Abstract
BACKGROUND AND PURPOSE:
Hippocampus volumetry is a useful surrogate marker for the diagnosis of Alzheimer disease, but it seems insufficiently sensitive for the aMCI stage. We postulated that some hippocampus subfields are specifically atrophic in aMCI and that measuring hippocampus subfield volumes will improve sensitivity of MR imaging to detect aMCI.
MATERIALS AND METHODS:
We evaluated episodic memory and hippocampus subfield volume in 15 patients with aMCI and 15 matched controls. After segmentation of the whole hippocampus from clinical MR imaging, we applied a new computational method allowing fully automated segmentation of the hippocampus subfields. This method used a Bayesian modeling approach to infer segmentations from the imaging data.
RESULTS:
In comparison with controls, subiculum and CA2–3 were significantly atrophic in patients with aMCI, whereas total hippocampus volume and other subfields were not. Total hippocampus volume in controls was age-related, whereas episodic memory was the main explanatory variable for both the total hippocampus volume and the subfields that were atrophic in patients with aMCI. Segmenting subfields increases sensitivity to diagnose aMCI from 40% to 73%.
CONCLUSIONS:
Measuring CA2–3 and subiculum volumes allows a better detection of aMCI.
aMCI is characterized by an EM deficit without significant impact in everyday life.1 Many consider aMCI as a transitional stage toward AD, but at least 25% of cases will not evolve to AD. Therefore, surrogate markers are needed to better define early AD stages.2
Mesiotemporal volumetry is a promising tool because it measures changes in the brain regions where the first AD lesions occur. Hippocampal atrophy has been demonstrated in AD and is also present in aMCI, though to a lesser extent: approximately 30% of MCI that will convert to AD in the next 18 months do not present significant hippocampal atrophy at baseline MR imaging.3 Segmenting hippocampus subfields seems to be an attractive approach that allows an earlier and more accurate diagnosis of AD. Postmortem studies have indeed shown that some subfields may be preferentially affected in AD.4
So far, only a few studies have addressed the issue, mostly by using a manual delineation of the hippocampus.5,6 In this way, it was suggested that AD patients display atrophy in CA1.5 In aMCI, atrophy was shown in the CA1-CA2 boundary.6 3D surface mapping of hippocampal atrophy also was performed in aMCI.7 This technique avoids manual delineation, though loses quantitative information. Atrophy was found in areas corresponding to the subiculum and CA1.7
Manual delineation is not applicable in clinical practice because this method requires considerable work (>2 days per subject!). Until recently, there was no automated method of segmentation applicable to the hippocampus substructures, but such a method has now been validated with high-resolution MR imaging.8 We wanted to show that this automated technique is fast enough (<3 hours for a single subject) to enable routine analysis of the hippocampal subfields from standard clinical MR imaging. We postulated that some hippocampus subfields are specifically atrophic in aMCI and that measuring hippocampus subfield volumes will improve sensitivity of MR imaging to detect aMCI. If due to the disease, atrophy should be related to a decrease in EM performance in aMCI, whereas in healthy ECs hippocampus volume should be age-related.
Materials and Methods
Subject Characteristics
Thirty subjects took part in this study (15 with aMCI and 15 EC). All aMCI subjects fulfilled Petersen criteria1: memory complaints corroborated by an informant, objective memory impairment for age (at least 1 SD below average normal scores on EM testing), essentially preserved general cognitive functioning, and intact functional activities (not demented). We excluded from the study subjects who had other neurologic or psychiatric conditions (including dementia and depression). Informed consent was obtained from each subject. The study was approved by the Ethical Committee of the Saint-Luc University Hospital (no. 2007/176). All but 1 subject in each group were right-handed. EM was evaluated with a French adaptation of the Consortium to Establish a Registry for Alzheimer's Disease Recall Test (sum of scores on 3 attempts to recall 10 words).9 General cognitive functioning was assessed through the MMSE (superior or equal to 25/30 in all subjects).
Age was not statistically different between groups (F1,29=2.61; P =.117): EC, 69.4 years (SD=4.8) and patients with aMCI, 72.3 years (7.3). Years of education from primary school were not different (F1,29=2.04; P =.165): EC, 14.9 years (2.4) and patients with aMCI, 13.7 years (2.7). Sex distribution did not significantly differ between groups (χ2=2.14, P =.143): EC, 66.7% of men and patients with aMCI, 40.0%]. Percentage of carriers of at least one allele ɛ4 of apolipoprotein E, a genetic risk factor for AD, was not different between EC and aMCI groups (χ2=2.22, P =.136): EC, 26.7% carriers and aMCI, 56.7% carriers. EM was statistically lower in patients with aMCI (F1,29=9.19, P =.005): EC, 19.9 (4.1) and patients with aMCI, 14.9 (4.9). MMSE was also lower in the aMCI than in the EC group (F1,29=6.47; P =.017): EC, 28.7 (1.5); aMCI, 27.3 (1.7).
Imaging Acquisition Parameters
Imaging was performed by using an Achieva 3T MR imaging scanner (Philips Medical Systems, Best, the Netherlands) and an 8-channel phased array head coil. Whole brain anatomy was provided by a 3D fast T1-weighted gradient-echo sequence with an inversion prepulse in a plane oriented along the longitudinal axis of the hippocampus (TR =9 ms, TE =4.6 ms, flip angle =8°, 150 sections, thickness =1 mm, resolution =0.81 × 0.95 mm2, scan time =491 s).
Data Analysis
Hippocampal Subfield Segmentation.
The hippocampal segmentation was carried out by using 2 successive methods. The whole hippocampus was initially segmented by completing the FreeSurfer image analysis pipeline (Martinos Center for Biomedical Imaging, Boston, Massachusetts) that is documented and freely available for download on-line (http://surfer.nmr.mgh.harvard.edu/). The technical details of these procedures were described in previous publications.10–13 In brief, the processing relevant to this work includes removal of nonbrain tissue by using a hybrid watershed/surface deformation procedure,12 automated Talairach transformation, and segmentation of the subcortical white matter and deep gray matter volumetric structures (including hippocampus, amygdala, caudate, putamen, and ventricles).13 In subjects (2 cases) with substantial anatomic differences with respect to the template, ie, enlarged ventricles, the resulting segmentation of the subcortical structures was improved by a pair-wise registration of the subject images to training images.14 We did not exclude these subjects to show that all clinical scans can be included in the second analysis part.
Next, automated segmentation of the hippocampus to its respective subfields was performed by using Bayesian inference and a statistical model of the medial temporal lobe. An atlas mesh has been previously built from the manual delineation of the right hippocampus of 10 control subjects.8 It was shown that the Dice overlap measures between manual and automated segmentation methods were approximately 0.7 for all the substructures (from CA2–3 and subiculum at 0.74 to CA1 at 0.62). For more details about this technique, and particularly about the borders used to define the different subfields, see Van Leemput et al.8
Statistical Analyses.
Because the atlas currently only includes right hippocampal subfields, we first evaluated left and right total hippocampus volumes in each group and we checked for the absence of significant lateralization effect. The right hippocampus was then segmented into 7 parts: CA1, CA2–3, CA4-DG, subiculum, presubiculum, fimbria, and hippocampal fissure. Statistical analyses were carried out by using STATISTICA version 9 (StatSoft France, Maisons-Alfort, France). We performed between-group comparisons with volumetric measurements as dependent variables and age and sex as covariates. We then evaluated in both groups which subfield volumes were explained by age or EM through a multiple linear regression, with sex introduced as a covariate in the model. Finally, we compared sensitivity and specificity of total hippocampus volume and the most relevant subfield volumes.
Results
The left and right hippocampus volumes were not significantly different in EC (F1,14=0.10; P =.752) and aMCI groups (F1,14=0.01; P =.996), allowing comparison of right hippocampal subfields.
Between-Groups Comparison of Volumetric Data
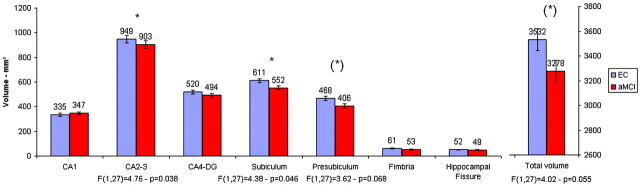
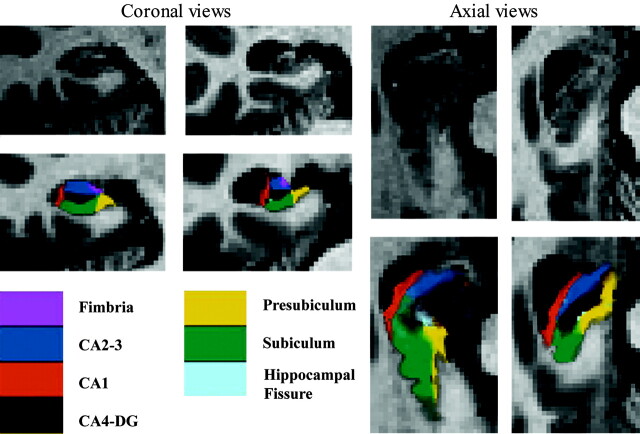
Total right hippocampus volume tended to be smaller in the aMCI than in EC group (P =.055). CA1 (P =.301), CA4-DG (P =.127), fimbria (P =.328), and hippocampal fissure (P =.706) volumes were not significantly different between the groups (Fig 1). By contrast, CA2–3 (P =.038) and subiculum (P =.046) were significantly smaller in the aMCI than in EC group, whereas presubiculum showed a similar trend (P =.068; Fig 1). A qualitative comparison between the hippocampus segmentation in 1 EC patient and in 1 patient with aMCI is presented in Fig 2.
Fig 1.
Volumetric data of EC (n =15) and patients with aMCI (n =15). Error bars represent SEM =SD/√n. *, Significant F-test (P < .050) given below the corresponding structure. (*), Trend toward a significant difference (P < .100). Age and sex are introduced as covariates in the model.
Fig 2.
Coronal and axial views of the right hippocampus. Left images are from a control; right images are from a patient with aMCI. The use of a huge zoom on a standard clinical MR imaging explains the low resolution.
Relationship between Volumetric Data, Age, and EM Performance
Total hippocampus volume was significantly explained by age in EC (P =.017) and by EM performance in aMCI groups (P =.027). Age tended to explain CA4-DG volume in ECs (P =.061). EM explained CA2–3 (P =.004), CA4-DG (P =.002), and subiculum (P =.006) volumes in the aMCI group (Table).
Results of the multiple linear regression between volumetric data, age, and EM in ECs and patients with aMCI
| Right Hippocampus | EC (n=15) |
aMCI (n=15) |
||||||
|---|---|---|---|---|---|---|---|---|
| β-age | P | β-EM | P | β-age | P | β-EM | P | |
| Total volume | −0.69 | .017 | −0.36 | .192 | −0.24 | .292 | 0.61 | .027 |
| CA1 | 0.16 | .555 | 0.10 | .735 | 0.44 | .107 | 0.35 | .222 |
| CA2–3 | −0.40 | .143 | −0.33 | .235 | 0.06 | .746 | 0.70 | .004 |
| CA4-DG | −0.56 | .061 | −0.29 | .321 | 0.04 | .849 | 0.80 | .002 |
| Subiculum | −0.50 | .126 | −0.30 | .368 | −0.23 | .246 | 0.69 | .006 |
| Presubiculum | −0.39 | .212 | −0.16 | .618 | −0.36 | .156 | 0.43 | .117 |
| Fimbria | −0.51 | .103 | −0.45 | .164 | −0.28 | .405 | 0.08 | .832 |
| Hippocampal fissure | <0.01 | .996 | −0.34 | .320 | −0.21 | .553 | −0.16 | .682 |
Note:—Sex has been introduced as a covariate in the model.
Bold type indicates significant values.
Discriminative Power of Hippocampus Subfield Volumetry
The discriminative power (correctly classified subjects) of total hippocampus volume was 60%, for a threshold of 3250 mm3, with a specificity of 80% (EC correctly classified) and a sensitivity of 40% (aMCI correctly classified). Subiculum volume offered a discriminative power of 77% (threshold, 575 mm3; specificity, 80%; sensitivity, 73%). Thus, 5 of 9 (56%) patients with aMCI without significant hippocampus atrophy were diagnosed by segmenting the subiculum. Similar results were obtained with the presubiculum (threshold, 440 mm3; specificity, 80%; sensitivity, 67%) and the CA2–3 (threshold, 865 mm3; specificity, 87%; sensitivity, 40%).
Discussion
To the best of our knowledge, this is the first study to propose automated segmentation of the hippocampus in aMCI. We consider it important to notify clinicians about the availability of this technique and the ease of its use in clinical routine (only a standard volumetric MR imaging is needed). Most of the processing time does not need any labor and people already familiar with FreeSurfer would not need any further training because these algorithms are directly implemented in the software.
It seemed from this study that segmenting subfields may allow improved sensitivity of MR imaging for early AD diagnoses. The present study is preliminary: follow-up of aMCI is ongoing to determine which proportion of convertors had early subiculum and CA2–3 atrophy. Larger groups of patients also will be necessary to confirm our results. Of note, although age is the main explanatory factor of hippocampus volume in EC; in patients with aMCI, it is the EM performance that better predicts both the total hippocampus volume and the subfields that were the most atrophic.
These results are somewhat different from the studies that applied a manual delineation6 or 3D surface mapping.7 This latest technique showed predominant subiculum atrophy in aMCI,7 though it suffers from an inherent lack of sensitivity in estimating the deepest subfields, such as CA2–3. The most important difference compared with manual delineation studies is that we found significant CA2–3 atrophy, whereas Mueller and Weiner found CA1–2 atrophy.6 In postmortem studies, it has been proposed that tau pathology first affects subfield boundaries,15 which stresses the importance of delineation boundary choice in segmentation (what we define as subiculum is sometimes included in CA1). Specifically, the reproducibility of our method will permit more consistency between studies than is possible with manual methods.8
Previous neuropathologic studies have shown that neuronal loss in the subiculum and CA1 are related to the severity of AD pathology.4 However, a recent postmortem study showed that preclinical AD displays CA1 neuronal hypertrophy, rather than atrophy.16 As we also studied predemential patients (aMCI), potentially presenting CA1 neuronal hypertrophy, it is not surprising that we did not find CA1 atropy on volumetric MR imaging. Besides, it has been proposed that CA3 was among the first subfields affected in aMCI due to the higher workload from synaptic plasticity requested by its involvement in associative memory.17
Conclusions
The involvement of the hippocampal subfields in predemential AD seems to be differential. We found a more pronounced atrophy in CA2–3 and in the subiculum. Automatically measuring hippocampus subfield volume seems to yield a promising surrogate marker with potential clinical and research applications. Segmenting hippocampal subfields is indeed feasible in clinical conditions (standard scanning conditions are sufficient; reasonable time of processing) and allows improved sensitivity to aMCI compared with total hippocampus volumetry.
Acknowledgments
We are grateful to Mert R. Sabuncu (Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Harvard Medical School) for help in processing subjects with anatomic anomalies (substantially enlarged ventricles).
Abbreviations
- AD
Alzheimer disease
- aMCI
amnestic mild cognitive impairment
- DG
dentate gyrus
- EC
elderly control
- EM
episodic memory
- MCI
mild cognitive impairment
- MMSE
Mini-Mental State Examination
Footnotes
Funding for this study was provided by the Belgian Foundation for Scientific Research (FNRS). K. Van Leemput was supported in part by the Academy of Finland (grant 133611), the National Institutes of Health (R01NS052585, R01EB006758, R01EB009051, P41RR014075), and Massachusetts General Hospital (Executive Committee on Research Formulaic Bridge Support/Proposal 2010A055663).
B.J.H. was the principal investigator of this study; he recruited and scanned the subjects and performed the neuropsychological evaluations and also performed the statistical analysis and wrote the text. K.V.L. performed the subfield segmentation and revised the text. M.K. performed the hippocampus segmentation, provided the picture, and revised the text. C.G. supervised the scanning of the subjects and revised the text. X.S. supervised the cognitive evaluations and revised the text. A.I. supervised the project, diagnosed the MCI patients, and revised the text.
Disclosures: Bernard Hanseeuw: Research Support (including provision of equipment or materials): Belgian National Foundation for Scientific Research (FNRS). Details: FNRS is paying the salary of the Principal Investigator (Bernard Hanseeuw-1820€/month). FNRS also paying the fees for MR imaging utilization (150€/hour). The grant received is a total amount of 27,000€.
References
- 1. Petersen RC. Mild cognitive impairment as a diagnostic entity. J Intern Med 2004; 256: 183–94 [DOI] [PubMed] [Google Scholar]
- 2. Dubois B, Feldman HH, Jacova C, et al. Research criteria for the diagnosis of Alzheimer's disease: revising the NINCDS-ADRDA criteria. Lancet Neurol 2007; 6: 734–46 [DOI] [PubMed] [Google Scholar]
- 3. Chupin M, Gerardin E, Cuingnet R, et al. Fully automatic hippocampus segmentation and classification in Alzheimer's disease and mild cognitive impairment applied on data from ADNI. Hippocampus 2009; 19: 579–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Giannakopoulos P, Kovari E, Gold G, et al. Pathological substrates of cognitive decline in Alzheimer's disease. Front Neurol Neurosci 2009; 24: 20–29 [DOI] [PubMed] [Google Scholar]
- 5. Frisoni GB, Ganzola R, Canu E, et al. Mapping local hippocampal changes in Alzheimer's disease and normal ageing with MRI at 3 Tesla. Brain 2008; 131: 3266–76 [DOI] [PubMed] [Google Scholar]
- 6. Mueller SG, Weiner MW. Selective effect of age Apo e4 and Alzheimer's disease on hippocampal subfields. Hippocampus 2009; 19: 558–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Apostolova LG, Thompson PM, Green A. 3D comparison of low intermediate and advanced hippocampal atrophy in MCI. Hum Brain Mapp 2010; 31: 786–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Van Leemput K, Bakkour A, Benner T, et al. Automated segmentation of hippocampal subfields from ultra-high resolution in vivo MRI. Hippocampus 2009; 19: 549–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Morris JC, Mohs RC, Rogers H, et al. Consortium to establish a registry for Alzheimer's disease (CERAD) clinical and neuropsychological assessment of Alzheimer's disease. Psychopharmacol Bull 1988; 24: 641–52 [PubMed] [Google Scholar]
- 10. Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage 1999; 9: 179–94 [DOI] [PubMed] [Google Scholar]
- 11. Fischl B, Salat DH, Busa E, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 2002; 33: 341–55 [DOI] [PubMed] [Google Scholar]
- 12. Segonne F, Dale AM, Busa E, et al. A hybrid approach to the skull stripping problem in MRI. Neuroimage 2004; 22: 1060–75 [DOI] [PubMed] [Google Scholar]
- 13. Fischl B, Salat DH, van der Kouwe AJ, et al. Sequence-independent segmentation of magnetic resonance images. Neuroimage 2004; 23 Suppl. 1: S69–84 [DOI] [PubMed] [Google Scholar]
- 14. Sabuncu MR, Thomas Yeo BT, Van Leemput K, et al. A generative model for image segmentation based on label fusion. IEEE Trans Med Imaging 2010; 29: 1714–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lace G, Savva GM, Forster G, et al. Hippocampal tau pathology is related to neuroanatomical connections: an ageing population-based study. Brain 2009; 132: 1324–34 [DOI] [PubMed] [Google Scholar]
- 16. Iacono D, Markesbery WR, Gross M, et al. The Nun study: clinically silent AD neuronal hypertrophy and linguistic skills in early life. Neurology 2009; 73: 665–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yassa MA, Stark SM, Bakker A, et al. High-resolution structural and functional MRI of hippocampal CA3 and dentate gyrus in patients with amnestic mild cognitive impairment. Neuroimage 2010; 51: 1242–52 [DOI] [PMC free article] [PubMed] [Google Scholar]