Abstract
eEF2K (eukaryotic elongation factor 2 kinase) is a Ca2+/CaM (calmodulin)-dependent protein kinase which regulates the translation elongation machinery. eEF2K belongs to the small group of so-called ‘α-kinases’ which are distinct from the main eukaryotic protein kinase superfamily. In addition to the α-kinase catalytic domain, other domains have been identified in eEF2K: a CaM-binding region, N-terminal to the kinase domain; a C-terminal region containing several predicted α-helices (resembling SEL1 domains); and a probably rather unstructured ‘linker’ region connecting them. In the present paper, we demonstrate: (i) that several highly conserved residues, implicated in binding ATP or metal ions, are critical for eEF2K activity; (ii) that Ca2+/CaM enhance the ability of eEF2K to bind to ATP, providing the first insight into the allosteric control of eEF2K; (iii) that the CaM-binding/α-kinase domain of eEF2K itself possesses autokinase activity, but is unable to phosphorylate substrates in trans; (iv) that phosphorylation of these substrates requires the SEL1-like domains of eEF2K; and (v) that highly conserved residues in the C-terminal tip of eEF2K are essential for the phosphorylation of eEF2, but not a peptide substrate. On the basis of these findings, we propose a model for the functional organization and control of eEF2K.
Keywords: calmodulin (CaM), eukaryotic elongation factor 2 (eEF2), α-kinase, SEL1 domain
Abbreviations: ATP-γS, adenosine 5′-[γ-thio]triphosphate; CaM, calmodulin; eEF2, eukaryotic elongation factor 2; eEF2K, eEF2 kinase; GST, glutathione transferase; HEK, human embryonic kidney; HRP, horseradish peroxidase; LB, Luria–Bertani; MHCKA, myosin heavy-chain kinase A; mTORC1, mammalian target of rapamycin, complex 1; Ni-NTA, Ni2+-nitrilotriacetic acid; STD, saturation transfer difference; TEV, tobacco etch virus; TRPM, transient receptor potential melastatin-like
INTRODUCTION
eEF2K [eEF2 (eukaryotic elongation factor 2) kinase] catalyses the phosphorylation of eEF2, which inactivates eEF2 by impairing its ability to bind to the ribosome [1,2]. eEF2K was first identified as a Ca2+/CaM (calmodulin)-dependent enzyme [3]. Following the cloning of cDNAs encoding eEF2K, it became clear that eEF2K does not belong to the main eukaryotic superfamily of protein kinases, but instead to a small group of genes dubbed ‘α-kinases’ [4,5]. The α-kinases display no clear sequence homology to other protein kinases [6]. eEF2K is a well-characterized member of this group; its substrate eEF2 is known and there is substantial information on the intracellular regulation of eEF2K activity (reviewed in [7,8]). In contrast, less information is available about these aspects for other members of this group, although MHCKA (myosin heavy-chain kinase A), in particular, has been studied in considerable detail (see for example [9–11]). The first crystal structure for an α-kinase (for the kinase domain of the ion-channel TRPM7 (transient receptor potential melastatin-like 7, also termed ChaK1 (channel-kinase 1) [12]) revealed that, despite the lack of sequence homology, the three-dimensional structure of this α-kinase displayed marked similarity to members of the main eukaryotic protein kinase superfamily. Structures have also now been determined for the catalytic domain of Dictyostelium MHCKA [13], and these show substantial similarity to that of the α-kinase domain of TRPM7. Both TRPM7 and the closely related channel TRPM6 have been studied in some detail (reviewed in [14]); however, the biological significance of their α-kinase domains remains to be elucidated since, for example, this domain of TRPM7 does not appear to be required for its channel function [15].
The activity of eEF2K is normally completely dependent upon Ca2+/CaM [3,16]. It can also be activated by phosphorylation by the AMP-activated protein kinase (at Ser398; [17]) or cAMP-dependent protein kinase (at Ser499; [18]). Conversely, its activity is impaired by signalling through mTORC1 (mammalian target of rapamycin, complex 1) [19], an effect which involves phosphorylation of eEF2K at one or more of three sites, Ser78 [20], Ser366 [21] and Ser359 [22].
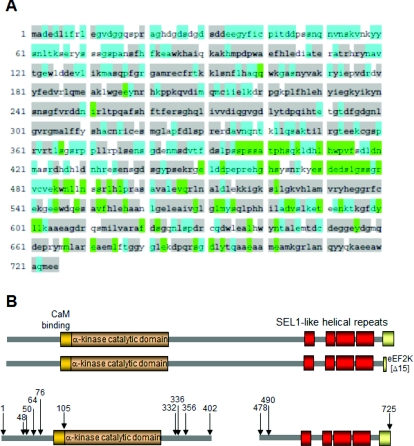
The overall layout of the structure of eEF2K is depicted in Figure 1(A). Its α-kinase catalytic domain lies towards the N-terminus, with the CaM-binding site immediately N-terminal to this [23,24]. The function of the region that is N-terminal of the CaM-binding site is not understood. Towards the C-terminus lie four predicted α-helical regions, which resemble SEL1-type repeats found in certain other proteins. Such features often provide a platform for protein–protein interactions [25]. Their role in eEF2K is unknown. At the extreme C-terminus is a region that is required for eEF2K to phosphorylate its substrate, eEF2 [24]. Between the catalytic and SEL1-type domains lies a region that is predicted to be unstructured and which may act as a linker between the two other major domains. Indeed, the fact that this region is highly conserved (Figure 1A) suggests that it is important for the function and/or regulation of eEF2K.
Figure 1. Structural organization and sequence conservation of eEF2 kinase.
(A) Sequence conservation between vertebrate eEF2K orthologues: grey, residues that are conserved from mammals to fish/reptiles; green, conserved across the five of the six species listed, or a conservative replacement in the sixth species; and blue, conserved in mammals. The sequences and GenBank® accession numbers on which the analysis is based are human (NP-037434), mouse (AAH55361), cow (NP_001179471), Anolis carolinensis (XP_003229274; the Carolina anole, a reptile), zebra finch (XP_002197529) and zebra fish (NP_001002740). (B) Upper panel: structural layout of full-length eEF2K. Lower panel: the main truncation mutants tested in the present study (data in Figure 4A). This panel is drawn approximately to scale.
The functional organization and regulation of eEF2K remain poorly characterized, although eEF2K is the best-understood member of this group in terms of both its physiological role and the inputs that modulate its activity. This prompted us to investigate the roles of the identified domains in eEF2K in its function and its control, in particular the possible interplay between the kinase domain and C-terminal regions of eEF2K.
The results of the present study show that the extreme N-terminal, linker and C-terminal ‘SEL1’ regions of eEF2K are not required for intrinsic activity or control by Ca2+/CaM. However, the C-terminal SEL1 region is indispensable for phosphorylation of substrates in trans, i.e. both the physiological substrate eEF2 and an artificial peptide substrate. Phosphorylation of eEF2 was much more strongly dependent upon the highly conserved C-terminal tip of the eEF2K polypeptide than phosphorylation of a peptide substrate. The present study shows that the catalytic and C-terminal domains, expressed separately, can interact with one another, leading to a model where the C-terminal part of eEF2K serves to recruit substrates to the catalytic domain for phosphorylation.
EXPERIMENTAL
Chemicals and other reagents
All chemicals and biochemicals were purchased from BDH-Merck and Sigma–Aldrich unless otherwise stated. Chromatography columns were purchased from GE Healthcare. [α-32P]ATP and [γ-32P]ATP were from PerkinElmer. Antisera against eEF2K and eEF2 (total and phospho-specific) were as described previously [17,20] unless stated otherwise. Plasmids for expression of eEF2K were as described previously [20,26] and were sequenced prior to use. eEF2 was purified from HeLa cell lysates (CilBiotech) as described by Knebel et al. [27].
Cloning and mutagenesis
For expression in Escherichia coli, the cDNA for human eEF2K was cloned between the BamHI and XhoI sites of the vector pGEX-6P (GE Healthcare), which yields a fusion protein containing GST (glutathione transferase) at the N-terminus. This can be removed by cleavage by the PreScission™ protease. Mutations were introduced by PCR mutagenesis using the QuikChange™ system (Stratagene). C-terminal truncations were created by introducing a termination codon at the appropriate position in the sequence, again using QuikChange™. Fragments of human eEF2K were created by amplifying the appropriate portion of the cDNA sequence (by PCR) and cloning this into the pGEX-6P vector. All vectors were resequenced prior to use.
For some experiments, eEF2K or mutants were expressed in HEK (human embryonic kidney)-293 cells. The cDNA for eEF2K was inserted into the vector pHM [28], which provides N-terminal His6 and Myc tags. Mutations were introduced by QuikChange™, and the vectors were resequenced.
To allow co-expression of two human eEF2K domain fragments, a different vector system was used. An N-terminal eEF2K fragment, eEF2K[48–336] was cloned into the T7 expression vector pNICZb, which provides N-terminal His6 and Z-basic [29] tags. A C-terminal eEF2K fragment, eEF2K[490–725], was cloned into the Duet expression vector pCDF, which provides an N-terminal His6 tag. The tags were removed by cleavage using the TEV (tobacco etch virus) protease (Invitrogen). Vectors were created using a ligation-independent cloning method [30,31].
Protein expression
For expression of GST-tagged eEF2K in bacteria, chemically competent E. coli Rosetta cells (Novagen) were transformed with the appropriate vector and grown at 37°C in LB (Luria–Bertani) medium supplemented with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol. When the attenuance at 600 nm had reached 0.5, expression of GST–eEF2K was induced by adding 0.5 mM isopropyl β-D-thiogalactoside. Cells were grown for a further 16 h at 18°C prior to harvesting. Although induction at 25°C gave a higher total yield of GST–eEF2K, much less of the protein was soluble. Cells were broken by sonication in buffer comprising 50 mM Tris/HCl, pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 0.5 mM EGTA, 5% (v/v) glycerol, 0.03% Brij 35, 14 mM 2-mercaptoethanol, PMSF (added just prior to use), 1 mM benzamidine, and 1 mM each of leupeptin, pepstatin and antipain. GST–eEF2K was purified on glutathione–Sepharose, being eluted with buffer containing 10 mM reduced glutathione.
The pair of recombinant constructs, pNICZb containing eEF2K[48–336] and pCDF containing eEF2K[490–725], were co-transformed into competent E. coli Rosetta cells and plated on to LB agar plates supplemented with 50 μg/ml kanamycin and 50 μg/ml spectinomycin. (It was necessary to express these two fragments together in order to obtain sufficient quantities of soluble protein). Transformants were grown in LB medium in the presence of both kanamycin and spectinomycin (to maintain selection for cells harbouring both plasmids) to a D600 of 0.6 at 37°C.
Protein expression was induced by the addition of isopropyl β-D-thiogalactopyranoside to a final concentration of 1 mM. Cells were grown for a further 16 h at 18°C prior to harvesting. Cells were resuspended in buffer A (50 mM Hepes, pH 7.5, 300 mM NaCl, 5% (v/v) glycerol, 1 mM 2-mercaptoethanol, PMSF added just prior to use and 10 mM imidazole) and then sonicated and clarified by centrifugation (16000 g for 15 min at 4°C). The lysate was loaded on to Ni-NTA (Ni2+-nitrilotriacetic acid) agarose resin (Qiagen). The column was washed with buffer A and eluted with buffer A containing 250 mM imidazole. The eluted protein was then applied to a HiTrap SP column equilibrated in 50 mM Hepes, pH 7.5, 50 mM NaCl and 5 mM 2-mercaptoethanol. The column was washed extensively with 50 mM Hepes, pH 7.5, 200 mM NaCl and 5 mM 2-mercaptoethanol, and the protein was eluted with 50 mM Hepes, pH 7.5, 700 mM NaCl and 5 mM 2-mercaptoethanol. The fractions containing the eEF2K complex were pooled. The protein was treated with TEV protease at 4°C overnight and concentrated. The concentrated protein was applied to a HiLoad Superdex 75 16/60 gel filtration column (Pharmacia) pre-equilibrated in 50 mM Hepes, pH 7.5, 150 mM KCl and 2 mM 2-mercaptoethanol. The purity of all recombinant proteins was confirmed by SDS/PAGE followed by staining with Coomassie Brilliant Blue.
For expression of eEF2K in mammalian cells, the appropriate pHM-eEF2K vectors were transfected into HEK-293 cells as described previously [32]. The cells were lysed 48 h later and eEF2K proteins were isolated by virtue of their His6 tag using Ni-NTA agarose according to the manufacturer's instructions.
Assays for eEF2 kinase activity
Assays of eEF2K activity were performed using buffer B [final concentrations: 50 mM Mops, pH 7.0 (unless pH stated otherwise), 20 μg/ml CaM (unless omitted), 5 mM MgCl2, 14 mM 2-mercaptoethanol, 0.67 mM CaCl2, 2 mM EDTA, 0.4 mM EGTA, 1 mM benzamidine, and 1 mM each of leupeptin, pepstatin and antipain]. Reactions, in a total volume of typically 20 μl, contained 1 μg of purified eEF2 and were initiated by adding [γ-32P]ATP (final concentration 0.1 mM, 1 μCi per reaction). Reactions were incubated at 30°C for the appropriate time (up to 30 min) and then SDS/PAGE sample buffer was added. The samples were immediately heated at 95°C for 5 min to denature the proteins and stop the reaction. Products were analysed by SDS/PAGE (10% gel) and, after staining with Coomassie Brilliant Blue, the gels were placed into destain/fixing solution [50% (v/v) methanol and 10% (v/v) acetic acid]. Gels were then placed on Whatman 3MM paper, covered with Saran wrap and then dried on a vacuum gel dryer. Radioactivity was detected either by autoradiography or using a phosphorimager (Typhoon, GE Healthcare).
Assays of eEF2K activity against the MH-1 peptide were performed in buffer B essentially as described above, except that analysis involved spotting samples from the reaction mixture on to phosphocellulose paper (P81, Whatman). Filters were immediately placed in approximately 300 ml (for up to 25 filters) of 75 mM orthophosphoric acid and washed three further times in a similar volume of 75 mM orthophosphoric acid. They were then rinsed in ethanol and dried in an oven at 100°C. Radioactivity was determined using Čerenkov counting.
UV cross-linking
The interaction between eEF2K and ATP was studied by UV cross-linking, using recombinant GST–eEF2K, in the presence or absence of CaM and CaCl2 in buffer B, but using [α-32P]ATP instead of [γ-32P]ATP. UV irradiation was performed using a hand-held light source (UVP Model UVGL-58, Mineralight lamp, emission wavelength maximum at 254 nm) held 2–3 cm above the open microfuge tube containing the sample for 60 min. Samples were then subjected to SDS/PAGE and autoradiography.
SDS/PAGE and immunoblotting
SDS/PAGE was performed using the BioRad Laboratories Protean 3 mini-slab gel system, using the procedure first described by Laemmli [33]. Immunoblotting was performed by electrotransferring proteins resolved by SDS/PAGE on to nitrocellulose membranes. Membranes were then blocked in PBS with 0.05% Tween 20 containing 5% (w/v) skimmed milk powder for 1 h at room temperature (20–22°C). Membranes were probed with the indicated primary antibody overnight at 4°C. After incubation with fluorescently tagged secondary antibody, signals were detected using a Li-Cor Odyssey® imaging system.
ELISA assays
To study the association of CaM with a biotinylated peptide corresponding to the CaM-binding region of human eEF2K (residues 76–95), or a variant which contains phosphoserine, not serine, at position 78, the wells of a Maxisorp ELISA tray (Nunc) were coated overnight at 4°C with CaM (0.2 μg/ml) in PBS. Wells were then washed with PBS containing 0.05% Tween 20, and blocked with 5% (w/v) BSA in the same buffer. The eEF2K[76–95] peptide was appropriately diluted in 50 mM Mops (at pH 6.5 or 7.5), 100 mM NaCl, 2 mM CaCl2, 0.05% Tween 20 and 0.5% BSA. The various dilutions of the peptide were added to wells in triplicate in 100 μl. After 1 h at room temperature, the wells were washed with buffer comprising 50 mM Mops, 100 mM NaCl and 0.05% Tween 20 at pH 6.5 or 7.5, and either 2 mM CaCl2 or 1 mM EGTA. Streptavidin–HRP (horseradish peroxidase; Pierce; 0.1 μg/ml) in the corresponding buffer was then added and plates were incubated for 30 min at room temperature, before being washed again. HRP activity was assessed using 3,3′,5,5′-tetramethylbenzamidine and absorbance was read at 450 nm.
To study the interaction of eEF2K with CaM by ELISA, a similar method was used, except that, after coating the plates with CaM, wells were probed with GST, GST–eEF2K or GST–eEF2K[W85G], and detection was performed using an anti-GST antibody followed by an anti-IgG–HRP antibody. When ELISA was used to study the association between the N- and C-terminal domains of eEF2K, the wells were first coated with GST–eEF2K[1–402] and then probed with GST–eEF2K[478–725] or, as a negative control, GST.
Analytical gel filtration
Analytical gel-filtration chromatography experiments were carried out using a Superdex 75 10/300 gel filtration column on an ÄKTA purifier (GE Healthcare). The column was pre-equilibrated in 50 mM Hepes, pH 7.5, 150 mM NaCl and 5 mM 2-mercaptoethanol in the absence or presence of 6 mM CaCl2. Purified eEF2K[48–336] or eEF2K[490–725] (0.6 mg/ml) in the absence or presence of CaM (1:1 ratio) was loaded on to a 100 μl sample loop and then injected into the gel filtration column at a flow rate of 0.5 ml/min. Fractions of 200 μl were collected and analysed by SDS/PAGE. CaM (0.4 mg/ml) on its own was analysed as a control. The column had previously been calibrated with proteins of known molecular mass. The elution profiles at 280 nm were recorded.
Analysis of binding to SP Sepharose
To study the interaction of the catalytic and SEL1 domain of eEF2K, we performed SP Sepharose-binding experiments utilizing the presence of two different tags. The His6–Zb-eEF2K[48–336] and His6–eEF2K[490-725] fragments in 50 mM Hepes (pH 7.5) and 75 mM NaCl were applied to pre-equilibrated SP Sepharose resin in 50 mM Hepes (pH 7.5) and 75 mM NaCl containing either 2 mM Ca2+ or 4 mM EDTA and incubated for 30 min at 4°C. The resin was washed three times with the same buffer containing either 2 mM CaCl2 or 4 mM EDTA. The protein was then eluted with 50 mM Hepes (pH 7.5) and 1 M NaCl. Samples of the flow-through (unbound material) and elution were analysed by SDS/PAGE. To study the interaction of eEF2K with CaM the same method was used, except that CaM was added. It was also verified that His6–eEF2K[490–725] and CaM did not bind to the SP Sepharose resin alone (see the Results section).
Analysis of binding of eEF2K to CaM–Sepharose
To study the interaction of eEF2K with CaM further, we performed CaM–Sepharose-binding experiments with wild-type GST–eEF2K. Each sample, in 50 mM Mops (pH 7.5) and 150 mM NaCl, was applied to CaM–Sepharose™ 4B (GE Healthcare) pre-equilibrated in 50 mM Mops (pH 6.5 or 7.5) and 150 mM NaCl; and either 2 mM CaCl2 or 4 mM EDTA and incubated for 30 min at 4°C. The resin was washed three times with the appropriate corresponding buffer. The protein was then eluted with 50 mM Mops, pH 6.5 or 7.5, 150 mM NaCl and 4 mM EDTA. Samples of the flow-through and elution were analysed by SDS/PAGE.
NMR sample preparation
Samples (six 0.5 ml aliquots) were prepared for NMR STD (saturation transfer difference) experiments. Sample 1: 0.5 mM ATP-γS (adenosine 5′-[thio]triphosphate; a non-hydrolysable ATP analogue) in 99.9% pure 2H2O. Sample 2: eEF2K[48–336] and eEF2K[490–725] at 9 μM in 20 mM Hepes, pH 7.5, 150 mM KCl, 2 mM 2-mercaptoethanol and 1 mM ATP-γS. Sample 3: Sample 2 without ATP-γS. Sample 4: 100 μM CaM in 20 mM Hepes, pH 7.5, 150 mM KCl, 1 mM CaCl2 and 1 mM ATP-γS. Sample 5: Sample 4 without ATP-γS. Sample 6: 9 μM eEF2K[48–336] and eEF2K[490–725] and 90 μM CaM in 20 mM Hepes, pH 7.5, 150 mM KCl, 1 mM CaCl2 and 1 mM ATP-γS.
Acquisition of NMR spectra
All NMR experiments were recorded at a temperature of 298 K with a spectral width of 10 kHz on a Varian INOVA-600 instrument with a Z-gradient room temperature triple resonance probe (Agilent). The 1H NMR chemical shifts were referenced to the residual HDO signal at 4.78 ppm. A standard Biopack STD experimental procedure was used. Selective saturation of the protein resonances was achieved by a train of Gaussian-shaped 50 ms pulses for 2 s at 1.5 ppm (on-resonance) and 30 ppm (off-resonance). Residual water suppression was achieved by excitation sculpting and protein signals were suppressed by a 40 ms trim pulse. The on- and off-resonance spectra were subtracted during the pulse programme execution via phase cycling, and the resulting spectra were processed using Vnmr 6.1c.
RESULTS
Conserved residues in the catalytic domain and the proposed metal ion-binding site are essential for eEF2K function
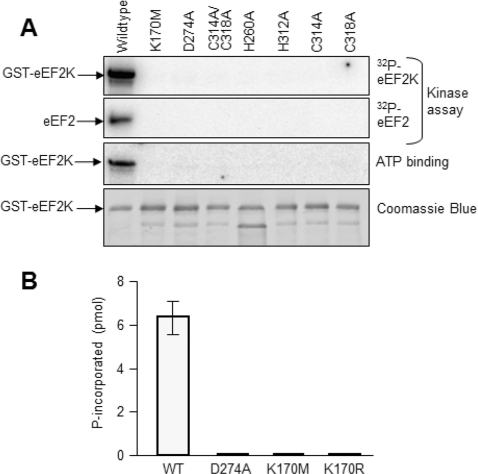
Many residues within the kinase domain are conserved between eEF2K sequences from disparate species (Figure 1A) and between these enzymes and other members of the α-kinase family [34]. The available crystal structures of the kinase domains of TRPM7 (ChaK1) [12] and the Dictyostelium MHCKA [13] reveal that some of the highly conserved residues may interact with the substrate ATP. Based on the three-dimensional structure of TRPM7 [12], Lys170 in eEF2K probably lies in a similar position to Lys72 in PKA (protein kinase A). This residue is strictly conserved in members of the main kinase superfamily and interacts with the α- and β-phosphates of ATP; mutating it strongly impairs kinase activity. We therefore tested the effect of mutating this residue in eEF2K to methionine (similar size but uncharged). The K170M mutant was essentially completely inactive both in autophosphorylation and in phosphorylating either eEF2 or the MH-1 peptide (Figures 2A and 2B). Even a more conservative replacement by arginine resulted in essentially complete loss of activity (K170R; Figure 2B). Assays conducted for longer time periods revealed that these Lys170 mutants do retain <0.5% of the wild-type activity (results not shown). Asp274 is universally conserved in α-kinases and is located close to the phosphates of the substrate ATP in the structure of TRPM7 [12]. Mutation of this residue to alanine caused a complete loss of detectable activity (Figure 2A).
Figure 2. Analysis of eEF2K mutants.
(A) Data for activity and ATP binding (UV cross-linking of [α-32P]ATP) to wild-type GST–eEF2K or the indicated point mutants: data for autophosphorylation of GST–eEF2K or activity against eEF2 are shown as a comparison (the Figure is an autoradiograph of an SDS/PAGE gel); the bottom section shows a Coomassie Brilliant Blue-stained gel of the GST–eEF2K proteins used. (B) Activities of selected point mutants against the MH-1 peptide. All assays were performed within the linear range of the assay. WT, wild-type.
The structures of both ChaK1 and MHCKA revealed the presence of a Zn2+ ion coordinated by two histidine and two cysteine residues [12,13]. Mutation of any of the corresponding residues in eEF2K to alanine (H260A, H213A, C314A and C318A mutants) led to complete loss of activity (Figure 2A), confirming that these residues, and presumably coordination of Zn2+, are also critical for the structural integrity and/or activity of eEF2K. Consistent with this, these mutations, and also those of Asp274 and Lys170, caused a loss of ability to bind ATP, as judged from UV cross-linking using [α-32P]ATP (Figure 2A).
Association of eEF2K with CaM
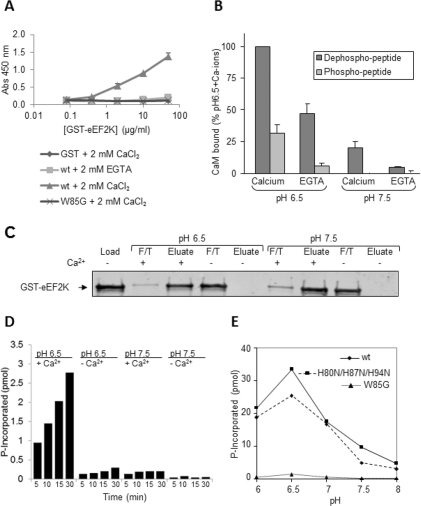
Some targets for CaM can bind it even in the absence of Ca2+ ions [35], e.g. ones containing a so-called ‘IQ’ (Ile-Gln) motif. The CaM-binding region of eEF2K does contain an Ile-Gln sequence, but the overall sequence does not conform closely to that of the standard IQ motif. To test the binding of eEF2K to CaM under different conditions, we first used an ELISA, in which CaM was immobilized on the plate, and then probed with differing quantities of GST–eEF2K, or as negative controls, GST itself or the eEF2K[W85A] mutant (in which a key residue involved in CaM binding is mutated [24]; Figure 3A). This was performed in the presence of Ca2+ ions or of the Ca2+ chelator EGTA. In the presence of Ca2+ ions, a clear interaction was seen with GST–eEF2K (Figure 3A), but not with GST or GST–eEF2K[W85G]. When EGTA rather than Ca2+ ions was present, almost no binding of CaM to GST–eEF2K was seen, except a trace at the highest concentration of GST–eEF2K used (Figure 3A). These data indicate that eEF2K only binds weakly, if at all, to CaM in the absence of Ca2+ ions.
Figure 3. Behaviour of eEF2K with CaM at different pH values.
(A) The association of CaM with eEF2K was tested by ELISA, using the indicated amounts of wild-type (wt) or W85G mutant GST–eEF2K, as described in the Experimental section. GST was used as a negative control, and binding was studied in the presence of CaCl2 or EGTA as noted. Results are means±S.D. Abs, absorbance. (B) The binding of biotinylated peptides based on the CaM-binding region of eEF2K to CaM was studied in the absence (EGTA) or presence of Ca2+ ions at pH 6.5 and pH 7.5. The peptides used correspond with the CaM-binding region of eEF2K or the same peptide with a phosphate on the equivalent of Ser78. Results are means±S.D. (C) The binding of wild-type GST–eEF2K to CaM Sepharose in the absence (EDTA) or presence of Ca2+ ions at pH 6.5 and 7.5. F/T, flow-through material. (D) Activities of wild-type eEF2K against the MH-1 peptide at pH 6.5 or 7.5 in the absence (EGTA) or presence of Ca2+ ions. (E) Activity of wild-type (wt) and the indicated mutants of GST–eEF2K against the MH-1 peptide over the indicated pH range. All procedures were performed as described in the Experimental section. Results are ??.
We also studied the binding of CaM to a peptide based on the sequence that includes the CaM-binding site within eEF2K which includes Trp85. At pH 7.5, which is close to physiological pH, little binding of CaM to this peptide was observed in the presence of EGTA, but much more association was seen when Ca2+ was present, as measured by an ELISA (Figure 3B). Interestingly, when the pH was lowered to 6.5, binding to CaM increased substantially in the presence of calcium ions and dramatically in their absence (Figure 3B). This enhanced binding to CaM could contribute to the activation of eEF2K that occurs at low pH [36]. The increased activity of eEF2K at pH values below 7 is not a general feature of α-kinases, as changing the pH did not affect the activities of MHCKA and TRPM7 in this way (Pigott, C.R. and Proud, C.G., unpublished work). Indeed, for MHCKA, a decrease in activity was observed at lower pH values [37].
We also studied the ability of full-length eEF2K to interact with CaM, in the absence or presence of Ca2+ ions, at pH 6.5 or 7.5. GST–eEF2K was applied to CaM–Sepharose beads under the appropriate conditions. After washing the beads with the corresponding buffer, bound proteins were eluted with buffer lacking Ca2+ ions. The loaded material and the non-bound and eluted fractions were analysed by SDS/PAGE and Western blotting (Figure 3C). The data clearly show that eEF2K cannot stably associate with CaM in the absence of Ca2+ ions at either pH in this type of assay.
We also examined the Ca2+ dependence of the activity of eEF2K against the MH-1 peptide at both pH values. Activity was much higher at pH 6.5 than at pH 7.5, in agreement with earlier data [36], but was still strongly dependent upon the presence of Ca2+ ions (Figure 3D), consistent with the requirement for Ca2+ ions for efficient binding of eEF2K to CaM at both pH values.
The fact that the CaM-binding region of eEF2K contains three histidine residues which can ionize around neutral pH prompted us to examine whether they played a role in the effects of pH on CaM binding, since their positive charge (a feature often involved in CaM binding) will change over this pH range. We conducted extensive studies using eEF2K in which one or more of these residues was replaced by asparagine (uncharged, but polar side chain) or lysine (charged at pH 7.5 as well as pH 6.5) and the corresponding peptides failed to reveal any influence of the histidine residues on binding to CaM (results not shown). As shown in Figure 3(E), even when all three histidine residues in this region were mutated to asparagine, the activity of the enzyme still responded to decreased pH in a similar way to the wild-type enzyme. Thus the effect of pH on eEF2K activity does not require the histidine residues in its CaM-binding region. Other features of eEF2K must therefore account for this.
The activity of eEF2K is negatively regulated by signalling through mTORC1 [19], in part through the mTORC1-promoted phosphorylation of Ser78, which lies immediately adjacent to the CaM-binding site. Phosphorylation here negatively influences the association of eEF2K with CaM [20]. Consistent with this, a version of the CaM-binding site peptide in which the equivalent of Ser78 has been replaced by a phosphoserine did not bind at all to CaM at pH 7.5, although some binding was observed at pH 6.5 (Figure 3B). This is consistent with previous data showing that the phosphorylation of Ser78 decreased binding of CaM to eEF2K in human cells [20].
Ca2+/CaM activate eEF2K by regulating eEF2K structure
Although eEF2K was first identified as a Ca2+/CaM-activated protein kinase [3], it has not so far been established how Ca2+/CaM actually bring about activation of eEF2K (or, conversely, why eEF2K is inactive in the absence of Ca2+/CaM). In other Ca2+/CaM-dependent kinases, regions outside the catalytic domain are involved in their control by Ca2+/CaM (for example see [38]). To study whether control of eEF2K by Ca2+/CaM also requires features outside its catalytic domain, we created a suite of truncations (Figure 1B).
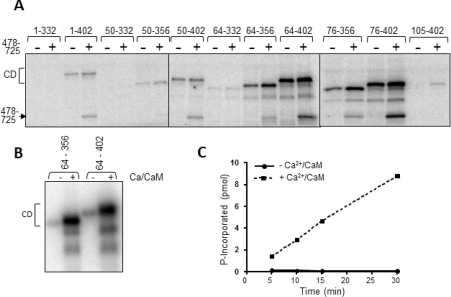
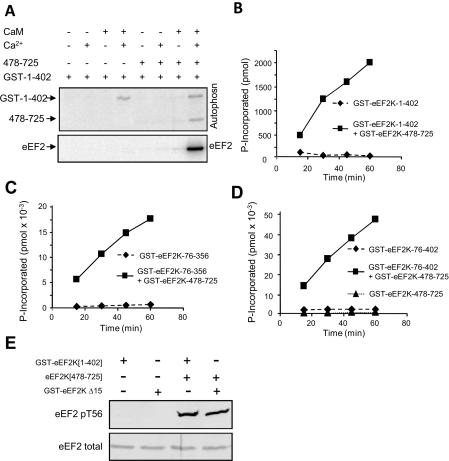
As shown in Figure 4(A), the fragment 1–402 (containing the N-terminus, CaM-binding motif, catalytic domain and ~65 additional residues) undergoes autophosphorylation in a Ca2+/CaM-dependent manner. This implies: (i) that the 1–402 fragment does possess catalytic activity and (ii) that it contains at least one site of autophosphorylation. eEF2K[1–402] also phosphorylated the eEF2K[478–725] fragment (Figure 4A), indicating that this C-terminal region contains at least one site of autophosphorylation.
Figure 4. Activities of a range of fragments of eEF2K.
(A) The indicated fragments, all GST fusions containing the catalytic domain of eEF2K, were incubated with [γ-32P]ATP in the presence of Ca2+/CaM and, where indicated, the eEF2K[478–725] fragment. The Figure is an autoradiograph of the stained gel. The positions of the catalytic domain fragments (CD) and eEF2K[478–725] are shown. (B) As (A), using the indicated GST–eEF2K fragments alone, or in the absence or presence of Ca2+/CaM. The faster-migrating radiolabelled species represent fragments created by proteolytic degradation. (C) GST–eEF2K[64–356] and eEF2K[478–725] fragments were incubated with the MH-1 peptide in the presence of [γ-32P]ATP with or without Ca2+ ions and CaM for the indicated times.
Of the shorter truncated proteins that we could express well in E. coli, fragments lacking residues 332 onwards showed very little activity in autophosphorylation (Figure 4A). Loss of activity could reflect misfolding, lack of the domains required for function or simply the removal of the autophosphorylated residues. Our other recent work has indeed identified residues that are absent from some of the ‘inactive’ fragments as autophosphorylation sites (Pyr Dit Ruys, S., Wang, X., Smith, E.W., Herinckx, G., Hussain, N., Rider, M.H., Vertommen, D. and Proud, C.P., unpublished results). Interestingly, these fragments (e.g. eEF2K[1–332], eEF2K[50–332] and eEF2K[64–332]; Figure 1B) were also unable to phosphorylate the C-terminal eEF2K[478–725] fragment, indicating that it is likely that they really are inactive, and do not simply lack an autophosphorylation site. In fact, as we have found (Pyr Dit Ruys, S., Wang, X., Smith, E.W., Herinckx, G., Hussain, N., Rider, M.H., Vertommen, D. and Proud, C.P., unpublished results), the 333–356 region does indeed contain two autophosphorylation sites, Thr348 and Thr353, the former playing a key role in the control of eEF2K activity.
Most of the other longer truncated proteins did show activity (Figure 4A). All of these fragments contain the previously identified CaM-binding site [23,24]. In contrast, a fragment that contains the region beyond position 332, but lacks the CaM-binding region (eEF2K[105–402]; Figure 4A), displayed very low autophosphorylation, much less than the 76–402 fragment that contains the same C-terminal part. This effect is not due to removing major autophosphorylation sites since the 76–104 region contains only one minor site, Ser78 (Pyr Dit Ruys, S., Wang, X., Smith, E.W., Herinckx, G., Hussain, N., Rider, M.H., Vertommen, D. and Proud, C.P., unpublished results). This concurs with the findings of Diggle et al. [24] who showed that removing the first 100 residues from eEF2K resulted in loss of activity. (We cannot, of course, rule out the possibility that some effects may be due to protein misfolding.)
The autophosphorylation of the 64–356 fragment (and the slightly longer 64–402 fragment) was strongly stimulated by Ca2+/CaM (Figure 4B). Thus control by Ca2+/CaM is an intrinsic property of the catalytic domain and adjacent flanking regions and does not require the extreme N-terminal region, the linker or the C-terminal SEL1 domains.
Shorter fragments were also capable of autophosphorylation. The smallest fragment which showed autophosphorylation consisted of residues 76–356, which contains about 30 residues C-terminal to the conserved α-kinase catalytic domain (Figure 4A, cf. Figure 1B). Interestingly, fragments lacking residues 1–64 or 1–76 actually tended to show higher activity than the 1–402 polypeptide (Figure 4A), indicating that these N-terminal regions negatively affect activity. These conclusions are borne out by data obtained using the MH-1 peptide as substrate (see below).
Regulation of ATP binding by Ca2+/CaM
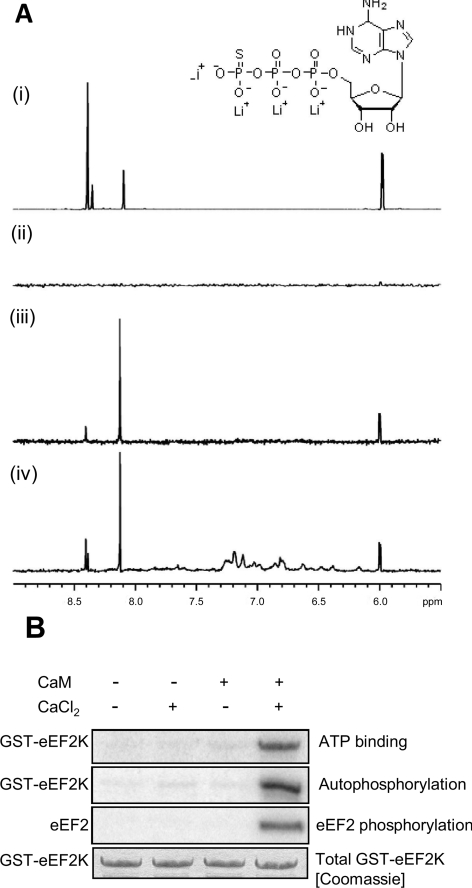
To study the effects of Ca2+/CaM on the ability of eEF2K to interact with its substrate ATP, we used STD NMR. To obtain sufficient protein for this analysis, we co-expressed the eEF2K[48–336] and [490–725] fragments. Its association with ATP was studied in the absence and presence of Ca2+/CaM. In this method, NMR signals of ligand protons are detected if the ligand is bound to the protein. In Figure 5(A), the same section of the ATP-γS spectrum is shown for four different conditions. The top spectrum shows the positions of the ATP-γS proton signals, whereas the absence of any signals in the spectrum below shows that the STD experiment does not generate a spurious signal that could be mistaken for bound ligand. The bottom two traces show proton signals that originate from the ATP-γS bound to eEF2K in the absence and presence of Ca2+/CaM, (it was shown in a separate STD experiment that ATP-γS does not bind to Ca2+/CaM in the absence of eEF2K; results not shown). This shows that ATP-γS can bind to eEF2K in the absence and presence of Ca2+/CaM, suggesting that the ATP-binding site of eEF2K is accessible in the Ca2+-free state.
Figure 5. Regulation of the binding of eEF2K to ATP studied by STD NMR.
(A) Binding of ATP-γS to eEF2K as a function of Ca2+/CaM. The same section of the 1H spectrum of a 1 mM ATP-γS solution is shown under four experimental conditions: (i) 1H NMR spectrum of ATP-γS; (ii) 1H STD spectrum of ATP-γS showing that the STD pulses do not saturate ATP-γS; (iii) same 1H STD spectrum of ATP-γS in the presence of 9 μM eEF2K[48–336] and eEF2K[490–725]; and (iv) in the presence of a mixture of 9 μM eEF2K[48–336] and eEF2K[490–725] and 1 mM CaCl2 and 100 μM CaM respectively. Binding of ATP-γS to Ca2+/CaM could be excluded because of the absence of STD signals in an ATP-γS Ca2+/CaM solution (results not shown). The inset shows the structure of ATP-γS. (B) Dependence of activity and ATP-cross-linking on Ca2+/CaM. To test ATP binding, recombinant GST–eEF2K was incubated with [α-32P]ATP in the absence or presence of Ca2+ ions or CaM, as indicated. Samples were subjected to UV irradiation as described in the Experimental section. In parallel, other samples were incubated with [γ-32P]ATP and eEF2 to examine autophosphorylation of eEF2K and activity against eEF2. Equal levels of eEF2K protein were confirmed by staining the gel with Coomassie Brilliant Blue (bottom panel). The other three panels show autoradiographs of SDS/PAGE.
We also studied the regulation of the ability of eEF2K to interact with ATP using UV cross-linking to [α-32P]ATP. As shown in Figure 5(B), eEF2K was only cross-linked to labelled ATP when both CaM and Ca2+ ions were present. Taken together, these results suggest that, whereas the ATP-binding site in eEF2K is accessible in the absence of Ca2+/CaM, Ca2+/CaM induces or stabilizes a conformation in the kinase domain of eEF2K which positions ATP in the active site in a way that brings it into closer proximity to residues involved in catalysis. Apparently this effect does not occur in the presence of CaM alone.
The catalytic and C-terminal domains of eEF2K cooperate in trans to promote substrate phosphorylation
Although the 1–402, 76–402 and 76–356 fragments all underwent autophosphorylation and phosphorylated the eEF2K[478–725] fragment (Figure 4A), they were completely unable to phosphorylate eEF2 (Figure 6A). Strikingly, addition of the eEF2K[478–725] fragment restored activity against eEF2 (Figure 6A). To test whether this property is limited to the physiological substrate (eEF2) or a more general property of eEF2K, we also tested the activities of catalytic domain fragments against the MH-1 peptide (Figures 6B–6D). All of the fragments tested were also unable to phosphorylate this substrate (Figure 6B and results not shown). Remarkably, addition of the C-terminal 478–725 fragment also restored activity against the MH-1 peptide (Figure 6B). Thus the N-terminal catalytic domain requires the C-terminal region containing the SEL1 domains in order to phosphorylate any known substrate in trans. The SEL1 region is not needed to impart catalytic activity itself since the catalytic domain efficiently undergoes autophosphorylation in its absence (Figure 4A). This finding is consistent with previous studies [23,24] which suggested that features of the C-terminal part of eEF2K are needed for phosphorylation of eEF2; importantly, the results of the present study reveal that the C-terminal fragment is also required for activity against the only other known eEF2K substrate, the MH-1 peptide.
Figure 6. Activities of catalytic fragments against eEF2 or the MH-1 peptide depends on the presence of the eEF2K[478–725] fragment.
(A) GST–eEF2K[1–402] was incubated with eEF2 in the presence of [γ-32P]ATP in the absence or presence of Ca2+ ions, CaM and/or the eEF2K[478–725] fragment as indicated. The Figure shows an autoradiograph of the resulting gels. (B–D) GST–eEF2K[1–402], [76–356] or [76–402] was incubated with the MH-1 peptide in the presence of [γ-32P]ATP in the presence of Ca2+ ions and CaM with or without the eEF2K[478–725] fragment. (E) The indicated proteins were incubated with eEF2 and non-radioactive ATP. Reaction products were analysed by SDS/PAGE and Western blotting with the anti-eEF2 (P)Thr56 antibody (pT56) or anti-eEF2 as a loading control.
Since both fragments are tagged with GST, and GST can form dimers [39], it was important to establish that the effects observed in the present study are an intrinsic property of eEF2K, not merely ones conferred by the GST tag. We were unable to remove the GST tag from the eEF2K[1–402] fragment without losing solubility, but did succeed in doing so for the C-terminal region without adverse effect. As shown in Figure 6(E), the untagged eEF2K[478–725] fragment was also able to restore activity to eEF2K[1–402], demonstrating that the effect is independent of its GST tag and is an intrinsic property of these regions of the eEF2K polypeptide.
These results are consistent with the idea that the C-terminal domain of eEF2K may recruit not only eEF2, but also the MH-1 peptide for phosphorylation by the catalytic domain; the present study shows that this works when the fragments are supplied in trans, also implying that the C- and N-terminal fragments may interact with one another even when not covalently linked as part of the same polypeptide chain and this was tested in subsequent experiments.
The discovery that addition of the eEF2K[478–725] fragment allowed catalytic domain fragments to phosphorylate the MH-1 peptide allowed us to test the dependence of such activity on Ca2+/CaM. The activity of the eEF2K[64–356] fragment against the MH-1 peptide (measured in the presence of the C-terminal eEF2K[478–725] fragment) was strongly dependent on Ca2+/CaM (Figure 4C), in agreement with data for the phosphorylation of eEF2 by the eEF2K[1–402] fragment in the presence of eEF2K[478–725] (Figure 6A).
Regions flanking the catalytic domain modify its activity
The discovery that the addition of the eEF2K[478–725] fragment restored the ability of the catalytic domain fragments to phosphorylate the MH-1 peptide allowed us to assess whether regions adjacent to the catalytic domain affect its activity in phosphorylating substrates in trans, rather than autophosphorylation (analysed in Figure 4A). Removal of residues 1–75 very strongly enhanced activity compared with that of the eEF2K[1–402] fragment (by >20-fold; compare 1–402 in Figure 6B with 76–402 in Figure 6D), whereas truncation of the latter fragment by removing residues 357–402 to create the 76–356 fragment decreased it by approximately 2.5 fold compared with the 76–402 polypeptide (Figure 6C). These results show that the N-terminal domain exerts an inhibitory function, whereas features of the 357–402 region may promote activity, both in autophosphorylation (Figure 4A) and against the MH-1 peptide (Figure 6B). The latter observation indicates that the reduced activity of the fragments lacking residues 357–402 is not simply due to removal of acceptor sites for autophosphorylation. The autophosphorylation of the eEF2K[64–356] and eEF2K[64–402] fragments was almost completely dependent upon Ca2+/CaM (Figure 4B).
The catalytic and SEL1 domains interact stably and directly
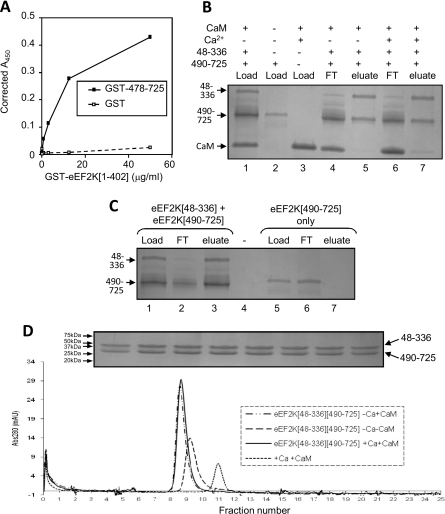
To examine whether the C- and N-terminal domains do indeed interact stably, we first used an ELISA-based approach coating the wells of the plate with either GST–eEF2K[478–725] or (as a negative control) GST. GST–eEF2K[1–402]–His6 was then added, and after extensive washing, detection was with Ni-NTA–HRP conjugate. As shown in Figure 7(A), a strong GST–eEF2K[1–402]–His6-dependent signal was observed in wells coated with GST–eEF2K[478–725], but not when GST itself was used. This confirms that the interaction between these domains is not mediated by GST and implies that the N- and C-terminal domains of eEF2K do indeed interact with each other in a rather stable manner.
Figure 7. The catalytic and SEL1 domains of eEF2K interact directly.
(A) An ELISA-based approach, described in the Experimental section, was used to study interactions between GST–eEF2K[1–402] and GST–eEF2K[478–725] (or, as a negative control, GST). (B and C) A SP Sepharose resin-binding assay was performed in the absence or presence of Ca2+ ions and CaM to study the interactions between His6–Zb–eEF2K[48–336] and His6–eEF2K[490–725]. CaM and His6–eEF2K[490–725] were loaded separately as controls and showed no binding. (D) Gel filtration was used to analyse the elution profile of eEF2K[48–336] and eEF2K[490–725] in the absence or presence of Ca2+ and CaM as indicated. CaM was loaded separately to indicate its elution profile on its own. The inset shows a Coomassie Brilliant Blue-stained SDS/PAGE gel of the fractions corresponding to the main protein peak (elution volumes 8.3–9.8 ml; lower panel) run in the absence of Ca2+ and CaM. This confirmed equal levels of eEF2K[48–336] and eEF2K[490–725] across the peak [their migration positions are indicated, as are those of the indicated molecular weight markers (in kDa)]. Abs, absorbance; FT, flow-through material.
We considered it important to confirm this interaction using additional approaches using fragments expressed without GST tags. The eEF2K[48–336] fragment was expressed with His6 and Z tags, and the C-terminal eEF2K[490–725] fragment with only a His6 tag. A mixture of the two eEF2K fragments, together with CaM, was applied to SP Sepharose, a cation exchange resin. The Z tag has a strong positive charge and thus the Z tag/eEF2K[48–336] fragment binds to SP Sepharose and is eluted with high levels of salt (Figure 7B, lane 5), whereas the C-terminal eEF2K[490–725] fragment was not itself retained on this resin (Figure 7C, lane 7). Excess eEF2K[490–725], which tends to be expressed at higher levels than the other fragment, flowed through the column when the two proteins were applied together (Figure 7B, lanes 4 and 6). The same behaviour was observed in the presence or absence of Ca2+ ions (Figure 7B, lanes 4–7) or in the absence of CaM (Figure 7C). These data indicate that the fragments containing the catalytic and SEL1 domains can interact directly, and that this interaction is not dependent on Ca2+/CaM. As expected, CaM was also retained on the resin, but only in the presence of Ca2+ (Figure 7B, compare lanes 5 and 7).
To test the interactions between the N- and C-terminal regions of eEF2K, we studied their behaviour by analytical gel-permeation chromatography. The purified eEF2K[48–336] and eEF2K[490–725] fragments were preincubated with or without Ca2+ ions and CaM, and then applied to the column. Under all conditions they eluted as a single peak (Figure 7D), and analysis of the fractions across the peak by SDS/PAGE revealed the presence of similar amounts of the two fragments in all fractions. Comparison with calibration standards revealed that the peak seen in the absence of Ca2+ and CaM corresponded with a molecular mass of 66 kDa, which is in good agreement with the expected size (60.4 kDa) of a heterodimer of the eEF2K[48–336] and eEF2K[490–725] fragments. This finding provides further evidence that the two fragments associate stably and reveals that they do so in the absence or presence of Ca2+/CaM. The peak was, as expected, shifted slightly to the left (higher mass) in the presence of CaM.
The extreme C-terminus of eEF2K is critical for its ability to phosphorylate eEF2
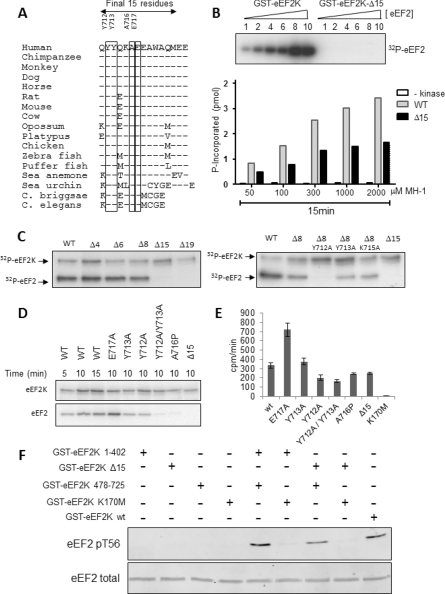
Residues within this extreme C-terminal region are highly conserved across all eEF2K sequences from nematodes and echinoderms to mammals (Figure 8A). It has previously been reported that removal of the extreme C-terminus of eEF2K abrogates its ability to phosphorylate eEF2, but not a peptide substrate (MH-1; [23]) and that loss of even only the last 19 residues abolishes the ability to phosphorylate eEF2 [24]. The fact that truncation of the C-terminus did not prevent autophosphorylation or phosphorylation of MH-1 indicates that this region is not actually required for kinase function, but rather for activity against eEF2, and perhaps for interaction with this substrate.
Figure 8. The C-terminal tip of eEF2K is required for phosphorylation of eEF2.
(A) Amino acid sequence alignment of the extreme C-termini of eEF2K from the indicated species. Sequence differences relative to human are noted in the single letter code; ‘-’ indicates identity. Highly conserved residues tested here are boxed. (B) The activities of wild-type (WT) GST–eEF2K and the Δ15 truncated protein (GST–eEF2K[1–710]) against various concentrations of eEF2 or the MH-1 peptide. The relative amounts of eEF2 are indicated above each lane (10 corresponds to a final eEF2 concentration of 0.22 mg/ml). Assays against the MH-1 peptide were also performed in the absence of GST–eEF2K (-kinase). The final concentrations are given in the Figure. Assays were performed as described in the Experimental section. (C–E) The activities of the indicated truncation and/or point mutated versions of eEF2K were determined against eEF2 (C and D) or the MH-1 peptide (E). The wild-type (WT) eEF2K and truncation mutants tested in (C) were expressed in HEK-293 cells. Other data used eEF2K proteins expressed in E. coli. (C and D) show autoradiographs of stained SDS/PAGE gels, with the positions of GST–eEF2K (eEF2K) and eEF2 being noted. The results in (E) are means±S.D. (F) The indicated combinations of recombinant GST–eEF2K-based proteins were incubated with eEF2 and non-radioactive ATP. Reaction products were analysed by SDS/PAGE and Western blotting with the anti-eEF2 (P)Thr56 (pT56) antibody or anti-eEF2 antibody as a loading control.
To test the importance of these residues, we assayed the activity of full length eEF2K and a truncated protein lacking the final 15 residues (Δ15-eEF2K) against eEF2 (Figure 8B). The full-length eEF2K readily phosphorylated eEF2, showing high activity against the highest concentrations of eEF2 tested in the assay; in contrast, the truncated protein showed no activity against eEF2 at any concentration tested. Thus the final 15 residues are essential for the ability of eEF2K to phosphorylate eEF2, and the fact that no activity is seen even at high substrate concentrations shows that this effect is not simply due to a change in the Km value (affinity) for eEF2. When tested against the MH-1 peptide, the Δ15 truncation showed substantial activity (about 50–60% of that of the full-length protein) at all of the peptide concentrations tested. These results indicate: (i) that the C-terminal region also aids phosphorylation of this substrate; but (ii) that, in distinction to eEF2, it is not essential for phosphorylation of the MH-1 peptide. The data also provide further evidence that the loss of activity of the Δ15 truncation against eEF2 is unlikely to be due to an increase in the Km value for this substrate, since, if that were the case, detectable activity should be observed at increasing substrate concentrations. However, we cannot rule out the possibility that the deletion of the last 15 residues causes such a drastic decrease in the affinity for eEF2 that even a 10-fold increase in eEF2 concentration is insufficient to allow detectable phosphorylation.
These data prompt a key question: which feature(s) within the far C-terminal region are required for phosphorylation of eEF2 by eEF2K? To address this, we first created a series of truncation mutants in which either the final four, six or eight residues, or 15 or 19 amino acids were removed (Figure 8C). The final four residues are absent from the nematode eEF2K sequences (Figure 8A), whereas the preceding four are not conserved in these sequences, but generally are in other eEF2K proteins. Loss of up to eight residues did not affect the activity of eEF2K against eEF2, but removal of the next seven (Δ15) completely eliminated detectable activity against this substrate. These residues are generally conserved among eEF2Ks from diverse species, some universally so (Figure 8A).
Having demonstrated the key importance of residues 711–718 for phosphorylation of eEF2, we then mutated several completely conserved residues within this region (Figure 8D). Mutation of either of the universally conserved tyrosine residues to alanine (the Y712A and Y713A mutants) led to a partial loss of activity against eEF2 (Figure 8C), but only had a modest (Y712A) or no (Y713A) effect on the ability to phosphorylate MH-1 (Figure 8E). The double mutant showed a complete loss of activity against eEF2 (Figure 8C), but retained considerable activity against MH-1 (Figure 8D). Ala716 is universally conserved and lies in a region predicted to adopt an α-helical conformation; to disrupt this, we mutated Ala716 to a proline residue. This drastically decreased activity against eEF2 (Figure 8D), but had little effect on the activity against the MH-1 peptide (Figure 8E). Glu717 is also universally conserved, but mutating it to alanine, to eliminate the negative charge, actually enhanced activity against eEF2 and, strikingly, also increased phosphorylation of the MH-1 peptide (Figures 8C and 8D).
Taken together, these data extend earlier findings by revealing that the conserved pair of tyrosine residues near the C-terminus of eEF2K is critical for phosphorylation of eEF2, and that they also play a role in the phosphorylation of the non-physiological MH-1 peptide substrate (albeit to a much lesser extent). As discussed above, the C-terminal part may recruit substrates for phosphorylation by the N-terminal domain. One might therefore expect that a truncated (e.g. Δ15) version of the C-terminal region would fail to restore activity to the 1–402 fragment; unfortunately, despite strenuous efforts, we were unable to express the 478–710 fragment or a version of 478–725 in which the two key tyrosine residues were mutated.
We therefore adopted an alternative approach where we first studied whether the intact C-terminal domain fragment of eEF2K could restore activity to eEF2K[Δ15]. As shown in Figure 8(F), it was indeed able to do so, yielding a level of activity only slightly below that seen for eEF2K itself or for the 1–402 plus 478–725 combination (Figure 8F). This implies that the 478–725 region can recruit eEF2 for phosphorylation by the catalytic domain of the eEF2K[Δ15] protein. This effect was independent of the GST tag on the C-terminal fragment (Figure 6E).
DISCUSSION
Two previous studies provided initial insights into the functions of the domains of eEF2K [23,24]. Pavur et al. [23] conducted a comprehensive deletion analysis of eEF2K, removing sections of roughly 50 residues from the protein. Although this approach carries a higher risk of causing misfolding of the internally deleted polypeptide, the authors' conclusions largely concur with those of the present study, in that we find that the regions between residues 1–75 and 357–477 are not required for function, whereas the section 76–356 shows activity, at least in autophosphorylation, and the C-terminal region 478–725 is required for phosphorylation of eEF2. Pavur et al. [23] found that, although some variants of eEF2K with deletions within the C-terminal region could still phosphorylate the MH-1 peptide, a number of others could not, and most did so at a greatly decreased rate. These findings are broadly consistent with the results of the present study. Diggle et al. [24] found that removal of the final 19 amino acids abolished the ability of eEF2K to phosphorylate eEF2, in line with the present findings, but also reported that removing the last 133 residues eliminated autophosphorylation. This is at odds with the results of the present study, which show that even fragments as short as 76–356 undergo robust autophosphorylation.
The CaM target database (http://calcium.uhnres.utoronto.ca/ctdb/ctdb/home.html) indicates a CaM-binding site at residues 514–526 in eEF2K (sequence LEKKIGKSILGKVHL). However, the results of the present study show that the entire C-terminal region is dispensable for the control of eEF2K activity by Ca2+/CaM. Our data also show that the eEF2K[478–725] fragment cannot bind CaM (C.R. Pigott, unpublished work) or CaM–Sepharose, whereas full-length eEF2K can. These data are consistent with previous work indicating that the CaM-binding site lies immediately N-terminal to the catalytic domain, and includes Trp85 (equivalent to Trp84 in rat eEF2K), which was shown to be essential for binding to CaM [24].
The findings of the present study extend previous work in a number of other ways. First, they imply that the N-terminal region (e.g. residues 1–75) exerts an inhibitory effect on eEF2K activity. Removal of this region enhances both the intrinsic autophosphorylation of the resulting fragment and the activity (in combination with the 478–725 region) against the MH-1 peptide.
Secondly, the results of the present study show that several residues which appear to be universally conserved among α-kinases are critical for activity. These data are in good agreement with the effects of mutating the corresponding conserved residues in TRPM7 and with deductions made from the crystal structures of the catalytic domains of other α-kinases, i.e. ChaK1/TRPM7 and MHCKA [12,13]. This extends to residues that are implicated in binding a metal ion; mutating these residues in eEF2K essentially abolishes kinase function. We tested whether incubation of eEF2K with metal ion-chelators such as N,N,N′,N′-tetrakis (2-pyridylmethyl)ethylenediamine might be able to strip out the bound metal; however, this had no effect on kinase activity (C.R. Pigott, unpublished work). It is possible that, within the folded structure of eEF2K, the ion is inaccessible to chelating agents.
Thirdly, our data identify specific residues in the extreme C-terminus which are required for phosphorylation of eEF2. Our data agree with those of Diggle et al. [24] who showed that removing the last 19, but not the final eight, residues from the C-terminus of rat eEF2K abolished its ability to phosphorylate eEF2 in vitro. This implies that the intervening residues, several of which are absolutely conserved among eEF2K sequences from species as diverse as nematodes and mammals, play a key role in substrate phosphorylation. Focusing on those residues, we show that the adjacent tyrosines (Tyr712/Tyr713 in human eEF2K) are critical for the ability to phosphorylate eEF2, although the eEF2K[Y712A/Y713A] mutant could still quite efficiently phosphorylate the MH-1 peptide. Ala716 is also crucial for this, since mutating it to a proline residue abolishes activity against eEF2. Interestingly, although Glu717 is also completely conserved, converting it into alanine actually enhanced activity, both against eEF2 and the MH-1 peptide.
Importantly, the requirements for activity against eEF2 and the MH-1 peptide clearly differ quite markedly. Although the eEF2K[478–725] region is essential for activity against both, the removal of the C-terminal region had little effect on the ability to phosphorylate the MH-1 peptide, while abolishing phosphorylation of eEF2 even at the highest substrate concentrations tested. Thus the region containing the SEL1 domains and the extreme C-terminus is essential for the phosphorylation of either substrate in trans, whereas the C-terminal tail is only required for phosphorylation of the physiological substrate eEF2. Mutating universally conserved residues within this region strongly decreased activity against eEF2 (e.g. the YY712/713AA and A716P mutants), but only modestly affected activity against the MH-1 peptide. This suggests that these residues might interact with eEF2 itself. However, despite strenuous efforts using several approaches, we were unable to demonstrate a stable interaction using wild-type eEF2K, so were unable to test the effect of these mutations.
Fourthly, it is striking that the N-terminal region containing the kinase domain and the C-terminal section of eEF2K can efficiently cooperate in trans to phosphorylate both eEF2 and the MH-1 peptide, i.e. they do not need to be part of the same polypeptide to do so. This is consistent with the finding that these two regions associate with one another (when expressed separately). This prompts the model shown in Figure 9 where the C-terminal region, including the SEL1 domains and the tip, recruit eEF2 to the catalytic domain for phosphorylation. Several of the regulatory phosphorylation sites in eEF2K (e.g. Ser359 [27], Ser366 [21], Ser377 [22] or Ser398 [17]) are located in the intervening ‘linker’ region. It is tempting to speculate that phosphorylation of these sites may influence the association of the N- and C-terminal domains and thus modulate substrate recruitment and activity. Further work is required to study this. It is interesting to note that in TRPM7, another α-kinase, a region outside the catalytic domain (immediately N-terminal to it) is again required for catalytic activity [10], although several aspects of the structural organization and control of this enzyme differ substantially from those of eEF2K.
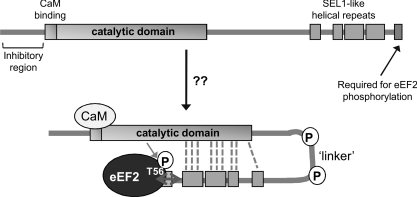
Figure 9. Summary scheme.
This diagram depicts a model for the structural organization of eEF2K which is described in the Discussion section. P indicates the presence of several phosphorylation sites in the linker region.
AUTHOR CONTRIBUTION
Craig Pigott, Halina Mikolajek, Claire Moore, Stephen Finn, Curtis Phippen and Jörn Werner conducted the experimental work; Craig Pigott, Halina Mikolajek, Claire Moore, Stephen Finn and Jörn Werner helped with the data analysis and preparation of the figures; the overall study was conceived by Christopher Proud, aided by Jörn Werner; the paper was written by Christopher Proud with assistance from Craig Pigott, Halina Mikolajek, Claire Moore and Jörn Werner.
ACKNOWLEDGEMENTS
We thank Stuart Findlow at the Southampton Centre for Biological NMR for support and access to the National NMR Facility at the National Institute for Medical Research (Mill Hill, London, U.K.). We thank Farnaz Taghizadeh for excellent technical support and Kailun Jiang for help with preparing some of the vectors used in the present study. Halina Mikolajek would like to thank Professor S. Knapp and the late Dr P. Rellos at the Structural Genomics Consortium (Oxford University, Oxford, U.K.) for generous support with the ligation-free cloning system as part of their visiting scientist programme.
FUNDING
This work was supported by grants from the Canadian Institutes of Health Research (to C.G.P.), the Wellcome Trust [grant number 086688/Z/08/Z (to C.G.P. and J.M.W.)] and the Royal Society (to C.G.P.).
References
- 1.Carlberg U., Nilsson A., Nygard O. Functional properties of phosphorylated elongation factor 2. Eur. J. Biochem. 1990;191:639–645. doi: 10.1111/j.1432-1033.1990.tb19169.x. [DOI] [PubMed] [Google Scholar]
- 2.Nairn A. C., Bhagat B., Palfrey H. C. Identification of calmodulin-dependent protein kinase III and its major Mr 100,000 substrate in mammalian tissues. Proc. Natl. Acad. Sci. U.S.A. 1985;82:7939–7943. doi: 10.1073/pnas.82.23.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryazanov A. G. Ca2+/calmodulin-dependent phosphorylation of elongation factor 2. FEBS Lett. 1987;214:331–334. doi: 10.1016/0014-5793(87)80081-9. [DOI] [PubMed] [Google Scholar]
- 4.Ryazanov A. G., Ward M. D., Mendola C. E., Pavur K. S., Dorovkov M. V., Wiedmann M., Erdjument-Bromage H., Tempst P., Parmer T. G., Prostko C. R., et al. Identification of a new class of protein kinases represented by eukaryotic elongation factor-2 kinase. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4884–4889. doi: 10.1073/pnas.94.10.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Middelbeek J., Clark K., Venselaar H., Huynen M. A., van Leeuwen F. N. The α-kinase family: an exceptional branch on the protein kinase tree. Cell Mol. Life Sci. 2010;67:875–890. doi: 10.1007/s00018-009-0215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryazanov A. G., Pavur K. S., Dorovkov M. V. Alpha kinases: a new class of protein kinases with a novel catalytic domain. Curr. Biol. 1999;9:R43–R45. doi: 10.1016/s0960-9822(99)80006-2. [DOI] [PubMed] [Google Scholar]
- 7.Browne G. J., Proud C. G. Regulation of peptide-chain elongation in mammalian cells. Eur. J. Biochem. 2002;269:5360–5368. doi: 10.1046/j.1432-1033.2002.03290.x. [DOI] [PubMed] [Google Scholar]
- 8.Herbert T. P., Proud C. G. Regulation of translation elongation and the cotranslational protein targeting pathway. In: Mathews M. B., Sonenberg N., Hershey J.W.B., editors. Translational Control in Biology and Medicine. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2006. pp. 601–624. [Google Scholar]
- 9.Crawley S. W., Cote G. P. Determinants for substrate phosphorylation by Dictyostelium myosin II heavy chain kinases A and B and eukaryotic elongation factor-2 kinase. Biochim. Biophys. Acta. 2008;1784:908–915. doi: 10.1016/j.bbapap.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Crawley S. W., Cote G. P. Identification of dimer interactions required for the catalytic activity of the TRPM7 α-kinase domain. Biochem. J. 2009;420:115–122. doi: 10.1042/BJ20081405. [DOI] [PubMed] [Google Scholar]
- 11.Crawley S. W., Gharaei M. S., Ye Q., Yang Y., Raveh B., London N., Schueler-Furman O., Jia Z., Cote G. P. Autophosphorylation activates Dictyostelium myosin II heavy chain kinase A by providing a ligand for an allosteric binding site in the α-kinase domain. J. Biol. Chem. 2011;286:2607–2616. doi: 10.1074/jbc.M110.177014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaguchi H., Matsushita M., Nairn A. C., Kuriyan J. Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity. Mol. Cell. 2001;7:1047–1057. doi: 10.1016/s1097-2765(01)00256-8. [DOI] [PubMed] [Google Scholar]
- 13.Ye Q., Crawley S. W., Yang Y., Cote G. P., Jia Z. Crystal structure of the α-kinase domain of Dictyostelium myosin heavy chain kinase A. Sci. Signal. 2010;3:ra17. doi: 10.1126/scisignal.2000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Runnels L. W. TRPM6 and TRPM7: a Mul-TRP-PLIK-cation of channel functions. Curr. Pharm. Biotechnol. 2011;12:42–53. doi: 10.2174/138920111793937880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsushita M., Kozak J. A., Shimizu Y., McLachlin D. T., Yamaguchi H., Wei F. Y., Tomizawa K., Matsui H., Chait B. T., Cahalan M. D., Nairn A. C. Channel function is dissociated from the intrinsic kinase activity and autophosphorylation of TRPM7/ChaK1. J. Biol. Chem. 2005;280:20793–20803. doi: 10.1074/jbc.M413671200. [DOI] [PubMed] [Google Scholar]
- 16.Nairn A. C., Palfrey H. C. Identification of the major Mr 100,000 substrate for calmodulin-dependent protein kinase III in mammalian cells as elongation factor-2. J. Biol. Chem. 1987;262:17299–17303. [PubMed] [Google Scholar]
- 17.Browne G. J., Finn S. G., Proud C. G. Stimulation of the AMP-activated protein kinase leads to activation of eukaryotic elongation factor 2 kinase and to its phosphorylation at a novel site, serine 398. J. Biol. Chem. 2004;279:12220–12231. doi: 10.1074/jbc.M309773200. [DOI] [PubMed] [Google Scholar]
- 18.Diggle T. A., Subkhankulova T., Lilley K. S., Shikotra N., Willis A. E., Redpath N. T. Phosphorylation of elongation factor-2 kinase on serine 499 by cAMP-dependent protein kinase induces Ca2+/calmodulin-independent activity. Biochem. J. 2001;353:621–626. doi: 10.1042/0264-6021:3530621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redpath N. T., Foulstone E. J., Proud C. G. Regulation of translation elongation factor-2 by insulin via a rapamycin-sensitive signalling pathway. EMBO J. 1996;15:2291–2297. [PMC free article] [PubMed] [Google Scholar]
- 20.Browne G. J., Proud C. G. A novel mTOR-regulated phosphorylation site in elongation factor 2 kinase modulates the activity of the kinase and its binding to calmodulin. Mol. Cell. Biol. 2004;24:2986–2997. doi: 10.1128/MCB.24.7.2986-2997.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X., Li W., Williams M., Terada N., Alessi D. R., Proud C. G. Regulation of elongation factor 2 kinase by p90RSK1 and p70 S6 kinase. EMBO J. 2001;20:4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knebel A., Haydon C. E., Morrice N., Cohen P. Stress-induced regulation of eEF2 kinase by SB203580-sensitive and -insensitive pathways. Biochem. J. 2002;367:525–532. doi: 10.1042/BJ20020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pavur K. S., Petrov A. N., Ryazanov A. G. Mapping the functional domains of elongation factor-2 kinase. Biochemistry. 2000;39:12216–12224. doi: 10.1021/bi0007270. [DOI] [PubMed] [Google Scholar]
- 24.Diggle T. A., Seehra C. K., Hase S., Redpath N. T. Analysis of the domain structure of elongation factor-2 kinase by mutagenesis. FEBS Lett. 1999;457:189–192. doi: 10.1016/s0014-5793(99)01034-0. [DOI] [PubMed] [Google Scholar]
- 25.Mittl P. R., Schneider-Brachert W. Sel1-like repeat proteins in signal transduction. Cell Signal. 2007;19:20–31. doi: 10.1016/j.cellsig.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 26.Smith E. M., Proud C. G. Cdc2-cyclin B regulates eEF2 kinase activity in a cell cycle- and amino acid-dependent manner. EMBO J. 2008;27:1005–1016. doi: 10.1038/emboj.2008.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knebel A., Morrice N., Cohen P. A novel method to identify protein kinase substrates: eEF2 kinase is phosphorylated and inhibited by SAPK4/p38δ. EMBO J. 2001;20:4360–4369. doi: 10.1093/emboj/20.16.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W., Wang X., van der Knaap M., Proud C. G. Mutations linked to leukoencephalopathy with vanishing white matter impair the function of the eukaryotic initiation factor 2B complex in diverse ways. Mol. Cell. Biol. 2004;24:3295–3306. doi: 10.1128/MCB.24.8.3295-3306.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hedhammar M., Hober S. Z(basic)-a novel purification tag for efficient protein recovery. J. Chromatogr. A. 2007;1161:22–28. doi: 10.1016/j.chroma.2007.05.091. [DOI] [PubMed] [Google Scholar]
- 30.Aslanidis C., de Jong P. J. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savitsky P., Bray J., Cooper C. D., Marsden B. D., Mahajan P., Burgess-Brown N. A., Gileadi O. High-throughput production of human proteins for crystallization: the SGC experience. J. Struct. Biol. 2010;172:3–13. doi: 10.1016/j.jsb.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall-Jackson C. A., Cross D. A., Morrice N., Smythe C. ATR is a caffeine-sensitive, DNA-activated protein kinase with a substrate specificity distinct from DNA-PK. Oncogene. 1999;18:6707–6713. doi: 10.1038/sj.onc.1203077. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Drennan D., Ryazanov A. G. α-kinases: analysis of the family and comparison with conventional protein kinases. Prog. Biophys. Mol. Biol. 2004;85:1–32. doi: 10.1016/S0079-6107(03)00060-9. [DOI] [PubMed] [Google Scholar]
- 35.Rhoads A. R., Friedberg F. Sequence motifs for calmodulin recognition. FASEB J. 1997;11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 36.Dorovkov M. V., Pavur K. S., Petrov A. N., Ryazanov A. G. Regulation of elongation factor-2 kinase by pH. Biochemistry. 2002;41:13444–13450. doi: 10.1021/bi026494p. [DOI] [PubMed] [Google Scholar]
- 37.Cote G. P., Bukiejko U. Purification and characterization of a myosin heavy chain kinase from Dictyostelium discoideum. J. Biol. Chem. 1987;262:1065–1072. [PubMed] [Google Scholar]
- 38.Rellos P., Pike A. C., Niesen F. H., Salah E., Lee W. H., von D. F., Knapp S. Structure of the CaMKIIδ/calmodulin complex reveals the molecular mechanism of CaMKII kinase activation. PLoS Biol. 2010;8:e1000426. doi: 10.1371/journal.pbio.1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fabrini R., De L. A., Stella L., Mei G., Orioni B., Ciccone S., Federici G., Lo B. M., Ricci G. Monomer-dimer equilibrium in glutathione transferases: a critical re-examination. Biochemistry. 2009;48:10473–10482. doi: 10.1021/bi901238t. [DOI] [PubMed] [Google Scholar]