 and AGT(PP1)-4 are 20-to-25 times more potent inhibitors of SRC than their unconjugated analogues 1 and 4, which demonstrates a consistent binding contribution from the SH3 domain ligand. For ABL, AGT(PP4)-4 is a 3-fold less potent inhibitor than AGT(PP4)-1. AGT(WT)-4 is at least 1.5 fold less potent inhibitor of ABL than AGT(WT)-1. The overall drop in potency demonstrated by the AGT(PP4)-4 conjugate compared to AGT(PP4)-1 and AGT(WT)-4 compared to AGT(WT)-1 mirrors the weaker inhibition exhibited by the unconjugated derivative 4 against ABL. However, both AGT(PP4) based protein-small molecule conjugates are at least 15-fold more potent inhibitors of ABL than the free BG-linked analogues 1 and 4. These data demonstrate that small differences in the affinity of the ATP-competitive ligand are directly correlated to the relative potencies of the corresponding bivalent inhibitors. Therefore, the affinity and selectivity of AGT-based bivalent inhibitors can rationally be tuned by modifying the ATP-competitive ligand.
The effectiveness of bivalent inhibitors that contain ligands with little or no affinity for the ATP-binding sites of the kinases being targeted was determined next. Gefitinib analogue 5 was conjugated to AGT(PP1) and AGT(PP4) and the subsequent bivalent inhibitors were tested for their ability to inhibit SRC and ABL (Figure 3B). Despite containing ligands that target the SH3 domains of SRC and ABL, AGT(PP1)-5 and AGT(PP4)-5 show no detectable inhibition at the highest concentration tested (2.5 μM). This observed lack of inhibition is similar to AGT(PP1) and AGT(PP4) constructs that are conjugated to simple alkyl moieties.15 Therefore, despite the similarity of analogue 5 to quinazolines 1 and 4, presentation from the AGT scaffold is not sufficient for kinase inhibition.
AGT constructs conjugated to ATP-competitive ligand 6 were generated (AGT(PP1)-6, AGT(WT)-6, and AGT(PP4)-6) and tested for their ability to inhibit SRC and ABL (Figure 3B). These protein-small molecule conjugates provide insight into the properties of bivalent inhibitors that contain ATP-competitive ligands targeting inactive forms of SRC and ABL (the DFG-out conformation). Despite unconjugated analogue 6 being an almost equally effective inhibitor of SRC and ABL as quinazoline 1, bivalent inhibitors displaying the pyrimidinepyridine ligand from their active sites show only a minimal increase in potency. Indeed, AGT(PP1)-6 and AGT(PP4)-6 are only 2-to-4-fold more potent inhibitors of SRC and ABL than unconjugated derivative 6. Furthermore, AGT(PP4)-6 is only 6-fold more potent inhibitor of ABL than AGT(WT)-6. To ensure that these results are not due to a difference in the binding kinetics of these slow-binding inhibitors, activity assays were repeated with increased pre-incubation times. We observed that the IC50s did not change with increased pre-incubation times (data not sown). There are several possible explanations for the observed lack of increased potency when compared to unconjugated derivative 6. Because the pyrimidinepyridine ligand binds to an inactive conformation of the ATP-biding sites of SRC and ABL, it is possible that inhibitor binding causes these kinases to form more favorable intra-molecular interactions with their own SH3 domain ligands. These increased intra-molecular interactions would reduce the accessibility of the SH3 domains of SRC and ABL to AGT constructs containing SH3 domain ligands. It is also possible that the binding orientation of the pyrimidinepyridine scaffold is not compatible with simultaneous engagement of the ATP-binding site and the SH3 domain, however, further biochemical and biophysical analysis is necessary to resolve the true cause of reduced potency.
SH3 Domain Ligand Affinity
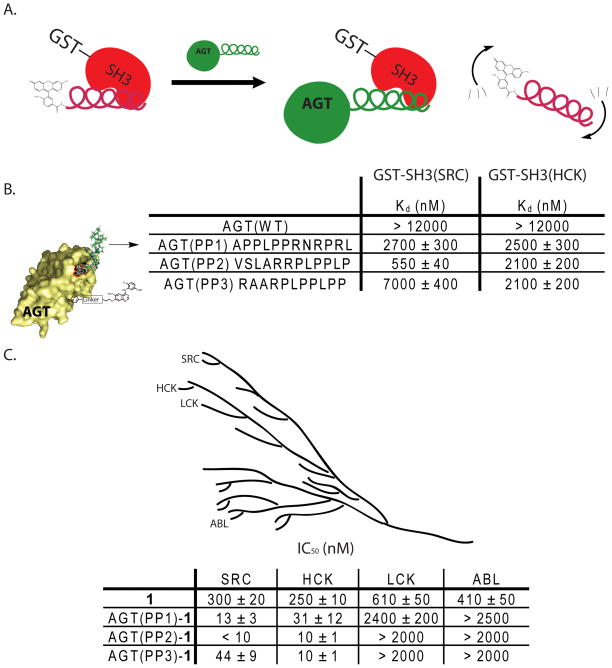
A number of PP motif-containing peptides that target the SH3 domains of SRC and ABL have been identified.19–22 These SH3 domain ligands tend to have dissociation constants in the low micromolar range and exhibit varying degrees of selectivity for their SH3 domain targets.22 We have utilized the ability of these peptides to discriminate between SH3 domains to generate selective bivalent inhibitors of kinases that have highly similar ATP-binding sites. To further dissect the contribution of SH3 domain ligands to overall bivalent inhibitor potency and selectivity, a small panel of AGT constructs containing peptide ligands with varying affinities for the SH3 domain of SRC were selected for study (Figure 4B). To characterize the affinities of these peptide ligands in the context of the AGT scaffolding protein, a fluorescence polarization competition assay was developed. This assay allows the affinity of each AGT fusion to be determined by observing the displacement of a fluorescein-labeled peptide (FAM-AAVSLARRPLPPLP-NH2) from the SH3 domain of SRC (GST-SH3(SRC)) (Figure 4A). As expected AGT(WT) shows no affinity for the SH3 domain of SRC as it contains no SH3 domain binding ligand. The observed Kd values of AGT(PP1)-AGT(PP3) range from 550 nM to 7000 nM, with AGT(PP2) being the highest affinity ligand for the SH3 domain of SRC and AGT(PP3) being the lowest (Figure 4B). Interestingly, the affinities of the AGT/PP motif fusion proteins for the SH3 domain of SRC are very similar to their corresponding free peptides.19–22 Therefore, displaying these ligands from AGT has only a small effect on their ability to access the SH3 domain of SRC. Having determined the affinity of each AGT fusion protein for the SH3 domain of SRC, the contribution of the AGT fusion protein/SH3 domain interaction to overall bivalent inhibitor potency was determined. AGT(PP1), AGT(PP2), and AGT(PP3) were conjugated to 1 and the subsequent protein-small molecule conjugates were tested for their ability to inhibit the catalytic activity of SRC (Figure 3B). The ability of each conjugate to inhibit SRC correlates well with the relative affinity of the SH3 domain peptide ligand displayed from AGT (Figure 4B). Indeed, AGT(PP2)-1 is the most potent inhibitor of SRC and AGT(PP3)-1 is the least. These results further highlight the modular nature of bivalent inhibitors based on the AGT scaffolding protein. Changes in the affinity of individual ligands directly correlate to overall bivalent inhibitor potency.
Figure 4.
Effect of PP motif affinity on bivalent inhibitor potency and selectivity. (A). The fluorescence polarization competition assay that was used to determine the affinities of AGT(WT), AGT(PP1), AGT(PP2), and AGT(PP3) for the SH3 domains of SRC and HCK. The SH3 domains of SRC and HCK were expressed as GST-fusion proteins (GST-SH3(SRC) and GST-SH3(HCK)) and used in a fluorescence polarization competition assay with the peptide FAM-AAVSLARRPLPPLP-NH2. In each assay, an AGT fusion protein was titrated against GST-SH3(SRC) or GST-SH3(HCK) in the presence of FAM-AAVSLARRPLPPLP-NH2 (100 nM when assaying GST-SH3(SRC), 300 nM when assaying GST-SH3(HCK)). (B). Affinities of AGT(WT), AGT(PP1), AGT(PP2), and AGT(PP3) for the SH3 domains of SRC and HCK. (C). Representation of the genetic relationship between the tyrosine kinases SRC, HCK, LCK and ABL. In vitro activities of unconjugated inhibitor 1 and protein-small molecule conjugates AGT(PP1)-1, AGT(PP2)-1, AGT(PP3)-1 against SRC-3D, HCK-3D, LCK-3D and ABL-3D.
Tyrosine Kinase Family Selectivity
We have previously demonstrated that selective bivalent inhibitors of tyrosine kinases with very similar ATP-binding sites can be generated by exploiting specific SH3 domain/PP motif ligand interactions.15 For example, even though ABL and CSK are sensitive to 4-anilinoquinazoline inhibitors and both tyrosine kinases contain SH3 domains, an AGT construct containing a PP motif that is selective for the SH3 domain of SRC can be used to generate a selective bivalent inhibitor of this kinase. Furthermore, the same bivalent inhibitor is able to discriminate between the closely related SRC-family kinases SRC and LCK, based on the higher affinity of SRC’s SH3 domain for the AGT fusion protein. To further characterize how specific SH3 domain ligands can contribute to bivalent inhibitor selectivity, we tested the abilities of AGT(PP1)-1, AGT(PP2)-1, AGT(PP3)-1 to inhibit the tyrosine kinases SRC, ABL, LCK and HCK (Figure 4C). All three bivalent conjugates are highly selective for SRC over ABL, with AGT(PP2)-1 demonstrating a greater than 200-fold lower IC50 for SRC (IC50 = < 10 nM) than ABL (IC50 = > 2000 nM). The selectivity observed for these bivalent inhibitors is consistent with previous studies demonstrating that the SH3 domains of SRC and ABL have distinct preferences for PP motif-containing peptides.21 AGT(PP1)-1, AGT(PP2)-1, AGT(PP3)-1 are also highly selective for SRC over the SRC-family kinase LCK. While it was not possible to directly determine the affinities of the PP motifs contained within these AGT constructs for the SH3 domain of LCK, these results are consistent with previous studies demonstrating that short PP motif peptides do not bind this kinase tightly. The SRC-family kinase HCK is very similar to SRC and is equally sensitive to inhibition by BG-linked derivative 1. AGT(PP1)-1, AGT(PP2)-1, and AGT(PP3)-1 are equipotent inhibitors of SRC and HCK. The potencies of these bivalent inhibitors against HCK suggest that the PP motifs in these AGT fusions have similar affinities for the SH3 domains of this kinase. To determine if this is the case, we performed a fluorescence polarization competition assay with the SH3 domain of HCK (GST-SH3(HCK)) and FAM-AAVSLARRPLPPLP-NH2. Indeed, AGT(PP1)-1, AGT(PP2)-1, AGT(PP3)-1 have Kds that are in the low micromolar range for the SH3 domain of HCK (Figure 4B). The similar affinities of AGT(PP1), AGT(PP2), and AGT(PP3) for the SH3 domains of SRC and HCK explains the lack of selectivity demonstrated by all three bivalent inhibitors. As expected AGT(WT) shows no affinity for the SH3 domain of HCK. To the best of our knowledge, peptides that are able to discriminate between the SH3 domains of SRC and HCK have not been described. If a ligand with a suitable selectivity profile can be identified, the modular nature of bivalent inhibitors based on the AGT scaffold should allow the rapid generation of a selective inhibitor.
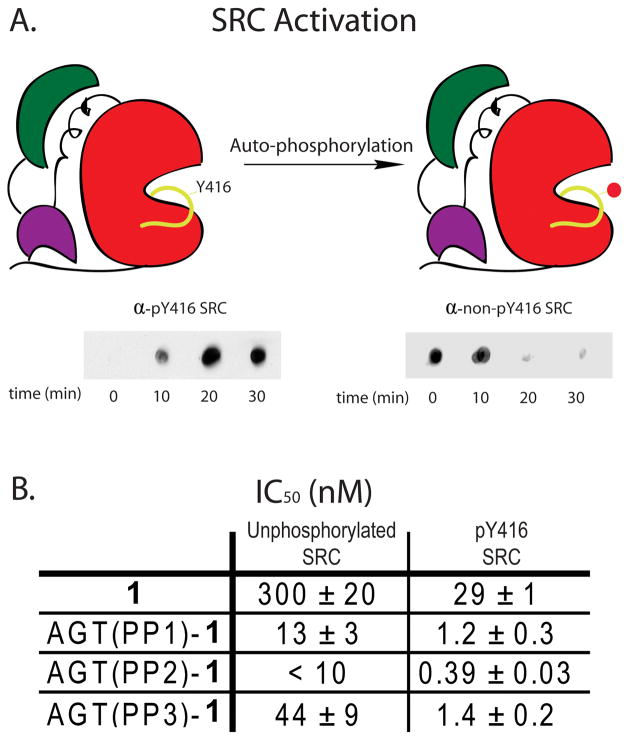
Activation Loop Phosphorylation
SRC kinase contains two tyrosine phosphorylation sites that regulate its catalytic activity. Phosphorylation of tyrosine 527, located at the C-terminus, down-regulates the catalytic activity of SRC through intra-molecular engagement of the SH2 domain. In contrast, phosphorylation of tyrosine 416 in the activation loop increases the catalytic activity of this kinase.31, 32 It is believed that these phosphorylation events affect the spatial relationship of the catalytic, SH2, and SH3 domains of SRC.31, 32 Because our inhibitors engage two binding sites that are distant in space, we were interested in determining if their potencies are sensitive to the activation state of SRC. To test this, conditions were developed for the generation of SRC that is quantitatively phosphorylated on tyrosine 416 (pY416 SRC). Incubation of 100 nM Src with 1 mM ATP for 30 minutes facilitated quantitative auto-phosphorylation of tyrosine 416 as determined by Dot Blot Analysis (Figure 5A). Due to the increased activity of the auto-phosphorylated product, we were able to perform inhibition assays at significantly lower enzyme concentrations. First, the potency of BG-linked derivative 1 was tested (Figure 5B). Surprisingly, 1 is 10-fold more potent inhibitor of activation loop phosphorylated SRC (pY416 SRC) (IC50 of 29 ± 1 nM) than the unactivated form (IC50 of 300 ± 20 nM). Next, AGT(PP1)-1, AGT(PP2)-1, and AGT(PP3)-1 were tested for their ability to inhibit pY416 SRC. Similar to BG-linked analogue 1, all three bivalent inhibitors are more potent against pY416 SRC. Because the IC50s observed for AGT(PP1)-1 (13 nM) and AGT(PP2)-1 (< 10 nM) against unactivated SRC are close to or less than the enzyme concentration used in the activity assays (10 nM), it is difficult to accurately determine the fold increase in potency of these conjugates for pY416 SRC. However, both conjugates appear to be about 10-fold more potent against pY416 SRC. More quantitatively, AGT(PP3)-1 is 31 times more potent against pY416 SRC (IC50 of 1.4 ± 0.2 nM), than the unphosphorylated form (44 ± 9 nM). Significantly, this construct shows a greater increase in potency than BG-linked analogue 1 alone. This suggests that activation loop phosphorylation not only increases the affinity of the quinazoline inhibitor to the ATP-binding site of SRC but also makes the SH3 domain of this kinase more accessible to ligands. This may be due to the SH3 domain of SRC being in a more favorable orientation for interacting with a bivalent inhibitor or result from a weakening of the intra-molecular interaction of the SH3 domain with the SH2 linker of this kinase. Unfortunately, due to the extremely low catalytic activity of SRC that is phosphorylated on tyrosine 527, the potencies of these bivalent inhibitors against this phospho-isoform were not able to be determined.
Figure 5.
Activity of bivalent inhibitors against specific phospho-isoforms of SRC. (A). SRC was auto-phosphorylated at tyrosine 416 by incubating SRC-3D (100 nM) with ATP (1 mM) and Na3VO4 (2.5 mM) in activation buffer for 30 minutes. Phosphorylation was monitored by immuno-blotting with antibodies specific for the pY416 and non-pY416 forms of SRC. (B). IC50 values of BG-linked analogue 1 and protein-small molecule conjugates AGT(PP1)-1, AGT(PP2)-1, AGT(PP3)-1 for pY416 and non-pY416 forms of SRC. All protein-small molecule conjugates were prepared in two independent labeling reactions, and values shown are the average of four assays ± SEM.
Conclusion
Identifying inhibitors that are able to discriminate between closely-related protein kinases is extremely challenging. One successful strategy for obtaining selectivity is the use of bivalent ligands that target two distinct binding sites. We have developed a bivalent inhibitor strategy that relies on the use of the self-labeling repair enzyme AGT to display a peptidyl ligand and an ATP-competitive inhibitor. Application of this methodology allowed the identification of potent and selective inhibitors of the tyrosine kinases SRC and ABL. In this study, we have profiled the molecular binding determinants of these bivalent ligands with the kinases SRC and ABL. First, we demonstrated that SRC and ABL have differential sensitivities to the orientation of the ATP-competitive inhibitor displayed from AGT. Next, we dissected how the affinity of each individual ligand affects overall bivalent inhibitor potency. These studies revealed that the interactions between AGT-based bivalent inhibitors and SRC and ABL are highly modular. Furthermore, they demonstrated that a single interaction outside of the ATP-binding site can be used to obtain high selectivity. Finally, the potency of bivalent inhibitors against distinct phospho-isoforms of SRC was determined. All inhibitors tested appear to be more potent against an activated form of SRC, with inhibitors that target two sites showing the largest increase overall. These data lay the foundation for a set of guidelines regarding development of bivalent inhibitors utilizing AGT as a scaffold. This method is attractive because of its modular design which allows for rapid generation of many permutations of inhibitors based on relatively small individual sets of monovalent ligands.
Experimental
Synthetic Methods
General
Unless otherwise noted, all reagents were obtained from commercial suppliers and used without purification. 1H-NMR spectra were obtained on a Bruker AV-300 or AV301 instrument at room temperature. Chemical shifts are reported in ppm, and coupling constants are reported in Hz. Mass spectrometry was performed on a Bruker Esquire Ion Trap MS instrument.
General HPLC Purification Conditions
Preparatory reverse-phase C18 column (250 × 21 mm), CH3CN/H2O–0.1% CF3CO2H gradient: 1:99 to 100:0 over 60 min; 8 mL/min; 220 and 254 nm detection for 65 min. All HPLC analyses were performed utilizing a Varian Microsorb-MV C18 reverse-phase analytical column (2.1 mm × 150 mm). The purity of each final compound was determined to be > 95% by analytical HPLC. Analytical conditions A: [C18 column (2.1 mm × 150 mm), CH3CN/H2O–0.1% CF3CO2H = 1:99 to 100:0 for 30 min; 1 mL/min; 220 and 254 nm detection for 30 min. Analytical conditions B: [C18 (2.1 mm × 150 mm), CH3OH/H2O–0.1% CF3CO2H = 1:99 to 100:0 over 30 min; 1 mL/min; 220 and 254 nm detection for 30 min].
1. 1 was synthesized using a previously published procedure.15
2. 2 was synthesized using a previously published procedure.15
3. To a mixture of O6-(4-Amino-methyl-benzyl)guanine (14.3 mg, 52.9 μmol, 1.0 equiv), 5-hexynoic acid (8.1 μL, 68.8 μmol, 1.3 equiv), HOBt·H2O (10.7 mg, 68.8 μmol, 1.3 equiv), and DIPEA ((30 μL, 158.7 μmol, 3.0 equiv)) in DMF (130 μL) was added EDCI·HCl (11.2 mg, 58.2 μmol, 1.1 equiv). The reaction mixture was stirred at rt for 24 h and then diluted with CH3CN/H2O (10 mL). The product was purified using General HPLC purification conditions to obtain 10.6 mg of pure N-((4-((2-amino-9H-purin-6-yloxy)methyl)phenyl)methyl)hex-5-ynamide (55% yield) 1H-NMR (MeOD) 1.77–1.88 (m, 2H), 2.20–2.28 (m, 3H), 2.38 (t, J = 7.2 Hz, 2H), 4.39 (s, 2H), 5.65 (s, 2H), 7.35 (d, J = 8.1 Hz, 2H), 7.53 (d, J = 8.1 Hz, 2H), 8.30 (s, 1H). Calcd for C19H20N6O2 (M+H+): 365.2 Found 365.2.
A mixture of N-((4-((2-amino-9H-purin-6-yloxy)methyl)phenyl)methyl)hex-5-ynamide (7.9 mg, 9.5 μmol, 3.3 equiv), 2-(2-(2-(2-(2-azidoethoxy)ethoxy)ethoxy)ethoxy)-1-(4-(3-(4-(2-chloro-5-methoxyphenylamino)-6-methoxyquinazolin-7-yloxy)propyl)piperazin-1-yl)ethanone26 (3.5 mg, 9.5 μmol, 3.3 equiv), DIPEA (25 μL, 143.8 μmol, 49.6 equiv) and CuI (0.6 mg, 2.9 μmol, 1.0 equiv) in MeOH (0.37 mL) was stirred at rt for 2 days. The product was purified using General HPLC purification conditions to obtain 4.2 mg of 3 (37% yield). 1H-NMR (MeOD) 1.15–1.34 (m, 2H), 1.79 (m, 2H), 2.13–2.74 (m, 10H), 3.00–4.26 (m, 30H), 4.46 (t, J = 5.1 Hz, 2H), 5.46 (s, 2H), 6.40 (m, 1H), 6.97(dd, J = 8.7, 3.0 Hz, 1H), 7.13–7.27 (m, 4H), 7.45 (m, 2H), 7.51 (s, 1H), 7.82 (s, 1H), 7.99 (m, 1H), 8.36 (m, 1H). Calcd for C52H65ClN14O10 ((M+2H+)/2): 541.2 Found 541.5.
4. To a mixture of 5-((4-((2-amino-9H-purin-6-yloxy)methyl)benzyl)amino)-5-oxopentanoic acid15 (2.5 mg, 6.7 μmol, 1.3 equiv), N-(6-chlorobenzo[d][1,3]dioxol-5-yl)-6-methoxy-7-(3-(piperazin-1-yl)propoxy)quinazolin-4-amine26 (5.2 μmol, 1 equiv), HOBt·H2O (1 mg, 6.7 μmol, 1.3 equiv), and DIPEA ((1.1 μL, 6.7 μmol, 1.3 equiv)) in DMF (50 μL), was added EDCI·HCl (1.3 mg, 6.7 μmol, 1.3 equiv). The reaction was stirred at rt for 24 h at which time the reaction was dissolved in CH3CN/H2O (mL) and purified using General HPLC conditions to obtain 0.6 mg of pure 4 (14% yield). Calcd for C41H44ClN11O7 ((M+2H+)/2): 419.66 Found 420.2
5. A mixture of N-Boc-1-(2-bromoethyl)-piperazine (40.5 mg, 138μmol, 1.2 equiv), 4-(3-chloro-4-fluoroanilino)-6-hydroxy-7-methoxyquinazoline33 (37 mg, 115 μmol, 1 equiv), and K2CO3 (127 mg, 922 μmol, 8 equiv) was stirred in DMF (280 μL) at 80 °C overnight. The reaction was concentrated in vacuo and purified using flash chromatography (MeOH/CH2Cl2) to yield 41 mg of 4-(3-chloro-4-fluoroanilino)-6-(2-(N-Boc-piperazin-1-yl)ethoxy)-7-methoxyquinazoline. (67 % yield) 1H-NMR (CDCl3) 1.43 (s, 9H), 2.52 (t, J = 4.8 Hz, 4H), 2.88 (t, J = 6 Hz, 2H), 3.42 (t, J = 4.8 Hz, 4H), 3.92 (s, 3H), 4.24 (t, J = 6 Hz, 2H), 7.09 (t, J = 8.7 Hz, 1H), 7.19 (s,1H), 7.55 (s, 1H), 7.60 (m, 1H), 7.87 (dd, J = 6.6, 2.7 Hz, 1H), 8.59 (s, 1H)
4-(3-chloro-4-fluoroanilino)-6-(2-(N-Boc-piperazin-1-yl)ethoxy)-7-methoxyquinazoline (10 mg, 19.1 μmol, 1 equiv) was stirred in 30% TFA/CH2Cl2 (191 μL) for 1.5 h at rt. Toluene was then added (l mL) and the reaction was concentrated in vacuo to afford crude 4-(3-chloro-4-fluoroanilino)-6-(2-(piperazin-1-yl)ethoxy)-7-methoxyquinazoline. The crude reaction product was carried on to the next step without further purification.
To a mixture of 5-((4-((2-amino-9H-purin-6-yloxy)methyl)benzyl)amino)-5-oxopentanoic acid (6 mg, 15 μmol, 1.3 equiv), 4-(3-chloro-4-fluoroanilino)-6-(2-(piperazin-1-yl)ethoxy)-7-methoxyquinazoline. (11 μmol, 1 equiv), HOBt·H2O (3 mg, 20 μmol, 1.7 equiv), and DIPEA ((2.5 μL, 15 μmol, 1.3 equiv)) in DMF (60 μL), was added EDCI·HCl (3 mg, 16 μmol, 1.4 equiv). The reaction was stirred at rt for 24 h at which time the reactions was dissolved in CH3CN/H2O (10 mL) and purified using General HPLC conditions to obtain 0.54 mg of pure 5 (6% yield). Calcd for C39H41ClFN11O5 ((M+2H+)/2): 399.7 Found 399.9.
6. To a mixture of 5-((4-((2-amino-9H-purin-6-yloxy)methyl)benzyl)amino)-5-oxopentanoic acid (3.5 mg, 9.1 μmol, 1.3 equiv), N-(3-(3-(6-(4-(2-aminoethoxy) phenylamino)pyrimidin-4-yl)pyridin-2-ylamino)-4-methylphenyl)-3-(trifluoromethyl) benzamide34 (7 μmol, 1 equiv), HOBt·H2O (1.4 mg, 9.1 μmol, 1.3 equiv), and DIPEA ((3.8 μL, 21 μmol, 3 equiv)) in DMF (35 μL), was added EDCI·HCl (1.7 mg, 9.1 μmol, 1.3 equiv). The reaction was stirred at rt for 24 h at which time the reaction was dissolved in CH3CN/H2O (10 mL) and purified using General HPLC conditions to obtain 1.2 mg of pure 6 (18% yield). Calcd for C50H46F3N13O5 ((M+2H+)/2): 483.7 Found 483.9.
Protein design, expression and purification
AGT fusion proteins
A mutant form of the human gene O6-alkylguanine-DNA-alkyltransferase (hAGT) was amplified from the pSS26b plasmid (Covalys) using the primers listed for Step 1 (Supporting Info). The fusions were amplified with a second round of PCR with the primers listed for Step 2 (Supporting Info). These constructs were incorporated into a pDEST™ 527 (Invitrogen) using Gateway® technology. All sequences of the final constructs were confirmed by sequencing of the entire gene.
Overlap extension PCR generated AGT constructs with the following N-terminal sequences: AGT(PP1) = (His6)-RSDITSLYKKAGF-(APPLPPRNRPRL)-SGSG-(DKDCE-AGT); AGT(PP2) = (His6)-RSDITSLYKKAGF-(VSLRRPLPPLPRL)-SGSGSGSGSG-(DKDCE-AGT); AGT(PP3) = (His6)-RSDITSLYKKAGF-(RAARPLPPLPP)-SGSG-(DKDCE-AGT); AGT(PP4) = (His6)-RSDITSLYKKAGF-(APTYSPPPPP)-SGSG-(DKDCE-AGT).
AGT(PP1), AGT(PP2), AGT(PP3), and AGT(PP4) were expressed and purified using a previously published procedure.15
Preparation of AGT-small molecule conjugates
AGT(WT), AGT(PP1), AGT(PP2), AGT(PP3), and AGT(PP4) were labeled with 1, 2, 3, 4, 5, or 6 using the following conditions. Purified AGT fusion protein (15 μM) was incubated with 1, 2, 3, 4, 5, or 6 (22.5 μM) in labeling buffer (50 mM Tris buffer, pH = 7.5, 100 mM NaCl, 0.1% Tween 20 and 1 mM DTT) for 1.5 h at 25 °C. The protein-small molecule conjugates were then separated from unconjugated BG derivatives (1, 2, 3, 4, 5, or 6) by running the reaction mixture over two Bio-Rad Micro Bio-Spin© columns equilibrated with Tris buffer. The concentration of each protein-small molecule conjugate was then determined using a Coomassie Plus – The Better Bradford™ Assay Kit. Purified AGT-small molecule conjugates were used directly in activity assays. Each protein-small molecule conjugate was prepared, purified and assayed in two independent labeling reactions.
Protein kinase, expression and purification
SRC that contains the SH1, SH2 and SH3 domains (SRC-3D, residues 83–533), ABL that contains the SH1, SH2 and SH3 domains (ABL-3D, residues 65–534), HCK that contains the SH1, SH2, and SH3 domains (HCK-3D, residues 78–527), and LCK that contains the SH1, SH2, and SH3 domains (LCK-3D, residues 76–520) were expressed and purified using a previously published procedure.24 This method generates non-phosphorylated forms of SRC, ABL, HCK, and LCK.
GST-SH3(SRC) and GST-SH3(HCK)
The SH3 domain of SRC (residues 83–139) was amplified from the chicken c-SRC gene using the primers listed in the Supporting Information. This construct was incorporated into a pDEST 15 bacterial expression vector (Invitrogen) using Gateway® technology. The N-terminal GST fusion of SH3(SRC) was expressed and purified using standard protocols.
The SH3 domain of HCK (residues 80–135,) was amplified from the Human HCK gene (isoform c) using the primers listed in the Supporting Information. This construct was incorporated into a pDEST 15 bacterial expression vector (Invitrogen) using Gateway® technology. The N-terminal GST fusion of SH3(HCK) was expressed and purified using standard protocols.
Assays
Preparation of activation loop-phosphorylated SRC (pY416 SRC)
SRC was auto-phosphorylated at tyrosine 416 by incubating SRC-3D (100 nM) with ATP (1 mM) and Na3VO4 (2.5 mM) in activation buffer (50 mM HEPES, pH = 7.5, 10 mM MgCl2, 2.5 mM EGTA, 100 mM NaCl). The reaction was incubated at rt for 30 min. Phosphorylation was monitored with antibodies that specifically recognize activation loop-phosphorylated SRC (pY416) (Cell Signaling Technology) and non-pY416 (non-pY416) (Cell Signaling Technology). SRC activated by this method was used directly in activity assays.
Activity inhibition assays
Inhibition assays of SRC-3D, ABL-3D, HCK-3D, and LCK-3D were preformed as previously described.15 ABL kinase was assayed at a final concentration of 6 nM. SRC kinase was assayed at a final concentration of 10 nM, except for pY416 SRC, which was assayed at 0.17 nM.
Synthesis of FAM-VLS12
To a mixture of the peptide AAVSLARRPLPPLP-CONH2 (4 mg, 2.7 μmol, 1 equiv), 5-(and-6)-carboxyfluorescein (1.3 mg, 3.5 μmol, 1.3 equiv), HOBt·H2O (0.5 mg, 3.5 μmol, 1.3 equiv), and DIPEA ((0.6 μL, 3.5 μmol, 1.3 equiv)) in DMF (28 μL), was added EDCI·HCl (0.68 mg, 3.5 μmol, 1.3 equiv). The reaction was stirred at room temperature for 24 h at which time the reactions was dissolved in CH3CN/H2O (10 mL) and purified using General HPLC conditions to obtain 0.89 mg of pure N- terminal labeled FAM-VLS12 (18% yield). Calcd for C88H127N21O21 ((M+3H+)/3): 605.65 Found 606.1.
Fluorescence polarization assays
The affinity of fluorescein-VLS12 for the SH3 domains of SRC (GST-SH3(SRC)) and HCK (GST-SH3(HCK)) was determined using a fluorescence polarization assay. 100 nM of fluorescein-VLS12 was incubated with varying concentrations of GST-SH3(SRC) or GST-SH3(HCK) (initial concentration, 7.33 μM, 3-fold serial dilution, 10 dilutions) in buffer containing 20 mM phosphate, pH 7.4, 1 mM EDTA, 50 mM NaCl, 0.2 mM NaN3, and 0.05% pluronic acid (final volume = 60 μL per well). The assay was incubated in the dark at rt for 30 min prior to reading. Data was analyzed using Prism Graphpad software and Kd values were determined using non-linear regression analysis.
Fluorescence polarization competition assays
The affinities of AGT fusion proteins (AGT(WT), AGT(PP1), AGT(PP2), and AGT(PP3)) for GST-SH3(SRC) and GST-SH3(HCK) were determined using a fluorescence polarization competition assay. An AGT fusion protein [(AGT(WT) (initial concentration = 12.75 μM; 3-fold serial dilution)) (AGT(PP1) (initial concentration = 13.3 μM; 3-fold serial dilution)) (AGT(PP2) (initial concentration = 1.9 μM, 3-fold serial dilution)) AGT (PP3) (initial concentration = 6.1 μM, 3-fold serial dilution))], fluorescein-VLS12 (100 nM when assaying GST-SH3(SRC) or 300 nM when assaying GST-SH3(HCK)), and GST-SH3(SRC) (500 nM) or GST-SH3(HCK) (900 nM) were incubated in buffer containing 20 mM phosphate, pH 7.4, 1 mM EDTA, 50 mM NaCl, 0.2 mM NaN3, and 0.05% pluronic acid (final volume = 60 μL). The assay was incubated in the dark at rt for 30 min prior to reading. Data was analyzed using Prism Graphpad software and Kd values were determined using non-linear regression analysis.
Supplementary Material
Acknowledgments
We thank J. Kuriyan (University of California, Berkeley) and M. Seeliger (State University of New York, Stony Brook) for providing expression plasmids for ABL-3D, SRC-3D, HCK-3D, and LCK-3D. This work was supported by the National Institute of General Medical Science (R01GM086858 to D.J.M.).
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Blume-Jensen P, Hunter T. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 3.Hunter T. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 4.Cohen P. Nat Rev Drug Discovery. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 5.Parang K, Cole PA. Pharmacol Ther. 2002;93:145–157. doi: 10.1016/s0163-7258(02)00184-5. [DOI] [PubMed] [Google Scholar]
- 6.Ricouart A, Gesquiere JC, Tartar A, Sergheraert C. J Med Chem. 1991;34:73–78. doi: 10.1021/jm00105a012. [DOI] [PubMed] [Google Scholar]
- 7.Hines AC, Cole PA. Bioorg Med Chem Lett. 2004;14:2951–2954. doi: 10.1016/j.bmcl.2004.03.039. [DOI] [PubMed] [Google Scholar]
- 8.Hines AC, Parang K, Kohanski RA, Hubbard SR, Cole PA. Bioorg Chem. 2005;33:285–297. doi: 10.1016/j.bioorg.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Schneider TL, Mathew RS, Rice KP, Tamaki K, Wood JL, Schepartz A. Org Lett. 2005;7:1695–1698. doi: 10.1021/ol050179o. [DOI] [PubMed] [Google Scholar]
- 10.Lee JH, Kumar S, Lawrence DS. ChemBioChem. 2008;9:507–509. doi: 10.1002/cbic.200700583. [DOI] [PubMed] [Google Scholar]
- 11.Meyer SC, Shomin CD, Gaj T, Ghosh I. J Am Chem Soc. 2007;129:13812–13813. doi: 10.1021/ja076197d. [DOI] [PubMed] [Google Scholar]
- 12.Shomin CD, Meyer SC, Ghosh I. Bioorg Med Chem. 2009;17:6196–6202. doi: 10.1016/j.bmc.2009.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hah JM, Sharma V, Li H, Lawrence DS. J Am Chem Soc. 2006;128:5996–5997. doi: 10.1021/ja060136i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Profit AA, Lee TR, Niu J, Lawrence DS. J Biol Chem. 2001;276:9446–9451. doi: 10.1074/jbc.M009262200. [DOI] [PubMed] [Google Scholar]
- 15.Hill ZB, Perera BG, Maly DJ. J Am Chem Soc. 2009;131:6686–6688. doi: 10.1021/ja900871y. [DOI] [PubMed] [Google Scholar]
- 16.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 17.Damoiseaux R, Keppler A, Johnsson K. ChemBioChem. 2001;2:285–287. doi: 10.1002/1439-7633(20010401)2:4<285::AID-CBIC285>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 18.Keppler A, Pick H, Arrivoli C, Vogel H, Johnsson K. Proc Natl Acad Sci U S A. 2004;101:9955–9959. doi: 10.1073/pnas.0401923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng S, Kasahara C, Rickles RJ, Schreiber SL. Proc Natl Acad Sci U S A. 1995;92:12408–12415. doi: 10.1073/pnas.92.26.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pisabarro MT, Serrano L. Biochemistry. 1996;35:10634–10640. doi: 10.1021/bi960203t. [DOI] [PubMed] [Google Scholar]
- 21.Tran T, Hoffmann S, Wiesehan K, Jonas E, Luge C, Aladag A, Willbold D. Biochemistry. 2005;44:15042–15052. doi: 10.1021/bi051403k. [DOI] [PubMed] [Google Scholar]
- 22.Zellefrow CD, Griffiths JS, Saha S, Hodges AM, Goodman JL, Paulk J, Kritzer JA, Schepartz A. J Am Chem Soc. 2006;128:16506–16507. doi: 10.1021/ja0672977. [DOI] [PubMed] [Google Scholar]
- 23.Martinelli G, Soverini S, Rosti G, Baccarani M. Leukemia. 2005;19:1872–1879. doi: 10.1038/sj.leu.2403950. [DOI] [PubMed] [Google Scholar]
- 24.Seeliger MA, Young M, Henderson MN, Pellicena P, King DS, Falick AM, Kuriyan J. Protein Sci. 2005;14:3135–3139. doi: 10.1110/ps.051750905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ple PA, Green TP, Hennequin LF, Curwen J, Fennell M, Allen J, Lambert-Van Der Brempt C, Costello G. J Med Chem. 2004;47:871–887. doi: 10.1021/jm030317k. [DOI] [PubMed] [Google Scholar]
- 26.Perera BG, Maly DJ. Mol Biosyst. 2008;4:542–550. doi: 10.1039/b720014e. [DOI] [PubMed] [Google Scholar]
- 27.Barker AJ, Gibson KH, Grundy W, Godfrey AA, Barlow JJ, Healy MP, Woodburn JR, Ashton SE, Curry BJ, Scarlett L, Henthorn L, Richards L. Bioorg Med Chem Lett. 2001;11:1911–1914. doi: 10.1016/s0960-894x(01)00344-4. [DOI] [PubMed] [Google Scholar]
- 28.Brehmer D, Greff Z, Godl K, Blencke S, Kurtenbach A, Weber M, Muller S, Klebl B, Cotten M, Keri G, Wissing J, Daub H. Cancer Res. 2005;65:379–382. [PubMed] [Google Scholar]
- 29.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, Floyd M, Hunt JP, Lockhart DJ, Milanov ZV, Morrison MJ, Pallares G, Patel HK, Pritchard S, Wodicka LM, Zarrinkar PP. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 30.Seeliger MA, Ranjitkar P, Kasap C, Shan Y, Shaw DE, Shah NP, Kuriyan J, Maly DJ. Cancer Res. 2009;69:2384–2392. doi: 10.1158/0008-5472.CAN-08-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porter M, Schindler T, Kuriyan J, Miller WT. J Biol Chem. 2000;275:2721–2726. doi: 10.1074/jbc.275.4.2721. [DOI] [PubMed] [Google Scholar]
- 32.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Mol Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 33.5 770 599. US Pat. 1998
- 34.Ranjitkar P, Brock AM, Maly DJ. Chem Biol. 2010;17:195–206. doi: 10.1016/j.chembiol.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.