Abstract
Large branched DNA structures are constructed by two-step reassociation of separated complementary strands from restriction fragments of different lengths. The displacement of DNA strands initially annealed to longer complementary DNA sequences, a process mediated by branch migration, is very rapid and has thus far been followed only under conditions which are second order, DNA reassociation rate limiting. The average lifetime of branched DNA leading to displacement of 1.6 Kb strands is estimated to be less than 10 seconds under conditions of DNA reassociation, Tm-25 degrees C. Several DNA-binding drugs, including intercalating dyes, have been tested to determine their influence, if any, on the kinetics of DNA strand displacements by branch migration. Only actinomycin D was found to have significant effect under the conditions we have described. The kinetics of the strand displacement in the presence of low concentrations of actinomycin D remain second order and slower rate of strand displacement must be attributed to decreased rate of reassociation of DNA strands to form the branched intermediates. Consideration is given to the potential manipulation of DNA structures at site-directed branches and the limitations due to rapid strand displacements. The feasibility of constructing sufficiently large branched DNA regions to approach first order, branch migration rate limiting kinetics is also discussed.
Full text
PDF




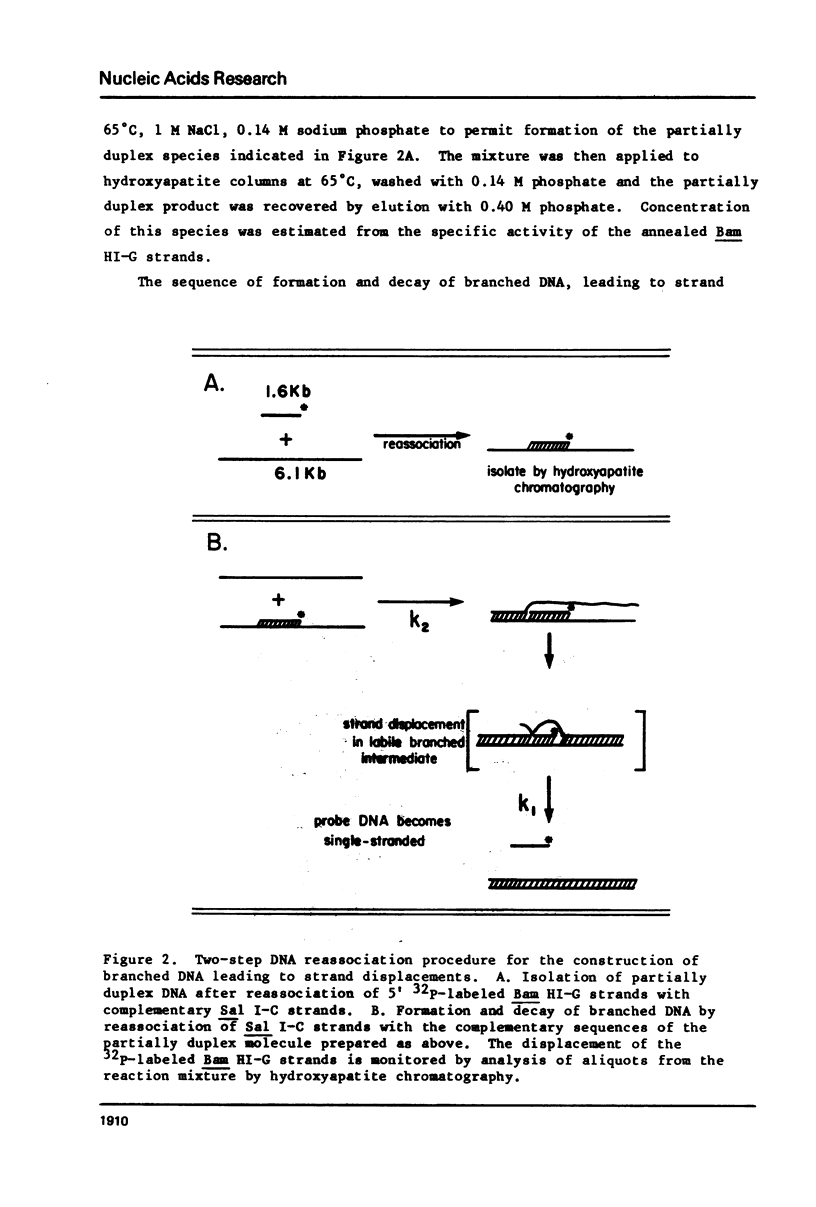
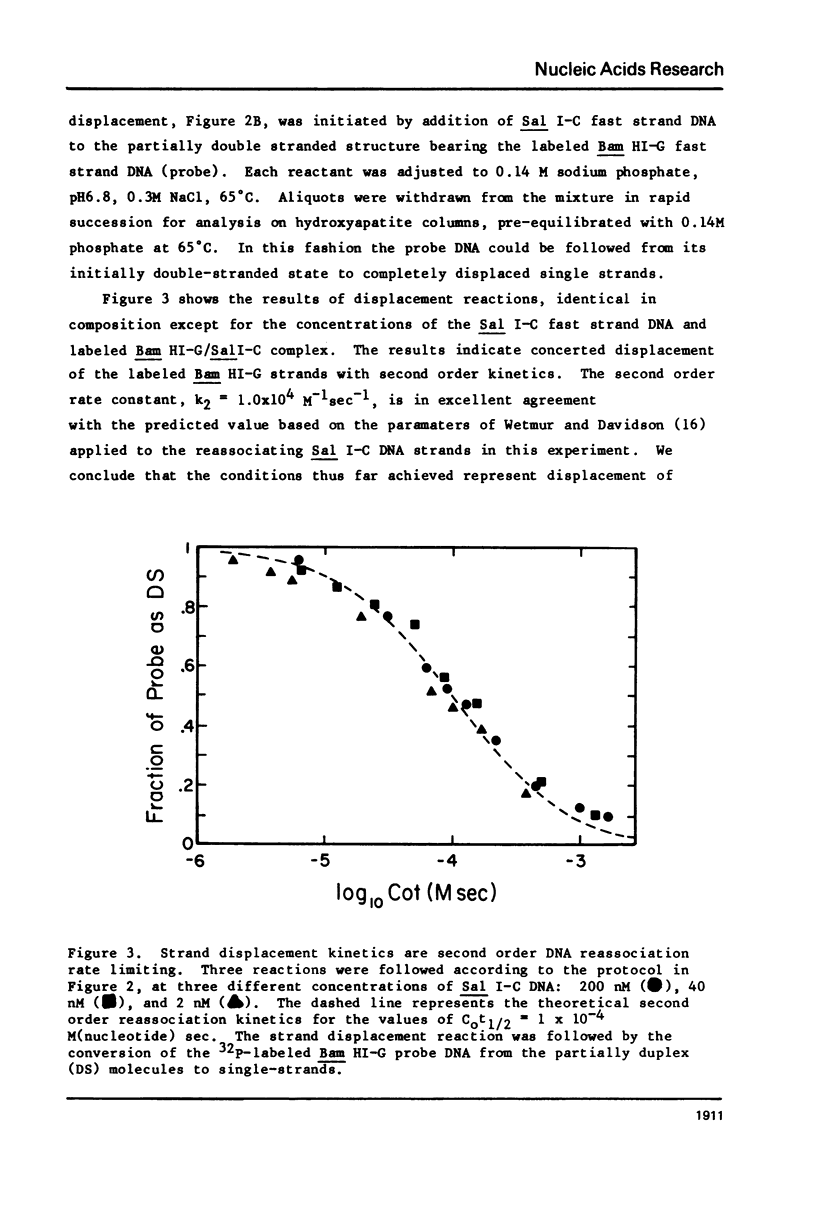

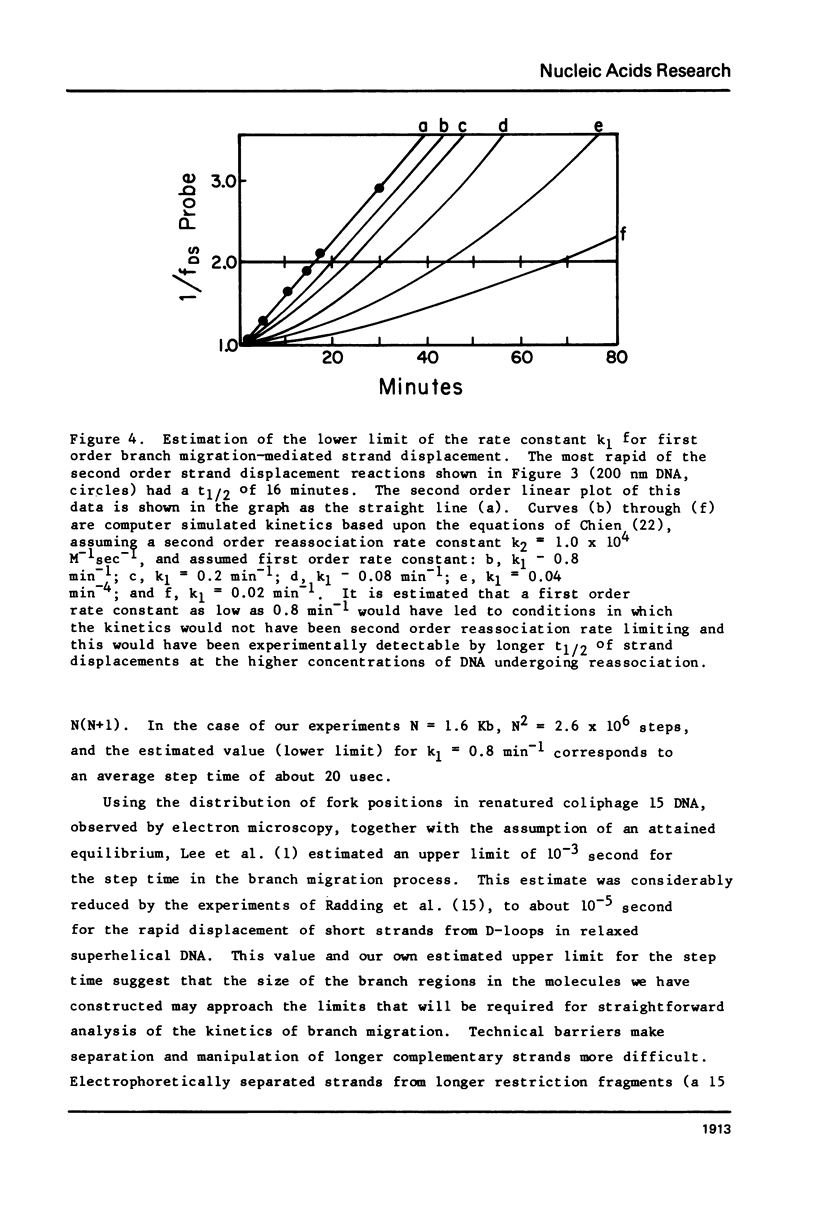
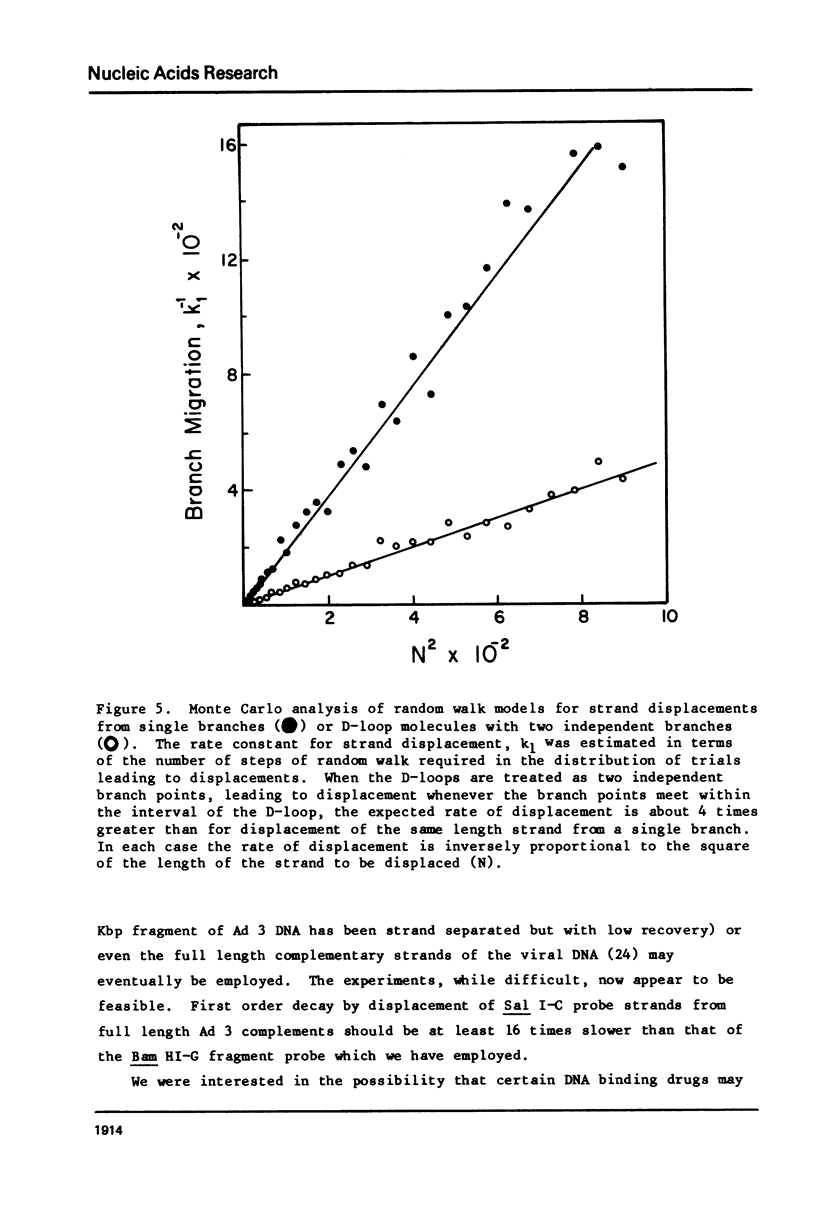
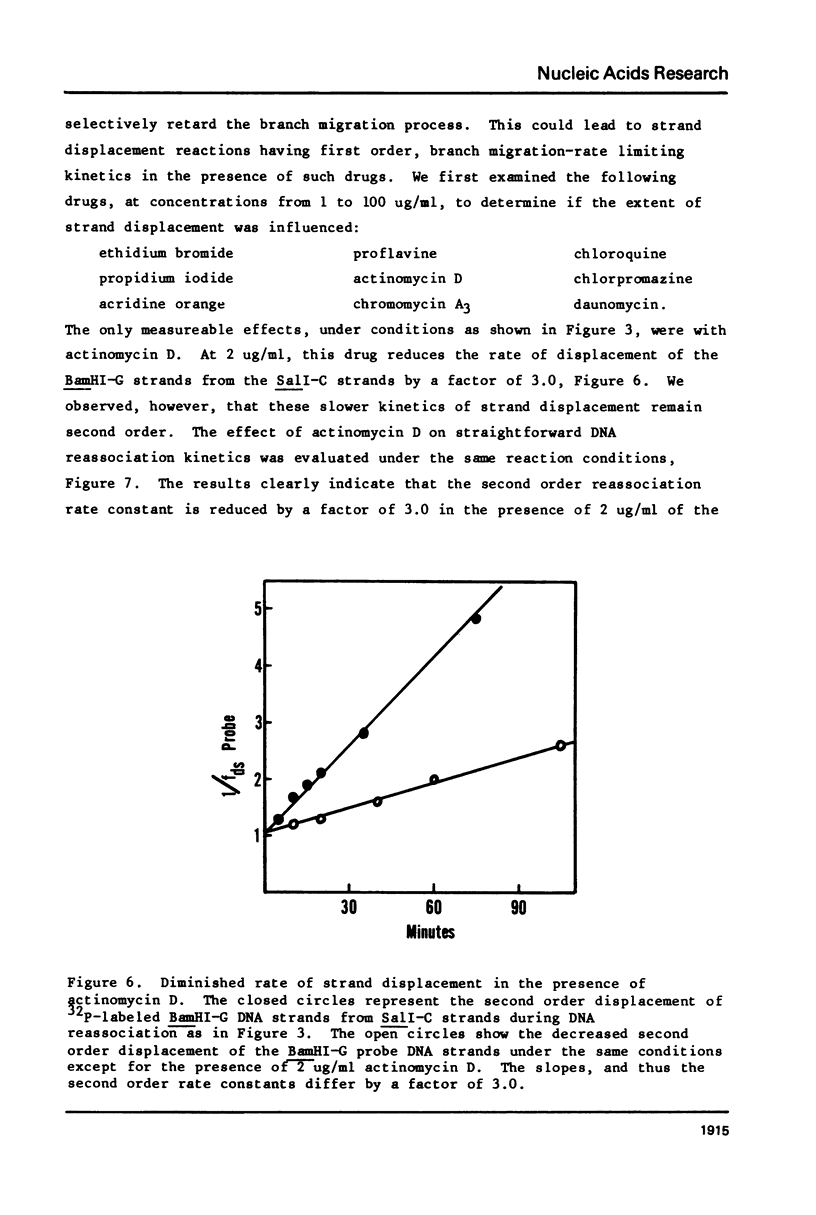
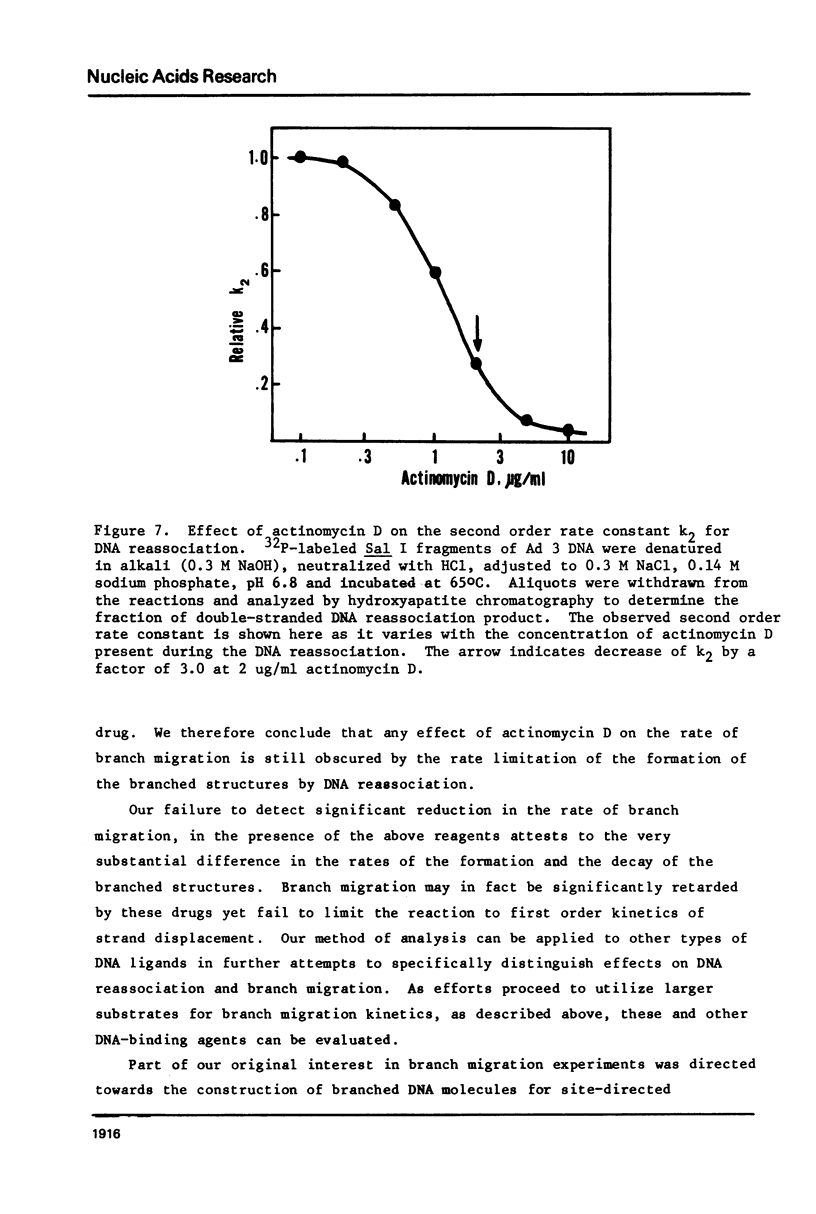


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beattie K. L., Wiegand R. C., Radding C. M. Uptake of homologous single-stranded fragments by superhelical DNA. II. Characterization of the reaction. J Mol Biol. 1977 Nov;116(4):783–803. doi: 10.1016/0022-2836(77)90271-6. [DOI] [PubMed] [Google Scholar]
- Broker T. R. An electron microscopic analysis of pathways for bacteriophage T4 DNA recombination. J Mol Biol. 1973 Nov 25;81(1):1–16. doi: 10.1016/0022-2836(73)90243-x. [DOI] [PubMed] [Google Scholar]
- Broker T. R., Lehman I. R. Branched DNA molecules: intermediates in T4 recombination. J Mol Biol. 1971 Aug 28;60(1):131–149. doi: 10.1016/0022-2836(71)90453-0. [DOI] [PubMed] [Google Scholar]
- Casey J., Davidson N. Rates of formation and thermal stabilities of RNA:DNA and DNA:DNA duplexes at high concentrations of formamide. Nucleic Acids Res. 1977;4(5):1539–1552. doi: 10.1093/nar/4.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefter M. L. DNA replication. Annu Rev Biochem. 1975;44:45–78. doi: 10.1146/annurev.bi.44.070175.000401. [DOI] [PubMed] [Google Scholar]
- Hayward G. S. Gel electrophoretic separation of the complementary strands of bacteriophage DNA. Virology. 1972 Jul;49(1):342–344. doi: 10.1016/s0042-6822(72)80042-4. [DOI] [PubMed] [Google Scholar]
- Lee C. S., Davis R. W., Davidson N. A physical study by electron microscopy of the terminally reptitious, circularly permuted DNA from the coliphage particles of Escherichia coli 15. J Mol Biol. 1970 Feb 28;48(1):1–22. doi: 10.1016/0022-2836(70)90215-9. [DOI] [PubMed] [Google Scholar]
- Oberg B., Philipson L. Replicative structures of poliovirus RNA in vivo. J Mol Biol. 1971 Jun 28;58(3):725–737. doi: 10.1016/0022-2836(71)90036-2. [DOI] [PubMed] [Google Scholar]
- Radding C. M., Beattie K. L., Holloman W. K., Wiegand R. C. Uptake of homologous single-stranded fragments by superhelical DNA. IV. Branch migration. J Mol Biol. 1977 Nov;116(4):825–839. doi: 10.1016/0022-2836(77)90273-x. [DOI] [PubMed] [Google Scholar]
- Robberson D. L., Kasamatsu H., Vinograd J. Replication of mitochondrial DNA. Circular replicative intermediates in mouse L cells. Proc Natl Acad Sci U S A. 1972 Mar;69(3):737–741. doi: 10.1073/pnas.69.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T., DasGupta C., Cunningham R. P., Radding C. M. Purified Escherichia coli recA protein catalyzes homologous pairing of superhelical DNA and single-stranded fragments. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1638–1642. doi: 10.1073/pnas.76.4.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B. J., Camien M. N., Warner R. C. Kinetics of branch migration in double-stranded DNA. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2299–2303. doi: 10.1073/pnas.73.7.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts C., Pettersson U., Johansson K., Philpson L. Relationship of mRNA from productively infected cells to the complementary strands of adenovirus type 2 DNA. J Virol. 1974 Feb;13(2):370–377. doi: 10.1128/jvi.13.2.370-377.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts C. Physical organization of subgroup B human adenovirus genomes. J Virol. 1977 Nov;24(2):564–579. doi: 10.1128/jvi.24.2.564-579.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J. I., Sakakibara Y., Kakefuda T. Replication of colicin E1 plasmid DNA added to cell extracts. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1050–1054. doi: 10.1073/pnas.72.3.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissmann C., Feix G., Slor H. In vitro synthesis of phage RNA: the nature of the intermediates. Cold Spring Harb Symp Quant Biol. 1968;33:83–100. doi: 10.1101/sqb.1968.033.01.014. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- Wiegand R. C., Beattie K. L., Holloman W. K., Radding C. M. Uptake of homologous single-stranded fragments by superhelical DNA. III. The product and its enzymic conversion to a recombinant molecule. J Mol Biol. 1977 Nov;116(4):805–824. doi: 10.1016/0022-2836(77)90272-8. [DOI] [PubMed] [Google Scholar]
- Winnacker E. L. Adenovirus DNA: structure and function of a novel replicon. Cell. 1978 Aug;14(4):761–773. doi: 10.1016/0092-8674(78)90332-x. [DOI] [PubMed] [Google Scholar]