Abstract
We describe implementation of a set-based method to assess the significance of findings from genome-wide association study data. Our method, implemented in PLINK, is based on theoretical approximation of Fisher’s statistics such that the combination of p-vales at a gene or across a pathway are done in a manner that accounts for the correlation structure, or linkage disequilibrium, between SNPs. We compare our method to a permutation based product of p-values approach and show a typical correlation in excess of 0.98 for a number of comparisons. The method gives Type I error rates that are less than or equal to the corresponding nominal significance levels, making it robust to the effects of false positives. We show that in broadly similar populations, reference datasets of markers are an appropriate substrate for deriving marker-marker LD, negating the need to access individual level genotypes, greatly facilitating its generic applicability. We show that the method is thus robust to LD-associated bias and has equivalent performance to permutation-based methods, with a significantly shorter runtime. This is particularly relevant at a time of increasing public availability of significantly larger genetic datasets and should go a long way to assist in the rapid analysis of these datasets.
Keywords: GWAS, set-based analysis, multiple dependent tests
Introduction
Gene-based, or more generally, set-based tests are often used to undertake secondary analyses of genome-wide association study (GWAS) data, the aim being to identify genes or pathways comprised of sets of genes that are associated with a phenotype . These tests aim to combine information from several single nucleotide polymorphism (SNP) markers and estimate significance of the set as a whole, rather than at every marker individually. The challenge here is to take into account the different number of markers in a set and the linkage disequilibrium (LD) structure between these markers to make the overall set-based p-value comparable across sets. There are several ways to assess the significance of a set of SNPs including rank/threshold truncated products of p methods [Zaykin et al., 2002, Dudbridge and Koeleman, 2003, Moskvina et al., 2008], set-based analysis (PLINK, [Purcell et al., 2007]), and direct calculations of the significance using association results and correlations e.g. Hotelling’s T2 test (see e.g. [Roeder et al., 2005]). Permutation analysis is one of the most widely used ways to account for LD. If one marker is significantly associated with the disease then other markers that are correlated by virtue of LD will also tend to be associated. Where this occurs, the set-based p-value will be inflated unless there is an adjustment for LD. However this requires individual genotypes which are not always available to analysts beyond the immediate research teams involved, and the permutation process is time-consuming on a genome-wide scale, particularly when datasets become larger and in doing so, include individuals from diverse ancestries. Liu et al., 2010 have introduced a method which makes use of publically available data (e.g. HapMap (http://hapmap.ncbi.nlm.nih.gov/) and 1000 Genomes databases (http://www.1000genomes.org/)) to estimate correlations between SNPs. They suggest estimating the empirical set-based p-value using the Monte Carlo approach where a large number of simulated multivariate normal vectors are drawn from the multivariate normal distribution with zero mean and variance matrix of pair-wise linkage disequilibrium (LD) values. In this way the individual genotype information is not required, however it is still necessary to perform a large number of simulations in order to estimate empirical significance for a set of SNPs.
In this paper we suggest an alternate approach to calculate the significance of a set of SNPs based on theoretical approximation [Brown, 1975] of Fisher’s statistics. If Fisher’s statistics combines the results of several tests when the tests are dependent, the method suggested here requires only the list of p-values for each SNP and knowledge of correlations between SNPs. The latter can be calculated from the data directly, or for those without access to the raw data, they can be estimated using publically available data, e.g. HapMap or 1000 Genomes databases. Thus, the approach we evaluate here has the potential to both permit analyses without access to all the raw GWAS data, and to avoid the need for permutation testing.
To evaluate the method, we compared set-based analyses results based on Brown’s approximation with a permutation based product of p (ProdP) method using individual genotypes generated in GWAS studies of schizophrenia, one restricted to a UK sample [O’Donovan et al., 2008] the other on a more complex dataset reported by the International Schizophrenia Consortium [ISC, 2008]. We use the UK Schizophrenia (SZ) dataset to evaluate the performance of Brown’s method for a relatively homogenous dataset that contains little population substructure, and the ISC data to see how this method performs in the presence of stratification as a result of including many European samples of diverse ancestry.
Materials and Methods
Approximation based method
Fisher’s method of combining probabilities is asymptotically optimal [Littell and Folks, 1971, 1973] for obtaining the overall significance of a set of p-values obtained from independent tests of the same null hypothesis (here, that each SNP is not associated with disease). The combined chi-square statistic
| (1) |
(where N is the number of markers (tests) and pi (i = 1,…, N) are the corresponding p-values) under the null hypothesis has a χ2 distribution with 2N degrees of freedom assuming that the performed tests are independent. Since markers, particularly those in close physical proximity (e.g. within a modestly sized gene), are often not independent as a result of LD, direct application of this test statistic is invalid, since the assumption of independence is violated. Therefore, instead of using the χ2 distribution for calculating an overall set-based p-value, permutations of case/control status are used to simulate the null hypothesis and assess the empirical distribution of the sum of the logarithms of p-values which is equivalent to the product of p-values. Modifications of this general approach are sometimes used in practice, in particular, calculating products of rank truncated [Dudbridge and Koeleman, 2003], or threshold truncated [Zaykin et al., 2002] p-values. The minimum p-value may also be used as the measure of significance of the set of SNPs, with permutation used to correct for performing multiple non-independent tests. However, the limitations of those approaches in the context of GWAS data are becoming clearer. The use of minimum p-value does not allow extraction of information from multiple quasi-independent signals from individual genes or gene pathways. As for truncation, it is now very clear that in typical GWAS datasets, much of the true association signal lies in extremely weakly associated variants that are not even nominally significant [ISC, 2009], making it unclear what threshold to apply for truncation. This leads to multiple threshold testing which then becomes problematic for interpreting significance. Also, truncation requires permutation testing, making it ineligible for the situations for which we propose Brown’s method.
Brown (1975) suggested a method for combining non-independent tests (see also [Kost & McDermott, 2002] and [Makambi, 2003]). If the tests are not independent, then the statistic in (1) has mean m = 2N and variance (σ2) where
| (2) |
and where pi and pj (i , j = 1,…, N) are the p-values for each test and covariance (cov) is calculated as
| (3) |
for non-negative correlation coefficients ρij between the two variables which we approximate by the correlation between two SNPs i and j, where SNPs are coded as 0,1,2 for genotypes AA, Aa, and aa accordingly. These covariances are evaluated from the Gaussian quadrature [Krylov, 1962]. It is also possible to evaluate the covariance (3) for negative correlation coefficients; however the sign of correlations between markers is defined by allele coding and can always be set as positive. Finally the overall significance of a set of non-independent tests is calculated using the statistic T which under the null hypothesis follows the central chi-square distribution
| (4) |
(see [Brown, 1975] for details).
Note that directionality of effects has no bearing on the correlation between SNPs, since this is the same regardless of which allele corresponds to increased risk of disease. The directionality of effects is only relevant if there is a prior hypothesis on the true direction of effect (e.g. replicating a previous result). If there is a prior hypothesis of directionality for multiple SNPs at a locus, or a subset of SNPs Brown’s method can be used without modification on that restricted set of SNPs using one-sided p-values, with the proviso that specification of SNPs and directionality are based upon a fully independent dataset. This proviso is required in order for the specified alleles to have a uniform distribution under the null hypothesis of no association.
Data
For the assessment of the performance of Brown’s method on genome-wide scale data, we used sets of SNPs annotated to genes. We then explored real GWAS datasets with different numbers of SNPs per set and different linkage disequilibrium (LD) patterns.
We assessed gene-wise significance using Brown’s method for all genes in the UK SZ dataset of 479 cases and 2938 controls. After the applied quality control (QC) criteria 377,742 SNPs were used in the analysis. Details of the QC measures for SNP and subject inclusion are described in the primary manuscript [O’Donovan et al., 2008]. SNPs were assigned to genes if they were located within the genomic sequence corresponding to the start of the first and the end of the last exon of any transcript corresponding to that gene. The chromosome and locations for all SNPs and genes and their identifiers were taken from the human genome assembly build 36.2 of the National Center for Biotechnology Information (NCBI) database as in [Moskvina et al., 2008]. In total, we obtained 11,791 unique genes (2 - 796 SNPs per gene). Single SNP association p-values were generated using the Armitage trend (1 df) test and then genomic control [Devlin and Roeder, 1999] adjusted based upon the observed λGC for the whole dataset of 1.08.
We also tested the robustness of Brown’s method on a second dataset consisting of 3,322 schizophrenic cases and 3,587 controls collected from 8 centres in Europe [ISC, 2008]. For the 739,995 SNPs which passed QC, as in the primary manuscript [ISC, 2008], p-values were obtained using a Cochran-Mantel-Haenszel (CMH) test as implemented in PLINK [Purcell et al., 2007]. As above we adjusted these p-values for the observed λGC of 1.1, and used these for gene-based analyses. For the SNP to gene annotation we used NCBI database, build 36.2 (as for UK Schizophrenia dataset). There were 13,981 genes, the number of SNPs per gene ranging between 2 and 1610.
To calculate empirical gene-wide significance for each gene, we performed 1000 genome-wide permutations for each GWAS data set. For the ISC data, we permuted each stratum separately and used the CMH test to calculate p-values for each permutation. For each gene in each permutation we obtained the product of all p-values in a gene as for the original dataset. We then calculated the empirical p-values for each gene in the observed data by determining the proportion of permuted datasets where the corresponding p-value obtained for each gene was equal to or smaller than was observed in the true dataset. For genes with gene-wise empirical p-values ≤ 0.005 we ran an additional 100,000 permutations to assess the significance with higher precision.
The primary analysis for most GWAS studies is based upon an additive model, which was therefore the focus of the present investigation. Generally speaking, it is not necessary for the individual p-values used in either the Fisher’s method for combining p-values (here implemented as ProdP) or Brown’s method to be calculated under the same model, provided that under the null hypothesis of no association, they have a uniform distribution. However, when non-additive models are tested, the covariance between the tests for each individual marker depends on the genetic model. For example, if the model is recessive, then for the Brown’s method, the correlation between tests must be calculated based upon genotypes rather than alleles, where genotypes are coded 0, 0, 1, the latter being the code for homozygosity for the putative recessive allele. For a dominant model, the genotypes must be coded 0, 1, 1, where 1 applies to heterozygous or homozygous carriers for the putative risk allele. To explore the applicability of the Brown’s approach, we have examined dominant and recessive disease models in the UK SZ dataset. Individual locus p-values were calculated using a χ2 test with 1df for 2×2 contingency tables of genotype counts under dominant and recessive disease models.
Results
UK Schizophrenia data
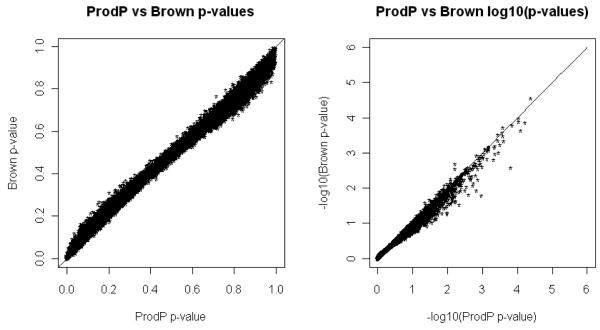
Figure 1 shows scatter plots of Brown’s and ProdP p-values in nominal and logarithmic scales for the UK Schizophrenia data. The correlation between the p-values obtained for each gene using the ProdP and Brown’s methods was 0.996. However, the shape of the scatter plot suggests that Brown’s approximation of the set test statistics is conservative relative to ProdP for results that have p-values < ~0.5 but anticonservative for results with p-values > ~0.5.
Figure 1.
Scatter plot of p-values calculated using ProdP method (the number of permutations is 1000) vs. Brown’s method for the UK SZ data. P-values are shown in nominal and logarithmic scale.
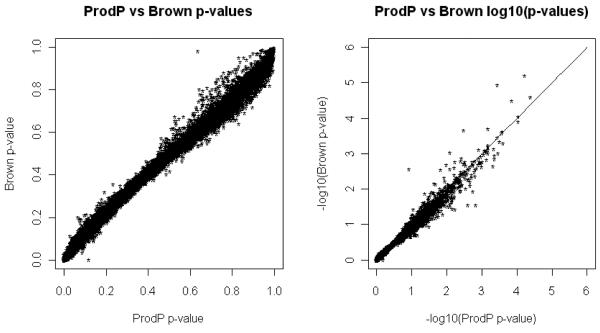
Next, we tested the performance of Brown’s method under the assumption that individual genotypes were not available for the UK study for estimating the correlations between markers. Instead, correlations were estimated using 60 unrelated subjects (parents) of European decent from HapMap database, HapMap2 Release. Brown’s p-values were still highly consistent with the ProdP p-values, with a correlation coefficient of 0.994 (Figure 2). There were 368,804 SNPs common to the UK and HapMap datasets. Note that when we restrict the analysis only to the genes where all SNPs in the gene are present in HapMap, the number of genes is reduced to 9,784.
Figure 2.
Scatter plot of p-values (in nominal and logarithmic scales) calculated using the ProdP method (the number of permutations is 1000) vs. Brown’s method where the LD between markers are estimated from HapMap2 data and include only genes where all SNPs are common to the UK SZ and HapMap data.
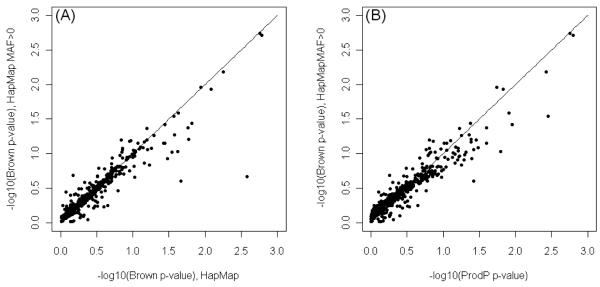
Another potential issue to consider is that when only 60 individuals from HapMap are used to estimate the LD, some SNPs with relatively small minor allele frequencies (MAF) in the test dataset (here the UK SZ dataset) are monomorphic in the HapMap. Since it is not possible to estimate covariance between monomorphic and polymorphic markers, there are two simple ways of dealing with this; 1) set the covariance to 0 or 2) exclude SNPs that are monomorphic in the HapMap data. In the analyses reported above, we used the first option. Setting covariance to 0 decreases both σ2 (2) and the constant c (4), which can lead to either an increase and decrease of p-values depending on the value of the chi-square statistic T0, the number of uncorrected degrees of freedom (1) and the value of the constant c (4). The other way of dealing with this problem is to exclude these markers from the calculations, although when markers with relatively small MAF are actually associated with disease, this may reduce power. In the UK SZ data there were 520 genes where one or more SNP had MAF=0 in HapMap. Figure 3 shows discrepancies in the gene wide p-values when these SNPs are included or excluded, although for future applications, this is less likely to be an issue given that there are many publically available GWAS data sets, and the sample sizes available through the HapMap and the 1000 Genomes Project are growing.
Figure 3.
Scatter plot of gene-wise p-values where the SNPs with MAF=0 in HapMap are included (x-axis) or excluded (y-axis) compared with Brown’s p-values (A) and ProdP p-values (B), both in logarithmic scales.
In Supplemental Figure 1 we compare the performance of the tests where SNP p-values were calculated under dominant and recessive disease models. The correlations between test statistics were calculated from the GWAS dataset itself using 0, 1, 1 and 0, 0, 1 to code the genotypes respectively for dominant and recessive models. For the dominant model, the correlation between ProdP and Brown’s methods was similar to that observed for the additive model (correlation = 0.996). However, when recessive disease model was used, the results were less well correlated (correlation = 0.965). This is to be expected because for many loci, there are small numbers of homozygotes for the minor alleles (e.g. even when MAF = 0.05, in our SZ UK sample we only expect a single case homozygote under HWE and the null hypothesis) which violates the assumptions of the χ2 test (minimum expected cell size ≥ 5), and it is therefore common practice to apply a more stringent MAF filter for testing recessive models. (Note that even if such alleles are excluded from the actual GWAS, examples will be generated during the permutation process). We re-filtered the data (MAF≥0.1) to ensure that in the observed data, the minimum cell size for any marker was 5 and re-ran the ProdP and Brown analyses. The result was an improvement in the correlation coefficient, which increased to 0.980 (see also Supplemental Figure 2).
ISC Schizophrenia data
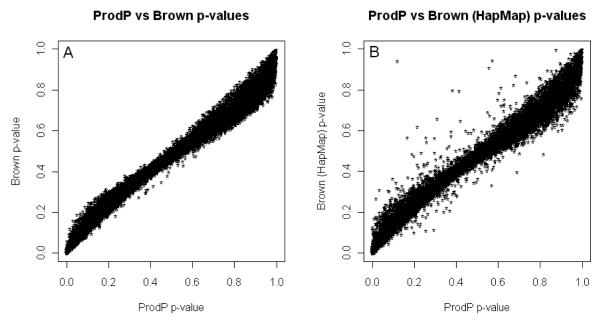
To investigate the robustness of this method in the presence of less homogenous GWAS datasets, we applied the same strategy of comparing Brown’s and ProdP methods using ISC GWAS data. Again, Brown’s method performs very well, the correlation between ProdP and Brown’s p-values being 0.990 (Figure 4 (A)). As before, where p-values are small, Brown’s approximation is conservative relative to ProdP, whereas where they are large, they are anticonservative. Also as for the UK data, if we use marker correlations estimated from the HapMap2 CEU data rather than the ISC genotypes, the results change only slightly, the correlation between ProdP and Brown’s p-values being 0.985 (Figure 4 (B)), or 0.990 after excluding markers with MAF=0 in the HapMap2 dataset.
Figure 4.
Scatter plot of gene-wise ProdP vs. Brown’s p-values in ISC data using correlations between SNPs from the ISC data (A) and from HapMap data (B).
To determine whether Brown’s method was conservative for significant results or whether it was ProdP that was anticonservative, we undertook an assessment of the type I error rates under the null. We selected one of the 1000 sets of permuted gene-wide ProdP statistics (of the UK SZ dataset) as “true (null) study” and assessed empirical significance of the ProdP results by bootstrapping from the remaining sets with replacement as in our previous paper [Moskvina et al., 2008]. This procedure was repeated 1000 times. For Brown’s method, we calculated the p-value for each gene directly in each of the 1000 replicates using correlations estimated from the UK SZ dataset. Then for each replicate we calculated the proportion of genes with p≤0.05 and 0.01. The proportions of genes with ProdP p-values ≤0.05 and ≤0.01 were 0.050 [SD=0.002], 0.01 [SD=0.001] respectively; the corresponding Brown’s results were 0.041 [SD=0.002] and 0.008 [SD=0.001]. The results were similar when we used the HapMap to estimate marker correlations for Brown’s method, these proportions were 0.038 [SD=0.002] and 0.007 [SD=0.0008] respectively. Thus, we can conclude that Brown’s method is slightly conservative in absolute terms rather than only relative to ProdP, which has the expected type I error rate for the thresholds. ProdP and Brown p-values for the 20 most significant genes are given for the UK SZ study and ISC data in Supplementary Tables 1 and 2. Brown’s p-values were calculated using correlations estimated from the relevant GWAS datasets and also from HapMap. Regardless of the datasets from which the correlations were derived, Brown’s p-values are similar to those based upon ProdP.
Discussion
In conclusion, we have evaluated the performance of Brown’s approximate method as applied to combining multiple SNPs for calculating gene-wide significance using two genome-wide association datasets, and using a widely used permutation-based product of p-values approach for each gene as a ‘gold-standard’ comparator. The advantage of the approximate method is that it does not require time-consuming permutations or individual genotype data. Compared to the product of p-values, the approximate method is consistently slightly conservative for significant results, that is it would miss some associations that could be detected through permutation (e.g. see LOC100129827 in Supplemental Table 2) and anticonservative for non-significant results, but overall, in terms of speed it represents a highly efficient alternative for mining GWAs datasets than more computationally intensive methods. Moreover, we show that in broadly similar populations, reference datasets of markers are an appropriate substrate for deriving marker-marker LD, thus negating the need to access individual level genotypes. Although a small number of errors arise due to markers that are of low frequency in the GWAS dataset, and which are monomorphic in the reference dataset, the impact of these at a genome wide level is not substantial, and can be mitigated by excluding markers that are monomorphic in the reference panel.
We should note that including markers with low MAF can influence the Brown’s method in two ways. Firstly, the theoretical distribution of the association test may not be valid in small samples and as a result, the p-values may not have a uniform distribution under the null. This will tend to result in an increased false-positive rate both for tests of individual SNPs and for the Brown’s method. This problem can potentially be alleviated by obtaining individual p-values for rare SNPs by Fisher’s exact method, although this is not applicable when the association analysis involves covariates. Secondly, estimating the correlation between pairs of SNPs when one or both have low MAFs is likely to be inaccurate, particularly when the sample being used to estimate correlation is small (e.g. HapMap 2). This will make the correlation estimates more variable, but should not bias them, provided the population used to estimate correlations is comparable to that in which the association testing is being performed. This will result in an increase in the variance between significance estimates from the Brown’s and ProdP methods, (Figure 4), although not an overall bias in the significance estimates. Nevertheless, as is routine in GWAS, it is sensible to apply a filtering criterion to remove SNPs with low MAF, with the exact threshold for filtering dependent on sample size. We also note that with ever increasing public datasets with detailed LD information and larger sample sizes, the estimates of marker LD are likely to become more precise, even for low frequency markers.
One consequence of the small inflation in significance for non-significant genes might arise if gene-wide p-values estimated using Brown’s method are themselves subsequently combined, as may happen if a researcher wishes to look at the distribution of gene-wide p-values among a set of genes comprising biological pathway, or indeed the distribution of gene-wide p-values for the same gene from different independent studies. Such an approach is again likely to be conservative for genes or pathways that are genuinely associated with a disease, but for those that are not, the inflation that occurs for non-significant genes may combine across many genes to generate a false positive result. Thus, we would caution against applying Brown’s method in this manner.
To facilitate application of this method, we have made gene-wise calculations implementable on the basis of HapMap data available in PLINK (plink --bfile hapmap_CEU_r23a --set-screen pvals.dat --make-set glist.dat), where “pvals.dat” is the file with SNP names and their p-values, and “glist.dat” is the file with genes’ names and their start and end positions.
Supplementary Material
Acknowledgements
This study makes use of data generated by the Wellcome Trust Case Control Consortium (www.wtccc.org.uk) and the International Schizophrenia Consortium. This research was supported by grants from the MRC (UK) and by a NIMH (USA) CONTE: 2 P50 MH066392-05A1.
References
- Zaykin DV, Zhivotovsky LA, Westfall PH, Weir BS. Truncated product method for combining p-values. Genet Epidemiol. 2002;22:170–185. doi: 10.1002/gepi.0042. [DOI] [PubMed] [Google Scholar]
- Dudbridge F, Koeleman BPC. Rank truncated product of p-values, with application to genomewide association scans. Genet Epidemiol. 2003;25:360–366. doi: 10.1002/gepi.10264. [DOI] [PubMed] [Google Scholar]
- Moskvina V, Craddock N, Holmans P, Nikolov I, Pahwa JS, Green E, Owen MJ, O’Donovan MC. Gene-wide analyses of genome-wide association data sets: evidence for multiple common risk alleles for schizophrenia and bipolar disorder and for overlap in genetic risk. Molecular Psychiatry. 2008;14:252–260. doi: 10.1038/mp.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am Jour of Hum Genet. 2007;81 doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder K, Bacanu SA, Sonpar V, Zhang X, Devlin B. Analysis of single-locus tests to detect gene/disease associations. Genet Epidemiol. 2005;28(3):207–219. doi: 10.1002/gepi.20050. [DOI] [PubMed] [Google Scholar]
- Liu JZ, Mcrae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, AMFS. Hayward NK, Montgomery GW, Visscher PM, et al. A Versatile Gene-Based Test for Genome-wide Association Studies. American Society of Human Genetics. 2010 doi: 10.1016/j.ajhg.2010.06.009. DOI10.1016/j.ajhg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MB. A method for combining non-independent, one-sided tests of significance. Biometrics. 1975;31:978–992. [Google Scholar]
- International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009 doi: 10.1038/nature08185. doi:10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- International Schizophrenia Consortium Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littell RC, Folks JL. Asymptotic optimality of Fisher’s method of combining independent tests. Journal of American Statistical Association. 1971;66:802–806. [Google Scholar]
- Littell RC, Folks JL. Asymptotic optimality of Fisher’s method of combining independent tests II. Journal of American Statistical Association. 1973;68:193–194. [Google Scholar]
- Kost JT, McDermott MP. Combining dependent p-values. Stat. Probab. Lett. 2002;60:183–190. [Google Scholar]
- Makambi K. Weighted inverse chi-square method for correlated significance tests. J. Appl. Stat. 2003;30:225–234. [Google Scholar]
- Krylov VI. Approximate calculation of integrals. McMillan; New York: 1962. [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55(4):997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.